Abstract
The main role of the immune system is to protect against infections caused by invading pathogens. The adaptive immune system is particularly important for protection against repeated exposure to pathogens, due to its ability to memorize antigens during an initial infection and then respond rapidly and strongly to subsequent antigen challenges from the same or a related pathogen. This system forms immunological memory. Most epigenetic studies in immunology have focused on analysis of differentiation of CD4 T subsets, key players in the adaptive immune system. However, the relationship between immunological memory and epigenetics has not been as well studied. In recent years, with the advancement of technology such as ChIP-seq or RNA-seq methods, the importance of epigenetic mechanisms in immunological memory is becoming apparent. This review outlines our understanding of how CD4 T cells acquire and maintain function during or after differentiation, using Th2 cells as a model. In addition, we summarize the general characteristics of memory T cells from the perspective of epigenetics and discuss the possibility of clinical application of epigenetic studies in immunology.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The immune system is an extremely important system for maintaining in vivo homeostasis, and small changes in the balance of the immune system can cause the onset of several diseases. Decreased immune function increases the risk of infectious disease and the development of malignant tumors, whereas excess immune responses cause allergy and autoimmune diseases. Immunological memory has been at the core of our understanding of disease protection for more than 200 years, since Edward Jenner first proved in 1796 that vaccination against smallpox could provide protection from disease. It is now becoming clear that immunological memory is regulated not only by immune processes but also by general cellular memory mechanisms (Zediak et al. 2011b). The ability to maintain function after differentiation into the different types of cells needed to make up our body can be referred to as cellular memory. “Epigenetics” is an important keyword in understanding the concept of the cellular memory (Goldberg et al. 2007). Histone modifications and DNA methylation form the molecular basis of the epigenetic regulation of cellular memory.
Epigenetic studies have made a substantial contribution to the elucidation of mechanisms controlling immunological memory, particularly in the field of T cells. This review aims to summarize the role that epigenetics plays in the establishment of immunological memory in T cells. In addition, we introduce the latest progress in the field of immunological memory, mainly based on our data in which Th2 cells, one of the CD4 T cell subsets, are used as a model.
2 How Do Epigenetic Approaches Contribute to Studies in the Immunological Memory Field?
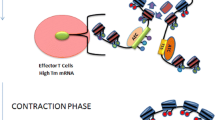
The immune system has the capacity to remember previous exposure to antigens that occurred during a primary infection. This memory enables the immune system to launch a more rapid and stronger response to the second antigenic challenge than to the first exposure. We will not describe the process of generation of immunological memory here due to length constraints. For a good review of this topic, see (Seder and Ahmed 2003). However, in very broad terms, immunological memory can be considered as being formed by a combination of both internal and external factors (Fig. 14.1). Internal factors include acquisition of high-affinity antigen receptors by gene rearrangement and high-level expression of certain transcription factors, cytokines, and cell surface molecules (Cuddapah et al. 2010; Zediak et al. 2011b). In addition, the external environment, such as the concentration of physiologically active substances including cytokines, intercellular interactions, and the extra-cellular matrix, can also influence memory cell function and formation (Shinoda et al. 2012; Tokoyoda et al. 2010). Epigenetic studies have focused on internal factors that regulate memory cells and have begun to elucidate the mechanisms controlling cellular memory of adaptive immune cells (Nakayama and Yamashita 2009). The concept of epigenetics is considered important because epigenetic information can dictate the transcriptional state of a gene (Goldberg et al. 2007). Typical examples of epigenetic changes are chemical modifications of histone proteins around which the genomic DNA is wound (histone modification) and DNA methylation involving the addition of a methyl group to the number 5 position of the cytosine pyrimidine ring (DNA methylation) (Turner 2002; Wu and Zhang 2010). As shown in Fig. 14.2a, methylation of histone H3K27, methylation of H3K9, and DNA methylation are indicators of transcriptional repression. On the other hand, methylation of H3K4 and acetylation H3K9 and H3K27 are indicators of transcriptional activation. Here, symbols such as K27 indicate the number of a particular amino acid in the N-terminus of histone H3. The transcriptional repression and transcriptional activation states are commonly referred to as closed chromatin structure and open chromatin structures, respectively. The term chromatin refers to a complex of DNA and protein existing in a eukaryotic cell nucleus.
Internal and external factors in immunological memory. Mechanisms of immunological memory are classified into internal factors and external factors. The internal factors are subdivided into genetically controlled mechanisms and epigenetically controlled mechanisms and shown with some examples (left). The external factors are subdivided into three factors: physiologically active substances, cell–cell interactions, and extracellular matrix (examples are shown in right panel)
Examples of epigenetic regulation. (a), Some typical examples of epigenetic information are shown. Histone modifications and DNA methylation are divided into two classes: active marks (left) and repressive marks (right). (b), Difference in epigenetic states at differentially expressed genes between two distinct cell types. In type A cells, the A gene has open chromatin structure and is highly expressed. On the other hand, in type B cells, the B gene has open chromatin structure and is highly expressed
The epigenetic concept can thus explain the question “how can the form and function of specific cells or tissues be different while they each have the same underlying DNA sequence?” Figure 14.2b shows two types of cells, cell type A and cell type B. The two cell types have the same genes (i.e., same genetic information), as the two cell types are of the same origin. However, gene A is expressed only in type A cells, and gene B is expressed only in type B cells. This mechanism can be explained as follows: in type A cells, the region of DNA encompassing the A gene has an open chromatin structure, in which active histone modifications such as H3K9ac and H3K4me3 are abundant. However, in type B cells, the chromatin structure at the A gene region contains repressive epigenetic marks such as H3K27me3 and DNA methylation and is closed. The open chromatin structure increases accessibility for molecules such as RNA polymerase and transcription factors that can control transcription. Thus, these molecules can easily accumulate at genes located in areas of open chromatin and activate transcription of these genes. Conversely, the closed chromatin structure decreases accessibility to transcriptional regulatory molecules and represses gene transcription. This also applies to the B gene. Chromatin structure around the B gene is closed in type A cells, whereas chromatin around the B gene contains open structure in type B cells. A complex combination of these epigenetic mechanisms allows cells with the same genetic information to acquire specific functions, according to the tissue each cell belongs to.
These internal factors regulating immunological memory are an example of how we can use our understanding of the concepts of genetics and epigenetics to understand how the immune system functions. In contrast to the epigenetic changes described above, the antigen receptor genes of lymphocytes such as B cells and the T cells are known to acquire high affinity to antigen by gene rearrangement (Hozumi and Tonegawa 1976), i.e., by direct changes to the genetic code within the cell that are largely irreversible. This process is one of the very few examples of normal physiological genetic rearrangements. Therefore, to avoid the need to directly rearrange the genetic code, epigenetic mechanisms are utilized for the acquisition of memory cell-specific gene expression profiles: high-level expression of certain transcription factors, cytokines, and cell surface molecules.
3 The Factors That Control Epigenetic States
The epigenetic modifications such as those described above are introduced and interpreted by epigenetic regulators which include Polycomb (PcG) and Trithorax (TrxG) proteins (Margueron and Reinberg 2011; Nakayama and Yamashita 2009; Schuettengruber et al. 2007, 2011). These complexes will only be described briefly here because they are discussed at length in other chapters in this volume. PcG and TrxG proteins were originally identified in Drosophila, but they also play major roles in defining mammalian gene expression programs during cell differentiation and in cancer cells. The Mammalian counterparts of TrxG proteins are the SET/MLL family of proteins. Figure 14.3 shows a schematic view of the principle PcG and TrxG complexes (Mohan et al. 2012). Previous studies report that the Polycomb-repressive complex 2 (PRC2) maintains a transcriptionally repressive state, primarily via EZH1/2 which has methyl transferase activity specific for H3K27. This state is then reinforced by the Polycomb-repressive complex 1 (PRC1) containing RING1 which has a repressive histone H2AK119 ubiquitin ligase activity. Conversely, the TrxG complex participates in the maintenance of a transcriptionally active state. The SET domain of MLL1/2 methylates H3K4 and induces open chromatin. In addition, some reports show that the TrxG complex interacts with histone acetyl transferase (HAT) and directly induces transcriptional activation (Schuettengruber et al. 2011). In these ways, it is thought that PcG and TrxG complexes antagonize each other and regulate the ON/OFF state of their target genes.
Polycomb (PcG) and Trithorax (TrxG) complex. A schematic representation of the Polycomb PRC2 complex (left) and the Trithorax MLL complex (right) is shown (Mohan et al. 2012). Menin protein is encoded by the MEN1 (multiple endocrine neoplasia 1) gene, whose mutation is involved in multiple endocrine neoplasia type 1 in human
4 Experimental Methods Used to Determine Epigenetic States
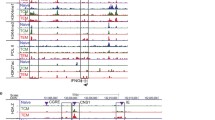
The chromatin immunoprecipitation (ChIP) assay is an experimental technique used to investigate the chromatin state (i.e., epigenetic state) within cells (Solomon et al. 1988). Firstly, target cells are collected and DNA–protein is cross-linked inside the cell using formaldehyde. Next, the DNA–protein complex is fragmented by sonication or a restriction enzyme. After performing immunoprecipitation by specific antibody to a target protein, DNA fragments are purified and collected. The collected DNA is analyzed by quantitative PCR using specific primers to detect the genomic regions of interest (Fig. 14.4a). Various kinds of histone modifications and transcription factor-binding patterns are thus detected by selecting protein-specific antibodies.
ChIP and ChIP-Seq method. (a–b), A summary of ChIP and ChIP-Seq method is shown. Performing ChIP by using a specific antibody against the protein a researcher wants to examine binding to DNA is common in both conventional ChIP and ChIP-Seq analysis. In conventional ChIP assay, purified DNA is quantified by quantitative PCR (a). In ChIP-Seq analysis, DNA sequences of purified and pretreated samples are read by next-generation sequencer and mapped on the reference genome (b). (c), An example of conventional ChIP analysis. Bmi1 (PcG)- and Menin (TrxG)-binding patterns and histone modifications were analyzed by ChIP with semiquantitative PCR. © 2010 Onodera et al. Journal of Experimental Medicine. 207:2493-2506.doi:10.1084/jem.20100760 Results in splenic B cells (navy), CD4 T cells (green) and fully developed Th2 cells (red) are shown
More recently, chromatin immunoprecipitation with high-throughput sequencing methods (ChIP-Seq) has been enabled due to the development of advanced next-generation sequencers (Fig. 14.4b) (Barski et al. 2007). ChIP-Seq is a method that can rapidly read the nucleotide sequences of the DNA fragments recovered from ChIP experiments, followed by mapping where these DNA fragments are derived from in the genome. Thus, this technique enables us to analyze histone modification states and binding sites of particular transcription factors on a genome-wide scale. Figure 14.4c shows an example of conventional ChIP analysis of Bmi1 (PcG)- and Menin (TrxG)-binding patterns in Splenic B, Naïve CD4 T, and fully developed Th2 cells, demonstrating that the activation of the Gata3 gene during Th2 cell differentiation is accompanied by decreased association of the Bmi1 PcG protein and increased association of the Menin TrxG protein.
5 Differentiation of CD4 T Cell Subsets
Naive CD4 T cells that receive antigen stimulation are known to differentiate into various subsets, depending on their surrounding environment and the type of cells that provide the antigenic signals for activation. The signals from cytokines, transmitted via their receptors expressed on the cell surface, are particularly important factors in deciding the type of differentiation that a cell will undergo (Fig. 14.5) (Kanno et al. 2012; Mosmann et al. 1986; Wilson et al. 2009; Zhou et al. 2009; Zhu et al. 2010). Th1 cells produce IFNγ and direct cell-mediated immunity against intracellular pathogens. The interleukin (IL)-12-STAT4 (signal transducer and activator of transcription 4) signaling pathway induces upregulation of the transcription factor T-bet and is required for Th1 cell differentiation. Th2 cells produce IL-4, IL-5, and IL-13 (Th2 cytokines) and are involved in humoral immunity and allergic reactions, which can include allergies and asthma. Upregulation of the IL-4-STAT6-dependent transcription factor GATA3 is crucial for Th2 cell differentiation. Th17 cells produce IL-17A, IL-17 F, and IL-22 and play an important role in immunity against bacteria and fungus. In mice, experimental autoimmune encephalomyelitis (EAE), a model of multiple sclerosis, is known as a Th17 cell-mediated disorder. The master transcription factor for Th17 cell differentiation is RORγt, whose expression is upregulated by cytokines including TGF-β, IL-6, IL-21, and IL-23. STAT3 is a signal transmission molecule located downstream of the receptors of IL-6, IL-21, and IL-23. Regulatory T cells (Treg), a subset with immune suppressive function, are characterized as cells highly expressing Foxp3 protein. Tregs are classified into two types: naturally occurring Treg (nTreg) that differentiate in the thymus, and inducible Treg (iTreg) whose differentiation is induced by TGF-β in peripheral tissues. In addition, other subsets have also been reported: Tfh cells assist in the maturation of B cells within the follicles of lymphoid organs, and Th9 cells have the ability to produce IL-9.
CD4 T cell subset differentiation and features. Naïve CD4 T cells are subjected to antigen presentation from antigen presenting cells (APC) and differentiate into Th1, Th2, Th17, iTreg, and Tfh subsets. Transcription factors and cytokines necessary for the differentiation of each subset are shown (Zhou et al. 2009)
6 Acquisition and Maintenance of Th2 Cell Identity
Here we briefly describe mechanisms of induction of Th2 cell differentiation and then outline data generated in mouse models showing how Th2 cell identity is maintained during the memory phase. Figure 14.6 (left) shows how the differentiation of Th2 cells is induced. Efficient Th2 cell differentiation requires two signaling pathways: the TCR (T cell receptor)-signaling pathway and the IL-4 and IL-4 receptor-signaling pathway (Nakayama and Yamashita 2010). GATA3, whose expression is upregulated by combined signaling through these two signaling pathways, is regarded as the master transcription factor responsible for forming active (accessible) chromatin at the Th2 cytokine gene loci (Amsen et al. 2009; Ho et al. 2009). STAT6-deficient CD4 T cells display impaired GATA3 upregulation, resulting in impaired Th2 cell differentiation and IL-4 production. This shows that the identity of Th2 cells is gained in an IL-4-STAT6-dependent manner along with the existence of signals from the TCR. However, until recently, the detailed mechanisms by which STAT6 induces the upregulation of Gata3 had been unknown. Recent research revealed that STAT6 directly bound to the Gata3 gene locus and induced epigenetic changes mediated by displacement of PcG proteins by TrxG proteins (Onodera et al. 2010). While there have been many reports demonstrating that high-level expression of GATA3 is indispensable for induction of Th2 cytokine genes, it was also unknown if GATA3 participated in the regulation of the expression of other Th2-specific genes. Recently, ChIP-Seq analysis was successfully utilized to identify genome-wide targets of GATA3 in Th2 cells (Horiuchi et al. 2011). Furthermore, by combining a Gata3 knockdown system with DNA microarray, the functional target genes of GATA3 were comprehensively identified (Sasaki et al. 2013). In addition, GATA3 function was shown to be controlled via protein–protein interactions. The Sox4 protein was also shown to antagonize GATA3 function (Kuwahara et al. 2012), and Chd4 was shown to coordinately regulate the formation of active and suppressive GATA3 complexes in T helper cells (Hosokawa et al. 2013).
Acquisition and maintenance of Th2 cell identity. Developing Th2 cells, represented in the left panel, refer to CD4 T cells cultured under the Th2 condition for <1 week. Developed Th2 cells, shown in the right panel, refer to Th2 cells cultured for 2–4 weeks in vitro or Th2 cells transferred into congenic mice and maintained for more than 4 weeks in vivo (memory Th2 cells). ChIP-Seq and DNA microarray datasets are deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE28292 and GSE46185
Another important issue to address is how the identity of developed Th2 cells is maintained after differentiation (Fig. 14.6, right). Interestingly, IL-4 is not necessary for the maintenance of developed Th2 cell function. This observation is true for in vitro-developed Th2 cells and in vivo-generated memory Th2 cells (Yamashita et al. 2004a). Th2 function is maintained in an IL-4-independent manner, and this maintenance mechanism requires GATA3 in vitro (Yamashita et al. 2004b). Additionally, Pai et al. have reported that GATA3 is required for the functional maintenance of Th2 cells in vitro (Pai et al. 2004). However, the IL-4-STAT6-independent mechanism for maintaining Gata3 expression in developed Th2 cells remains unclear. Two reports clearly proved that MLL and Menin, both members of the TrxG complex, were important for the functional maintenance of developed Th2 cells (Onodera et al. 2010; Yamashita et al. 2006). The same mechanism was also recently reported in human Th2 cells (Nakata et al. 2010). On the other hand, Bmi1 and Ring1B, which are members of the PcG complex (the counterpart of the TrxG complex), are important for survival of Th2 cells by controlling apoptosis (Suzuki et al. 2010; Yamashita et al. 2008). In developed Th2 cells, GATA3 was also indispensable to maintain the ability to produce Th2 cytokines. However, it was not known whether GATA3 is required for the maintenance of expression of Th2-specific genes other than Th2 cytokines. From the results of our in vitro and in vivo experiments, we found that maintenance of high-level expression GATA3 was indispensable for keeping many Th2-specific genes active in developed Th2 cells (Sasaki et al. 2013). Furthermore, we reported that pathogenic memory Th2 cells produced IL-5 in vivo and were involved in the pathogenesis of airway inflammation (Endo et al. 2011) and that NKT cells controlled the pool size of memory Th2 cells (Iwamura et al. 2012). In these and other similar ways, researchers are currently expending considerable effort trying to unravel the mechanisms of immune memory from the viewpoint of maintenance of Th2 cell identity.
7 General Transcriptional and Epigenetic Features of Memory T Cells
7.1 Memory T Cell Classification and Their Transcriptional Features
We have discussed immunological memory in terms of maintenance of Th2 cell phenotype until this section. An increasing number of groups are beginning to investigate epigenetic regulation of immune cell memory, most of which are using the approach of separating and classifying cells by surface molecules and then analyzing the epigenetic states of these populations. In this section, we describe the currently known epigenetic characteristics of immune memory cells defined by cell surface molecules, focusing on T cells.
In murine T cells, high expression of the cell surface molecule CD44 is used as a marker of memory cells (Dutton et al. 1998). Memory T cells can be further subdivided into central memory T cells (TCM; CD62L high) and effector memory T cells (TEM; CD62L low) according to the expression levels of CD62L (Sallusto et al. 2004). CD45RA and CD45RO are often used as markers of naïve and memory human T cells, respectively (Dutton et al. 1998), and like murine memory cells, human memory cells can also be subdivided into TCM and TEM based on expression of CD62L. T cells are classified into helper T cells expressing CD4 and cytotoxic T cells expressing CD8; however, memory cells generated from these two populations display a high degree of transcriptional similarity (Seder and Ahmed 2003), as do TCM and TEM cells (Weng et al. 2012). Therefore, we will discuss here general gene expression profiles and epigenetic signatures that are shared by all memory T cells. Naïve and memory T cells show less than a 5 % difference in their overall gene expression profiles when assessing the number of genes that are differentially expressed (Araki et al. 2009; Kaech et al. 2002). Weng et al. reported a list of highly expressed genes in human memory T cells (Weng et al. 2012). Kinetic analysis before and after T cell activation has revealed that there are activation-induced genes that are upregulated more rapidly in activated memory T cells than in activated naïve T cells, in addition to genes that are expressed at higher levels by activated memory T cells than by activated naïve T cells (Araki et al. 2009). When comparing memory T cells to naïve T cells, genes that are highly expressed in either resting or activated memory T cells are likely to be important for memory T cell function. Genes that are highly expressed by memory cells include those with immune functions, such as cytokines, chemokines, and receptors (Weng et al. 2012). Memory T cells also express genes that promote T cell survival and homeostasis and other genes with multiple or undefined functions (Weng et al. 2012).
7.2 Epigenetic Features of Memory T Cells
Although it is easy to imagine that epigenetic mechanisms are important for the control of the expression of these genes, there are technical limitations in our ability to efficiently analyze memory T cell populations. First, the number of memory T cells is so small in vivo that it is difficult to collect sufficient cell number for analysis. Several detailed studies have overcome this problem and found direct evidence that epigenetic processes are involved. Analyses of histone modifications in memory T cells have revealed that the more rapid response of memory T cells is associated with histone H3 acetylation and H3K4 methylation (Araki et al. 2008; Fann et al. 2006; Zediak et al. 2011b). For example, histone acetylation at the Ifng gene locus provides a molecular basis for the enhanced responsiveness of memory CD8 T cells (Northrop et al. 2006). Functions of memory T cells are also associated with reduced histone H3K27 methylation at numerous genes (Araki et al. 2009; Zediak et al. 2011a). In addition, Bernstein et al. have reported that in embryonic stem (ES) cells, many key developmental genes display both histone H3K4me3 and H3K27me3 marks, displaying a so-called bivalent phenotype (Bernstein et al. 2006). Resolution of these bivalent genes was also observed in differentiated cells (Mikkelsen et al. 2007). This is also the case in memory T cells, indicating that bivalent domains mark and regulate a select group of important genes in T cells (Araki et al. 2009). The second difficulty encountered when analyzing epigenetic modifications in memory T cells is that this group of cells exist as a heterogeneous population, so population analysis is not sufficient to fully understand the mechanisms regulating the phenotype of these cells. Dispirito and Shen (2010) have shown a global increase in histone H3 acetylation levels in single cells using a flow cytometric assay. The development of techniques that enable single-cell genome-wide analysis should shed light on this complex issue (Kalisky et al. 2011). Finally, the timing of when the features of memory T cells are completely established is unclear. Are memory T cells programmed during the effector phase, in which T cells encounter a pathogen for the first time, or during the contraction phase, while the levels of pathogen is rapidly declining and the infection is being eliminated? Additionally, it is possible that epigenetic states in T cells change during the memory phase. Many questions about the process of memory T cell formation remain unsolved. Some studies provide evidence for stable epigenetic marks that are established during the effector phase and persist in the memory phase (Fann et al. 2006; Mirabella et al. 2010; Yamashita et al. 2004a). The persistence of an active chromatin signature at relevant gene loci is observed in resting memory cells, even when those genes are transcriptionally inactive (Zediak et al. 2011b). The epigenetic study of memory T cells is at a nascent stage, and further research development along with progress in technology to analyze rare cell populations will continue to advance this field.
8 Perspective
We are conducting epigenetic research of immune cell memory using Th2 cells as a model. Th2 cells are considered to be responsible for causing allergic diseases, so elucidating the mechanisms controlling Th2 cell differentiation and maintenance of their function is relevant to the development of potential therapies. In particular, it is thought that the maintenance of functions after development of Th2 function is closely related to the pathogenesis of chronic disease and repetitive allergic reactions. For example, several studies have found that in pollinosis patients, antigen-specific immune cells remain a long time and repeatedly respond to cedar pollen scattered in early spring each year (Horiguchi et al. 2008). That is, as a treatment for patients who suffer from allergic diseases, suppressing the initial differentiation of Th2 cells is not sufficient. Instead, approaches to suppress the function of developed Th2 cells or to change them into other subsets are required. Of course, most memory T cells are not pathogenic and are instead important for defense against infection. Th2 cells assist in antibody production by B cells and are crucial for the elimination of pathogens by humoral immunity. Th1 cells are indispensable to the cell-mediated immunity mediated mainly by CD8 T cells. The specific defense mechanisms against infection that are regulated by memory T cells remain unclear and is one of the major unresolved issues related to immunological memory. For many immunology researchers, a major goal is to unravel the mystery of the immune memory mechanism and to establish new and efficient methods for vaccine development (Pulendran and Ahmed 2011). In today’s modern society, preventive medicine is highly desired. However, there are many kinds of pathogens against which we cannot successfully generate preventive immune responses using current vaccine technology. In addition, the establishment of a rapid vaccine development system for emerging infectious diseases that pose a threat to humans such as avian influenza (Gao et al. 2013) and MERS (Middle East respiratory syndrome) (Memish et al. 2013) is an urgent unmet need. Suppressing pathogenic memory T cells and increasing beneficial memory T cells are two essential aspects of immunological memory study when considering the application to clinical medicine. Moreover, it is hoped that we can apply the knowledge acquired using Th2 cells as a model to further studies of immunological memory in other CD4 T cell subsets, CD8 T cells, and B cells.
As discussed in this review, immunological memory is an important concept that forms the basis of long-lasting immunity. However, the mechanisms controlling memory cell development and functional maintenance have not yet been fully elucidated. Certainly, the nature of immunological memory is controlled by epigenetic mechanisms. However, there are few immunology researchers who have studied it from such a viewpoint. Elucidation of the molecular mechanism controlling immunological memory in terms of epigenetic regulation will lead to further development of immunology and new applications to clinical medicine. We consider there to be at least two important points with respect to investigations of the factors regulating immunological memory. The first is delineation of the mechanisms regulating memory cell survival, and the second is to determine the regulatory pathways by which memory cells maintain their phenotype. Survival of nonfunctional memory cells is likely avoided to conserve essential energy and nutrients. In contrast, survival of functional memory cells that retain the ability to react quickly upon secondary antigen encounter is central to immunological memory. With respect to the survival of memory cells, is likely that only the cells able to provide the most efficient responses to secondary antigen encounter are allowed to survive and become memory cells. However, how this process is regulated is almost completely unknown (Mueller et al. 2013). If the general concept of immunological memory was investigated in the same manner as the memory in a brain, research of functional maintenance would be similar to investigating the mechanism regulating “how can we remember.” In contrast, research of memory cell survival would be more similar to investigating “why only a part of memory remains” or “why do we have selective memory.”
Ultimately, through our epigenetic research on Th2 cells, we would like to find the fundamental principles that can be applied generally to cell differentiation and maintenance of differentiated cell identity. Recent reports about the generation of induced pluripotent stem (iPS) cells reversed the old thinking that once cells underwent functional specialization that they could not return to an undifferentiated state (Takahashi et al. 2007; Takahashi and Yamanaka 2006). Research on the differentiation of specific tissues from iPS cells for the purpose of regenerative medicine is a good example of the study of cell differentiation that relies on our understanding of epigenetic regulation. The epigenetic mechanisms defined using Th2 cell differentiation systems combined with analytic methods of ChIP-Seq and bioinformatics technology that we have developed and utilized may therefore also benefit the development of new regenerative medicine studies. In the near future, technical progress will enable us to perform single-cell ChIP-Seq analysis or high-throughput ChIP-Seq analysis with clinical samples (Northrup and Zhao 2011). The bioinformatic techniques for ChIP-Seq analysis which we and others have developed will also be useful when analyzing these samples.
Finally, the effects of radiation on epigenetic states should be described. Since the accident at the Fukushima nuclear power plant in 2011, people have made arguments about how radiation influences the human body (Christodouleas et al. 2011). It is likely that not only genetic changes but also epigenetic changes are induced by radiation. Thus, epigenetic research will come to take on more and more importance in the future.
References
Amsen D, Spilianakis CG, Flavell RA (2009) How are T(H)1 and T(H)2 effector cells made? Curr Opin Immunol 21:153–160
Araki Y, Fann M, Wersto R et al (2008) Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B). J Immunol 180:8102–8108
Araki Y, Wang Z, Zang C et al (2009) Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30:912–925
Barski A, Cuddapah S, Cui K et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Bernstein BE, Mikkelsen TS, Xie X et al (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326
Christodouleas JP, Forrest RD, Ainsley CG et al (2011) Short-term and long-term health risks of nuclear-power-plant accidents. N Engl J Med 364:2334–2341
Cuddapah S, Barski A, Zhao K (2010) Epigenomics of T cell activation, differentiation, and memory. Curr Opin Immunol 22:341–347
Dispirito JR, Shen H (2010) Histone acetylation at the single-cell level: a marker of memory CD8+ T cell differentiation and functionality. J Immunol 184:4631–4636
Dutton RW, Bradley LM, Swain SL (1998) T cell memory. Annu Rev Immunol 16:201–223
Endo Y, Iwamura C, Kuwahara M et al (2011) Eomesodermin controls interleukin-5 production in memory T helper 2 cells through inhibition of activity of the transcription factor GATA3. Immunity 35:733–745
Fann M, Godlove JM, Catalfamo M et al (2006) Histone acetylation is associated with differential gene expression in the rapid and robust memory CD8(+) T-cell response. Blood 108:3363–3370
Gao R, Cao B, Hu Y et al (2013) Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897
Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128:635–638
Ho IC, Tai TS, Pai SY (2009) GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol 9:125–135
Horiguchi S, Tanaka Y, Uchida T et al (2008) Seasonal changes in antigen-specific T-helper clone sizes in patients with Japanese cedar pollinosis: a 2-year study. Clin Exp Allergy 38:405–412
Horiuchi S, Onodera A, Hosokawa H et al (2011) Genome-wide analysis reveals unique regulation of transcription of Th2-specific genes by GATA3. J Immunol 186:6378–6389
Hosokawa H, Tanaka T, Suzuki Y et al (2013) Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc Natl Acad Sci USA 110:4691–4696
Hozumi N, Tonegawa S (1976) Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci USA 73:3628–3632
Iwamura C, Shinoda K, Endo Y et al (2012) Regulation of memory CD4 T-cell pool size and function by natural killer T cells in vivo. Proc Natl Acad Sci USA 109:16992–16997
Kaech SM, Hemby S, Kersh E et al (2002) Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111:837–851
Kalisky T, Blainey P, Quake SR (2011) Genomic analysis at the single-cell level. Annu Rev Genet 45:431–445
Kanno Y, Vahedi G, Hirahara K et al (2012) Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol 30:707–731
Kuwahara M, Yamashita M, Shinoda K et al (2012) The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-beta and suppresses T(H)2 differentiation. Nat Immunol 13:778–786
Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349
Memish ZA, Zumla AI, Al-Hakeem RF et al (2013) Family cluster of middle east respiratory syndrome coronavirus infections. N Engl J Med 368(26):2487–2494
Mikkelsen TS, Ku M, Jaffe DB et al (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560
Mirabella F, Baxter EW, Boissinot M et al (2010) The human IL-3/granulocyte-macrophage colony-stimulating factor locus is epigenetically silent in immature thymocytes and is progressively activated during T cell development. J Immunol 184:3043–3054
Mohan M, Herz HM, Shilatifard A (2012) SnapShot: Histone lysine methylase complexes. Cell 149(498–498):e1
Mosmann TR, Cherwinski H, Bond MW et al (1986) Two types of murine helper T cell clone I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol 136:2348–2357
Mueller SN, Gebhardt T, Carbone FR et al (2013) Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol 31:137–161
Nakata Y, Brignier AC, Jin S et al (2010) c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. Blood 116:1280–1290
Nakayama T, Yamashita M (2009) Critical role of the Polycomb and Trithorax complexes in the maintenance of CD4 T cell memory. Semin Immunol 21:78–83
Nakayama T, Yamashita M (2010) The TCR-mediated signaling pathways that control the direction of helper T cell differentiation. Semin Immunol 22:303–309
Northrop JK, Thomas RM, Wells AD et al (2006) Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol 177:1062–1069
Northrup DL, Zhao K (2011) Application of ChIP-Seq and related techniques to the study of immune function. Immunity 34:830–842
Onodera A, Yamashita M, Endo Y et al (2010) STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J Exp Med 207:2493–2506
Pai SY, Truitt ML, Ho IC (2004) GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci USA 101:1993–1998
Pulendran B, Ahmed R (2011) Immunological mechanisms of vaccination. Nat Immunol 12:509–517
Sallusto F, Geginat J, Lanzavecchia A (2004) Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol 22:745–763
Sasaki T, Onodera A, Hosokawa H et al (2013) Genome-wide gene expression profiling revealed a critical role for GATA3 in the maintenance of the Th2 cell identity. PLoS One 8:e66468
Schuettengruber B, Chourrout D, Vervoort M et al (2007) Genome regulation by polycomb and trithorax proteins. Cell 128:735–745
Schuettengruber B, Martinez AM, Iovino N et al (2011) Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol 12:799–814
Seder RA, Ahmed R (2003) Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol 4:835–842
Shinoda K, Tokoyoda K, Hanazawa A et al (2012) Type II membrane protein CD69 regulates the formation of resting T-helper memory. Proc Natl Acad Sci USA 109:7409–7414
Solomon MJ, Larsen PL, Varshavsky A (1988) Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53:937–947
Suzuki A, Iwamura C, Shinoda K et al (2010) Polycomb group gene product Ring1B regulates Th2-driven airway inflammation through the inhibition of Bim-mediated apoptosis of effector Th2 cells in the lung. J Immunol 184:4510–4520
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Takahashi K, Tanabe K, Ohnuki M et al (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872
Tokoyoda K, Hauser AE, Nakayama T et al (2010) Organization of immunological memory by bone marrow stroma. Nat Rev Immunol 10:193–200
Turner BM (2002) Cellular memory and the histone code. Cell 111:285–291
Weng NP, Araki Y, Subedi K (2012) The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nat Rev Immunol 12:306–315
Wilson CB, Rowell E, Sekimata M (2009) Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol 9:91–105
Wu SC, Zhang Y (2010) Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11:607–620
Yamashita M, Shinnakasu R, Nigo Y et al (2004a) Interleukin (IL)-4-independent maintenance of histone modification of the IL-4 gene loci in memory Th2 cells. J Biol Chem 279:39454–39464
Yamashita M, Ukai-Tadenuma M, Miyamoto T et al (2004b) Essential role of GATA3 for the maintenance of type 2 helper T (Th2) cytokine production and chromatin remodeling at the Th2 cytokine gene loci. J Biol Chem 279:26983–26990
Yamashita M, Hirahara K, Shinnakasu R et al (2006) Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity 24:611–622
Yamashita M, Kuwahara M, Suzuki A et al (2008) Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J Exp Med 205:1109–1120
Zediak VP, Johnnidis JB, Wherry EJ et al (2011a) Cutting edge: persistently open chromatin at effector gene loci in resting memory CD8+ T cells independent of transcriptional status. J Immunol 186:2705–2709
Zediak VP, Wherry EJ, Berger SL (2011b) The contribution of epigenetic memory to immunologic memory. Curr Opin Genet Dev 21:154–159
Zhou L, Chong MM, Littman DR (2009) Plasticity of CD4+ T cell lineage differentiation. Immunity 30:646–655
Zhu J, Yamane H, Paul WE (2010) Differentiation of effector CD4 T cell populations. Annu Rev Immunol 28:445–489
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Onodera, A., Tumes, D.J., Nakayama, T. (2014). Epigenetic Control of Immune T Cell Memory. In: Bonifer, C., Cockerill, P. (eds) Transcriptional and Epigenetic Mechanisms Regulating Normal and Aberrant Blood Cell Development. Epigenetics and Human Health. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-45198-0_14
Download citation
DOI: https://doi.org/10.1007/978-3-642-45198-0_14
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-45197-3
Online ISBN: 978-3-642-45198-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)