Abstract
Bdellovibrio and like organisms (BALOs) are gram-negative, obligate predators of other gram-negative bacteria. These small bacteria interact with their prey as highly motile attack phase cells, attaching to the outer membrane and consuming the prey extracellularly (epibiotic predation) or penetrating their periplasm (periplasmic predation). The former divides in a binary fashion, while the latter grows as a polynucleotide filament to finally split as progeny attack cells. High-resolution microscopy, molecular genetics, genomics, and functional genomics have been applied to study the cell cycle of BALOs, revealing functions required for predation and for cellular organization. Until recently, Bdellovibrio bacteriovorus was the only recognized species of BALOs. Culture-dependent and culture-independent approaches have shown that these predators form diverse monophyletic groups, including the three families Bdellovibrionaceae, Bacteriovoraceae, and Peridibacteraceae in the δ-proteobacteria, and the genus Micavibrio in the α-proteobacteria. Based on this detailed taxonomical knowledge, it has become possible to track predator and prey interactions in natural systems, providing first evaluations of the impact of bacterial predation on community structure.
An erratum to this chapter can be found at http://dx.doi.org/10.1007/978-3-642-39044-9_405
Access provided by Autonomous University of Puebla. Download reference work entry PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Predation is an important factor affecting both the ecology and evolution of organisms. While predatory interactions are common and the subject of numerous investigations in the animal world, much less is known in the bacterial realm. A number of reasons may explain the paucity of knowledge on predatory bacteria: It is difficult and time consuming to search for predatory interactions between bacteria in vivo by examining natural samples; predatory bacteria vary in their prey range, and thus, their isolation is limited by the number of possible prey that can be experimentally manipulated – most potential prey in the environment, as other bacteria, may not be amenable to cultivation and thus their predators remain unknown; until recently (Pasternak et al. 2012) genome data could not be used to identify novel predators.
That said, the field of predatory bacterial interactions has had many significant contributions since serendipitous discovery of the first obligate predatory bacterium Bdellovibrio by Stolp and Petzold (1962). This was followed by numerous groundbreaking researches on the physiology, ecology, taxonomy, interactions with prey, and cell cycle of Bdellovibrio in the 1960s, 1970s, and into the 1980s, mainly by the groups of Conti, Diedrich, Hespell, Rittenberg, Ruby, Shilo, Stolp and Starr, Thomashow, Tudor, and Varon. More recently, the field has greatly benefited from the introduction of modern molecular biology into microbial ecology and genetics brought by the groups of Jurkevitch, Kadouri, Koval, Linscheid, Sockett, Strauch, and Williams. Most, if not all, of these works were performed on Bdellovibrio and like organisms (BALOs), the “taxonomical progeny” of the original single Bdellovibrio bacteriovorus taxon. These bacteria can be described as predators, parasites or symbionts (Starr 1975), or parasitoids, and the consumed bacterium as prey or host. In this review, the terms predator–prey and predator-host will be used interchangeably.
The BALOs’ Life Cycle
The life cycle of BALOs is concomitant to its cell cycle and is composed of two main and distinct phases, i.e., an attack phase (AP) and a growth and division phase (GP). Further subdivision of these stages depends upon the predatory strategy adopted: BALOs are whether epibiotic or periplasmic predators. Epibiotic predators like Bdellovibrio exovorus (Fig. 1.1a ) and Micavibrio aeruginosavorus (Fig. 1.1b ) (Davidov et al. 2006b; Koval et al. 2012) attach onto the prey cell, digesting its content while remaining extracellular to finally divide in a binary fashion. Most of the knowledge being on periplasmic predators (Fig. 1.1c ), the description of BALOs’ life cycle will center on this particular predatory strategy (Fig. 1.2 ), with an emphasis on the physiological features of each stage.
The life cycle of Bdellovibrio bacteriovorus. Eight stages are depicted. 1 free swimming, attack phase, 2 attachment to prey cell, 3 penetration of the prey periplasm, 4 establishment and initiation of growth, 5 filamentous growth and depletion of prey cytoplasm, 6 division to progeny cells, 7 lysis of ghost prey cell and release of progeny, 8 host-independent mutant
Attack Phase
-
I.
Motility and prey detection. Attack phase (AP) cells are small (0.7–1.5 × 0.5 mm), mostly vibrioid, highly motile non-replicative cells (Fig. 1.2 , stage 1). Cell shape is maintained by the cytoskeleton protein MreB2 (Butan et al. 2011; Fenton et al. 2010c;) with the coiled-coil-repeat protein CcrP probably acting as an underlying scaffold (Fenton et al. 2010a). The nucleus is tightly packed, and electron-dense granules resembling acidocalcisomes, enriched in phosphorus, calcium, and oxygen can be found in the cytoplasm (Borgnia et al. 2008).
The AP cell is endowed with a single sheathed flagellum composed of six different flagellin proteins (FliC1-6) that propels it to velocities as high as 160 μm s−1 (Iida et al. 2009; Lambert et al. 2006; Seidler and Starr 1968; Thomashow and Rittenberg 1985). None of the flagellin genes appears to be essential except for fliC3 which is required for predation in suspension cultures (Lambert et al. 2006). Likewise, three pairs of MotAB flagellar motor proteins contribute unevenly to flagellar rotation, none being essential (Morehouse et al. 2011). Flagellar motility is crucial for encountering prey but is neither required for prey penetration nor for slow surface-associated gliding motility (15–20 μm h−1). Gliding appears to be advantageous in predation of biofilms and in low-moisture environments (Abram et al. 1974; Lambert et al. 2006, 2011; Medina et al. 2008).
Multiple methyl-accepting chemotaxis proteins (MCP) sense various ligands, providing chemotactic cues towards inorganic ions, organic acids and amino acids, and oxygen (LaMarre et al. 1977; Sourjik and Wingreen 2012; Straley et al. 1979). Although attraction towards prey bacteria is only noticeable at high prey concentrations (Straley and Conti 1977), deletion of mcp2 reduces predation, suggesting chemotaxis is involved in prey detection (Lambert et al. 2003).
-
II.
Attachment to prey. Attachment of BALO to prey (Fig. 1.2 , stage 2) is affected by many factors such as the composition and the pH of the medium, oxygen tension, and temperature (Varon and Shilo 1968). At first attachment is reversible – as seen when the predator encounters non-prey cells (Shemesh et al. 2003) – but within minutes, it becomes irreversible. The basis of prey recognition by BALOs is still obscure. Core oligosaccharides of the prey’s lipopolysaccharide layer (LPS) are sensed by B. bacteriovorus, yet when they are depleted, attachment stalls but is not abolished. In contrast, attachment by Bacteriolyticum stolpii is indifferent to prey LPS composition but is reduced in the absence of particular porins (outer membrane proteins, OMPs) such as OmpF and OmpC in E. coli (Schelling and Conti 1986; Varon and Shilo 1969). BALOs synthesize unique membrane lipid structures that greatly vary between taxa: in B. bacteriovorus, the lipid A is completely devoid of negatively charged groups and possesses α-d-mannopyranose residues instead of phosphate groups; B. stolpii contains sphingophospholipids with unique 2-amino-3-phosphonopropanate heads; and Peridibacter starrii’s lipids include phosphatidylethanolamine structures with an additional N-glutamyl residue (Muller et al. 2011; Nguyen et al. 2008). The varied composition of lipids implies that predator’s membrane fluidity and permeability are altered and, in turn, its interaction with the prey surface. However, the ability of the different predators to use the same prey (e.g., E. coli or pseudomonads) while bearing diverse lipid structures in their outer membranes suggests that these structures whether have no meaningful interaction with the prey, interact with different components of the prey’s cell wall, or interact with similar components of the prey through different mechanisms.
Type IVa pili are present at the nonflagellated pole of AP cells and are essential for prey attachment and penetration of periplasmic and probably for attachment of B. exovorus to its prey as well (Evans et al. 2007; Mahmoud and Koval 2010). Anchoring of the pilus onto the prey envelope enables the invasion of the prey periplasm. This appendage is kept during GP, when it is found adhering to the prey cytoplasmic membrane. The machinery for its assembly is expressed throughout the entire growth cycle, suggesting that the pilus also plays a role during replication. In addition, a type IVb Flp pilus promotes B. bacteriovorus predation in biofilm (Medina et al. 2008).
-
III.
Prey invasion and bdelloplast formation. Irreversible attachment triggers local lysis of the prey envelope (Abram et al. 1974). Penetration of the periplasm by B. bacteriovorus is achieved by the predator squeezing through a pore (Evans et al. 2007) (Fig. 1.2 , stage 3), a process that may also involve the use of pili (Evans et al. 2007; Mahmoud and Koval 2010). The process is completed within 5–20 min after attachment and is sensitive to streptomycin (Varon and Shilo 1968), suggesting the production of enzymes in the formation of the pore. All the while, the prey peptidoglycan is modified, producing a bdelloplast. At that stage, damage to the prey’s cytoplasmic membrane leads to a rapid drop in prey respiration (Rittenberg and Shilo, 1970).
Bdelloplast construction is brought about by the activity of glycanase(s) and the solubilization of 10–15 % of the cell wall’s N-acetylglucosamine (Thomashow 1978a). To prevent premature prey cell lysis, N-deacetylase(s) controls glycanase activity immediately after penetration (Thomashow 1978b). Acylation of the prey peptidoglycan by long-chain fatty acids (Thomashow 1978c) and solubilization of 25 % of the LPS glucosamine by a lipopolysaccharidase activity (Thomashow 1978a) increase bdelloplast hydrophobicity, potentially stabilizing the outer membrane, which now acts as an osmotic barrier (Cover 1984). The growth chamber is further altered by the action of peptidases that actively cleave Braun’s lipoprotein (Thomashow 1978b), the release of diaminopimelic acid (DAP) from the peptidoglycan during penetration and latter during prey cell lysis (Thomashow 1978a), and the re-incorporation of DAP into the prey peptidoglycan during penetration and bdelloplast stabilization (Araki 1988; Ruby 1984).
After predator invasion, a bdelloplast is immune to superinfection. This was initially thought to result from N-deacetylation of the peptidoglycan (Thomashow 1978c). However, (Tudor et al. 1990) observed that in Bdellovibrio sp. strain W both glycanase and N-deacetylase activities are lacking and the non-spherical bdelloplasts generated are not superinfected. This and other data suggested that peptidase and not glycanase activity enables prey penetration and that bdelloplast rounding is a by-product caused by host autolytic muramidases (Tudor et al. 1990). However, (Lerner et al. 2012) showed that a double mutation in two homologs of the PBP4 DD-endo/carboxypeptidase that are mostly expressed during invasion leads to non-spherical bdelloplasts. The mutants were also slower to penetrate the prey, demonstrating that peptidase activity is a non-exclusive enzymatic requirement for invasion; further, single mutants in each of the encoding genes and more so the double mutant were sensitive to superinfection.
-
IV.
Growth and division. The intraperiplasmic B. bacteriovorus predator grows in a filamentous manner (Fig. 1.2 , stages 4 and 5) at the expanse of the prey cytoplasmic content. It incorporates up to 70 % of the prey’s DNA material by sequentially digesting it with dedicated enzymes (Rosson and Rittenberg 1979). The predator also degrades 20–40 % of the prey’s RNA ribonucleotides into the base and the ribose-l-phosphate moieties. The sugar phosphate is then used for energy production and for the biosynthesis of non-nucleic acid cell material (Hespell and Odelson 1978). Yet, BALOs encode the full complement of genes for purine and pyrimidine metabolism (Pasternak et al. 2012; Rendulic et al. 2004; Wang et al. 2011). In contrast, BALOs lack the ability to synthesize and degrade various amino acids and riboflavin, which should be acquired from the host (Pasternak et al. 2012; Rendulic et al. 2004). Other prey cell components were thought to be imported and utilized as building blocks by the predator, such as lipopolysaccharide moieties (Kuenen and Rittenberg 1975; Stein et al. 1992) and OMPs (Diedrich et al. 1984). It was shown that in fact, B. bacteriovorus synthesizes its own lipopolysaccharides (Schwudke et al. 2003) and does not import OMPs (Barel et al. 2005; Beck et al. 2004). Strikingly, and for hitherto unknown reasons, most BALOs use the mevalonate pathway instead of the common DOXP bacterial pathway (Pasternak et al. 2012).
The cytoskeleton is affected during GP: the MreB eukaryotic actin homologue MreB1 is essential, as hampering polymerization of the protein leads to arrested growth early in bdelloplast formation (Fenton et al. 2010c). Localization studies of MreB2-mTFP in AP cells showed it to be intimately connected to the spiral-shaped nucleoid. Also, the position of the nucleoid at approximately equal distances from the cell poles suggested that a parallel pattern of extension with cell length during cell division (Butan et al. 2011). Finally, the filament divides into progeny, the number of which is proportional to the size of the prey (Kessel and Shilo 1976). The number of progeny varies, so that odd and even numbers of Bdellovibrio are produced (Fenton et al. 2010b): an average of 5.7 progeny per prey in E. coli (Seidler and Starr 1969a) and up to 30 in Aquaspirillum serpens (Stolp 1967) (Fig 1.2 , stage 6). Division exhibits particular features as septation events occur synchronously along the filamentous Bdellovibrio cell, even in doubly infected prey (Fenton et al. 2010b). The resulting attack phase cells escape from the bdelloplast through discrete pores (Fig. 1.2 , stage 7). AP cells then mature and increase in length (Fenton et al. 2010b).
Marine BALOs were shown to produce stable bdelloplasts under nutrient deprivation, synchronous infection of stationary phase prey, and final low concentration of bdelloplasts. These structures remained viable for months, in contrast to attack phase cells that died rapidly but were as sensitive to environmental challenges (Sanchez Amat and Torrella 1990). They lysed in the presence of yeast extract, releasing AP cells.
-
V.
Bdellocysts. A few Bdellovibrio strains have been reported to enter a cyst-like stage under low-nutrient conditions and multiplicity of infection per prey cell (Tudor and Conti 1977). Bdellocysts occur in an infected prey. The predatory cell enlarges into a kidney-shaped cell enclosed by a structureless, amorphous outer layer. A finely particulate inner layer surrounds the more particulate plasma membrane of the predatory cell. Structures resembling storage granules are present. Bdellocysts are more resistant than vegetative cells to desiccation, high temperatures, and sonication (Tudor and Conti 1977), and their germination is favored by l-glutamate, K+, and NH4+ (Tudor and Conti 1978).
Host Independence
Soon after the discovery of Bdellovibrio bacteriovorus, saprophytic variants capable of growing in the absence of prey were isolated by plating concentrated suspensions of WT cells on a standard complete medium (Stolp and Petzold 1962; Stolp and Starr 1963). To date, all the periplasmatically growing B. bacteriovorus strains and Bacteriovorax species tested are able to generate saprophytic derivatives under laboratory conditions (Baer et al. 2000, 2004; Schwudke et al. 2001; Seidler and Starr 1969b). These variants, coined host-independent (H-I), manifest the archetypical dimorphic life cycle and retain a predatory potential (Fig. 1.2 , stage 8). They are, hence, facultative predators. Yet, predation is less efficient than in the parental strain (Cotter and Thomashow 1992a, b). Additionally, sequential transfers on complete medium without prey result in the loss of predatory ability (Roschanski et al. 2011; Varon and Seijffers 1975; Wurtzel et al. 2010). H-I BALOs may also occur in the environment as such strains were isolated on several occasions (Diedrich et al. 1970; Doskina 1973; Hobley et al. 2012b). Unique characteristics of H-I variants, in comparison to wild-type (WT) progenitors, suggest that host independence might be a genuine stage of the BALOs’ lifestyle. Unlike colorless WT cells, H-I isolates produce a yellowish pigment, protective against photooxidative damage (Friedberg 1977). H-I derivatives of B. bacteriovorus utilize a broader variety of carbon sources (Ishiguro 1974), synthesize different LPS structures (Schwudke et al. 2003), and, perhaps most intriguingly, form tenacious biofilms (Medina and Kadouri 2009).
H-I variants are isolated on a standard complete medium where the vast majority of the isolates form small colonies called “type I” (Seidler and Starr 1969b; Varon and Seijffers 1975). These colonies cannot be sub-cultured after initial development unless a large inoculum is streaked to form tight and small growing colonies or if the medium is supplemented with an extract of prey cells (Gray and Ruby 1990). Under such conditions, about 1 % of the isolates will forms large colonies that can be regrown in a density-independent manner on standard, un-supplemented medium, forming “type II” mutants (Thomashow and Cotter 1992). “Type I” H-I mutants are cell-extract dependent (they are saprophytic) and result from a single mutation; “type II” H-I mutants do not require cell extract (they are axenic) and result from an additional mutation, i.e., they are double mutants. Type I mutants arise at a frequency of 10−6 to 10−7 (Seidler and Starr 1969b; Varon and Seijffers 1975); type II H-I mutants are selected from type I at a frequency of 10−2 to 10−3 (Thomashow and Cotter 1992). Strikingly, it has been reported that H-I derivatives (type undefined) can be obtained at a frequency of up to 10−2 (Dashiff and Kadouri 2009). These data suggest that at least part of the pathway leading to the axenic phenotype is mutation prone and not based on single, random events. Genetic studies in B. bacteriovorus addressed the genetic background for this gradual acquisition of host independence, identifying the genomic loci implicated in it (Cotter and Thomashow 1992a, b; Roschanski et al. 2011; Wurtzel et al. 2010). Deleterious mutations in bd0108, a gene with no known function, lead to the type I phenotype. Moreover, not all BALO forming H-I variants contain bd0108 homologs in their genomes, and H-I mutants with a WT bd0108 allele were isolated, thus indicating that other gene products may underlie this phenotype (Lambert et al. 2010a; Schwudke et al. 2001; Wurtzel et al. 2010). Type II H-I mutants result from alterations in rhlB (bd3461) or pcnB (bd3464). These two genes encode for distinct components of the degradosome machinery, which is a multiprotein complex involved in RNA turnover. A loss of function of each enables progression from a type I to a type II H-I mutant (Roschanski et al. 2011).
The identities of the prey molecules necessary for WT or saprophytic growth are still not known. It has been shown that prey extract is required for initiation of DNA synthesis in saprophytic H-I mutants and in WT cells released from bdelloplasts. In contrast, prey extract cannot promote de novo proliferation of WT AP cells (Gray and Ruby 1990; Ruby and Rittenberg 1983; Thomashow and Cotter 1992), suggesting that replication of WT cells relies on two cues from the prey: one leading to a physiological transition from AP to GP and another one activating DNA synthesis (Gray and Ruby 1991). To date, the nature of the first cue is not known. The second cue is soluble, heat stable, resistant to RNase or DNase treatments, and fractionated over a wide range of molecular masses (10 to >200 kDa) (Gray and Ruby 1990). Saprophytic mutants overcome the need to sense the first cue; axenic mutants surmount the requirement for the second cue as well (Roschanski et al. 2011; Thomashow and Cotter 1992).
Cell Cycle Genetics
-
I.
Phase transition. Each phase is characterized by different gene and protein expression patterns and by characteristic activities (Karunker et al. 2013; Lambert et al. 2010a; McCann et al. 1998; Roschanski et al. 2011; Thomashow and Cotter 1992) Work on H-I mutants and prematurely released cells from bdelloplasts showed that the transition between AP to GP necessitates sensing of two prey cues (Gray and Ruby 1991; Roschanski et al. 2011; Thomashow and Cotter 1992): the first being activated during attachment (Thomashow and Cotter 1992), the second during growth (Gray and Ruby 1990). Such programming is most likely governed by distinct, master regulators whose modulations afford a swift transition between the phases (Lambert et al. 2010a; McCann et al. 1998). In B. bacteriovorus FliA (sigma 28) promoters are over represented upstream of AP-specific genes. Flia by itself is overexpressed during AP. It is thus reasonable to assume that FliA acts as an AP master regulator (Karunker et al. 2013), been shown that RpoE-like sigma factors in B. bacteriovorus are not essential but affect predatory efficiency or regulate chaperonin levels (Lambert et al. 2012). The heat shock response might be involved in phase transition, as heat shock promotes axenic growth (Gordon et al. 1993; Wang et al. 2011).
The prevalent second messenger molecule cyclic di-GMP is implicated in lifestyle determination in many bacteria (Mills et al. 2011) and BALOs are no exception. BALO genomes encode for a plethora of cyclic di-GMP synthesizing and degrading enzymes (diguanylate cyclases and phosphodiesterases, respectively) as well as cyclic di-GMP and a single cyclic di-GMP type I riboswitch (Karunker et al. 2013). Cyclic di-GMP signaling has been shown to be essential in determining WT and H-I phenotypes, as well as to affect flagellar and gliding motility in B. bacteriovorus (Hobley et al. 2012a). Such phenotypic differentiation is achieved by different sets of specific diguanylate cyclases (Hobley et al. 2012a) The cyclic di-GMP riboswitch is abnormally expressed during AP and is down regulated during GP. Its function is yet to be deciphered (Karunker et al. 2013).
-
II.
Genetics, genomics, and post-genomics. High-throughput transcriptome analyses were performed in the periplasmic predator B. bacteriovorus and in the epibiotic predator M. aeruginosavorus (Karunker et al. 2013; Lambert et al. 2010a; Wang et al. 2011). In all, global transcriptional changes correlated with phase transition. Lambert et al. 2010a compared transcription profiles of AP, predatory (i.e., 30 min post-prey infection) and host-independent (H-I) B. bacteriovorus, and identified exclusively overexpressed genes in each state. This enabled them to confine subsets of genes (most of which unannotated) and functions to the AP, the invasion phase and the GP. Motility and taxis genes were overexpressed in the AP. Cell wall metabolism, activation of transport systems, and early replication functions were expressed during the early stages of predation. Macromolecule degradation, massive transport, biosynthesis pathways, and DNA replication were expressed in the H-I samples and inferred to be expressed in the GP. Clearly, H-I mutants represented a transcriptional mosaic, mixing profiles unique to the AP, to attachment and, probably, to the GP. These data thus support the hypothesis that the mutations underlying the H-I phenotype essentially lead to a cell cycle freed from regulatory constraints (Dori-Bachash et al. 2008; Roschanski et al. 2011; Thomashow and Cotter 1992) Karunker et al. (2013) utilized whole transcriptome sequencing to find large subset of B. bacteriovorus genome exclusively expressed over AP and GP. Here again, genes encoding for motility, chemotaxis and cell surface proteins were upregulated in AP. Genes overexpressed during GP are related to cell growth, including ribosome biogenesis, cell division, DNA polymerase and chromosome partitioning proteins, and energy metabolism (Karunker et al. 2013). In the epibiotic a-proteobacterium M. aeruginosavorus expression profiles of AP and growth phase corresponded to those of the periplasmic, δ-proteobacterium B. bacteriovorus. Flagellar and chemotaxis genes were upregulated in the AP, while replication-associated genes and transport-related genes were upregulated during attachment. Surprisingly, hydrolase coding genes were found to be expressed constitutively (Wang et al. 2011).
Transport
Secretion by BALOs relies on type I and type II (sec) systems (not to be confounded with type I and type II H-I mutants) and include the twin arginine targeting protein translocation (Tat) system (Rendulic et al. 2004). Type III and type IV secretion systems are absent. Protein secretion into the prey cytoplasm is probably first accomplished via secretion into the prey periplasm and then by retrotranslocation into the host cytoplasm (Barabote et al. 2007). Another peculiar mechanism is the embedding by B. bacteriovorus of predator OMPs onto the prey cytoplasmic membrane, probably permeabilizing it to small hydrophilic molecules (Barel et al. 2005; Beck et al. 2004).
In B. bacteriovorus the Tat system is essential for growth of WT and of H-I strains. Some of the system’s components are specifically expressed during particular phases of the cell cycle and appear to promote the transfer of proteins to the prey cytoplasm (Chang et al. 2011).
BALO genomes encode for unusually large inventories of transport systems (Barabote et al. 2007). The δ-proteobacteria Bdellovibrionaceae and Bacteriovoracaceae bear numerous uptake systems for amino acids and peptides; in contrast, none is detected in the epibiotic α-proteobacteria Micavibrio aeruginosavorus (Hobley et al. 2012b; Rendulic et al. 2004; Wang et al. 2011). BALOs have few sugar transporters and depend on noncarbohydrate metabolism for carbon and energy. B. bacteriovorus has three sugar permeases, enabling the import of ribose, glycosides, maltose, and malto-oligosaccharides (Hespell et al. 1973). In contrast, many efflux pumps are found (Barabote et al. 2007). The phosphoenolpyruvate-dependent sugar transporting phosphotransferase system (PTS) is absent. Nucleotide uptake is a rare trait in bacteria, mostly found in obligate parasites. B. bacteriovorus is able to take up nucleotides, possibly through two different systems (Barabote et al. 2007; Ruby and McCabe 1986; Ruby et al. 1985). The M. aeruginosavorus genome lacks nucleotide transporter coding genes (Wang et al. 2011).
Energy Metabolism
In culture, BALOs do not enter stationary phase, and they are continuously using energy at high rates, whether for high-speed swimming or for growth and replication. During both phases, endogenous and substrate respiration rates, which are seven times higher than in E. coli (Hespell et al. 1973), lead to the saturation of the functional capacity of either the tricarboxylic acid cycle or the electron-transport chain (Hespell et al. 1973; Rittenberg and Shilo 1970). BALOs are incapable of fermentation and de facto are unable to use sugars (Seidler and Starr 1969a). They obtain energy (ATP) from amino acids, some organic acids (acetate and α-ketoglutarate), and polyhydroxyalcanoates (PHA) (Hespell et al. 1973; Martinez et al. 2012). ATP turnover in B. bacteriovorus during endogenous respiration is five times higher than in E. coli but similar during substrate respiration, while P/O ratios under both conditions are similar to these of E. coli (Gadkari and Stolp 1976). The ability of BALOs to recycle prey material renders them extremely energy efficient: GP B. bacteriovorus displays a YATP (biomass formed per ATP consumed) of 26, compared with 10.5 for bacteria cultivated in rich medium (Rittenberg and Hespell 1975).
The high respiration rates result in rapid energy depletion and a typically short half-life for BALOs, e.g., a 95 % loss in viability of a cell suspension of B. bacteriovorus in 20 h Hespell et al. 1973). This is due, at least in part, to BALOs’ energetically costly vigorous swimming which uses 20–40 % of the total available energy (Hespell et al. 1974). B. bacteriovorus exhibits a peculiar mode of oscillatory energy production in the absence of an exogenous substrate, by degrading its own cellular materials (Gadkari and Stolp 1975). This pattern fits the observed pattern of varying intensity – in contrast to constant rate – of RNA degradation under starvation (Hespell et al. 1974).
Survival is extended by using respirable substrates like amino acids, some organic acids, and PHAs. Lately, it has been shown that B. bacteriovorus is able to depolymerize PHA made by its prey and to use it to produce ATP (Hespell et al. 1973; Martinez et al. 2012).
Some BALOs are able to overcome harsh environmental conditions by entering a cyst-like state or, as shown with marine BALOs, to transform the bdelloplast into a dormant state (see above).
Taxonomy
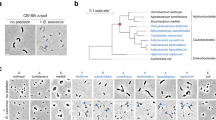
Bdellovibrio and like organisms form a polyphyletic taxon which is so defined for ease: the term describes all known obligate predatory bacteria, endowed with high motility, and having a basically two-phase life cycle composed of a search phase and of a growth and division phase. BALOs are found in the α-proteobacteria, where they form the genus Micavibrio and in the δ-proteobacteria where they form three families: the Bdellovibrionaceae, the Bacteriovoracaceae, and the Peridibacteriaceae (Fig. 1.3 ).
All long (>1,200 bp), cultured rDNA 16S sequences of the order Bdellovibrionales were retrieved from the RDP-II database, and the 375 sequences were aligned using MUSCLE. The alignment was trimmed at both ends to eliminate artificial gaps and overhangs, resulting in 1,248 bp. Pairwise distances between the aligned sequences were calculated using MOTHUR (Schloss et al. 2009), with consequent clustering which resulted in 38 operational taxonomic units (OTUs) at >97 % similarity between all sequences within each OTU. A representative sequence was chosen for each OTU, either a type sequence or the non-type sequence which had the lowest average pairwise distance to all the other members of the OTU. A phylogenetic tree containing the 38 representatives was inferred by the Maximum Likelihood method based on the Tamura-Nei model (Tamura and Nei 1993) in MEGA5 (Tamura et al. 2011). Sequence names are comprised of GenBank accession number, species name, strain (if available), and (T) (if a type sequence). The bootstrap consensus tree inferred from 200 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50 % bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (200 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of nucleotide substitutions per site
The Bdellovibrionaceae contains one species, Bdellovibrio, and two defined species, B. bacteriovorus and B. exovorus. B. bacteriovorus HD100T and B. exovorus JSST differ in % mol G+C content (50 % and 41 %, respectively) and diverge by 7 % (i.e., they are 93 % similar) in their 16S rRNA gene sequence (Koval et al. 2012). Yet, the major difference between the two is in their predatory strategy: B. bacteriovorus is a periplasmic predator, i.e., it penetrates and settles in the periplasmic space of its gram-negative prey, but B. exovorus is epibiotic, remaining attached to the prey’s cell wall (for details see the section “The BALOs’ Life Cycle”). The Bdellovibrionaceae was defined as encompassing nine clusters (Davidov and Jurkevitch 2004). The advent of large-scale sequencing of environmental samples revealed that the Bdellovibrionaceae tree splits into two clusters: one encompassing all the sequences from cultured strains as well as sequences from uncultured bacteria and another cluster without any cultured representative. This latter group may thus represent organisms that are different from the “classic” Bdellovibrio, maybe due to their inability to use the organisms used so far as prey to isolate the predators (Koval et al. 2012).
The Bacteriovoracaceae is composed of 10 phylogenetic clusters based on 96.5 % gene similarity in the 16S rRNA gene sequence of environmental isolates (Pineiro et al. 2007). These clusters are robust, as shown by further analysis of the rpoB gene that enables finer subdivisions but retains the same clusters (Pineiro et al. 2008). The family encompasses nine clusters of the saltwater Bacteriovorax, with the defined species Bacteriovorax marinus and Bacteriovorax litoralis (Baer et al. 2004), as well as one cluster of the freshwater/soil Bacteriolyticum stolpii (Pineiro et al. 2008). The former has a % mol G+C content of 37.7–38.3, and the latter of 41.8.
The Peridibacteriaceae were recently split as a monophyletic offshoot of the family Bacteriovoracaceae based on a % mol G+C content of 43.5 %, on its presence in freshwater and soil but not in saline environments, and in marked differences in 16S rRNA gene sequence with both Bdellovibrionaceae and Bacteriovoracaceae (Pineiro et al. 2008) (Fig. 1.3 ).
Few, yet striking, relationships between phylogeny and ecological parameters have been discovered: as mentioned above, Bacteriovorax are found in salt waters, while Bdellovibrio, Bacteriolyticum, and Peridibacter are freshwater and soil isolates. More specifically, Bacteriovorax clusters are widely distributed, but cluster V has been exclusively found in estuarine environments (Pineiro et al. 2007). Further, particular taxa appear to be differentially associated with prey specificity: in prey spiking experiments of natural water samples, it was shown that Bacteriovorax cluster IX is a versatile predator, able to prey as efficiently on various prey that other separate clusters specialize on (Chen et al. 2011). It was further discovered that cluster IV, consisting of predators that are predominantly isolated from low-salt waters, is selected for by the addition of prey bacteria originating from freshwater (Chen et al. 2012). In the Bdellovibrionaceae, predators isolated using Agrobacterium tumefaciens as prey, and originating from sources such as the tomato rhizosphere and soils in Israel and soils in Germany and India, formed discrete clusters, separated from clusters containing isolates from many sources and using both various enterobacteria and pseudomonads as prey (Davidov and Jurkevitch 2004). These data strongly suggest that both prey and environmental parameters shape BALO communities in the environment.
Micavibrio is defined as two species: M. admirantus (% mol G+C content 57.1 %), which only grows on Stenotrophomonas (Pseudomonas) maltophilia (Lambina et al. 1982), and M. aeruginosavorus (% GC 54.7 M) which grows on various enterobacteria and pseudomonads. Micavibrio forms a deep branching clade in the α-proteobacteria, sister to the Rhodospirillales (Davidov et al. 2006b; Wang et al. 2011). Very few isolates and environmental sequences of Micavibrio are available.
Habitat and Ecology
BALOs are widely distributed in marine, freshwater, and terrestrial ecosystems, including estuaries, seacoasts and oceans, rivers, sewage, fish ponds, runoff of irrigation water, and man-made water supplies (Davidov and Jurkevitch 2004; Framatico and Cooke 1996; Fry and Staples 1976; Pineiro et al. 2007; Schoeffield and Williams 1990; Snyder et al. 2002). BALOs have been isolated from the gills of crabs, sediments, submerged surfaces, soils, rice paddies, the rhizosphere of plants, and fish ponds (Chu and Zhu 2009; Kelley et al. 1997; Uematsu 1980; Williams et al. 1995) (Jurkevitch et al. 2000; Stolp and Starr 1963). They have been isolated from animal feces (Schwudke et al. 2003) and detected as forming dominant populations in the leech Hirudo verbana (Kikuchi et al. 2009). BALOs colonize biofilms that form in aquatic habitats (Kelley et al. 1997), and under lab conditions, they efficiently clear biofilms (Kadouri and O’Toole 2005; Kadouri et al. 2007). As mentioned above (see taxonomy section), salinity and, less so, prey range are ecological factors that can be used to characterize BALO clades.
The abundance and diversity of BALO populations as appraised by culture-based approaches appear to have been largely underestimated. Indeed, first estimates of population sizes using specific primers targeting the different BALO clades have shown their abundance to be more than 2.5 orders of magnitude higher than that detected by counting plaques (Van Essche et al. 2009b; Zheng et al. 2008). A limited evaluation of the relative abundance of BALOs, based on 16S rRNA gene sequence distributions in a wide range of environments, showed that the predators account in average for close to 0.2 % of total bacteria (Table 1.1 ). Similarly, culture-independent technologies reveal that BALO diversity is much larger than the one detected by the characterization of isolates (Davidov et al. 2006a).
BALOs are able to consume the majority of gram-negative cells present in natural water bodies (Rice et al. 1998). They were shown to move to spots of concentrated native aquatic bacteria (Chauhan et al. 2009) and to respond to sudden increases or “spikes” in numbers of specific prey bacteria or consortia of native microbial communities (Chauhan et al. 2009; Chen et al. 2011, 2012).
BALOs are considered as obligate aerobes but they are commonly found in sediments (Williams 1988). Recently, Monnappa et al. (2013) showed that B. bacteriovorus successfully preyed under complete anoxic conditions as long as nitrate was supplemented. Nitrite reductase and nitrite oxide reductase but not bona fide nitrate reductase genes are found in B. bacteriovorus (Rendulic et al. 2004). In Micavibrio, M. aeruginosavorus EPB but not strain ARL-13 encodes for a nitrate reductase complex (Pasternak et al. 2012).
Prey acquisition of resistance to BALO predation was shown by Varon (1979), using a chemostat containing B. bacteriovorus with its prey Photobacterium leiognathi (Varon 1979). Also, Gallet et al. (2007, 2009) demonstrated that predation pressure can lead to the selection of resistant prey but that the type of resistance (moderate or extreme) depended upon the ecological conditions under which selection occurred. In addition to such genetic resistance, it appears that prey may also exhibit transient plastic responses in prey populations thereby eradication preventing eradiction. However, resistance falters as prey populations expend (Shemesh and Jurkevitch 2004). Defense mechanisms may be triggered in predatory cultures: Lambert et al. (2010b) reported a futile transcriptional “scream” of genes as a response to predation-induced osmotic stress in E. coli cultures, 15 min postinfection with B. bacteriovorus.
Applications
The BALOs’ characteristic lifestyle makes them attractive candidates for a number of applications: BALOs have been proposed as living alternatives to chemical antibiotics(Sockett and Lambert 2004), as agents for improving water quality in aquaculture (Qi et al. 2009), as a means to control dental plaque bacteria (Dashiff and Kadouri 2011; Van Essche et al. 2009a), and as treatment of Salmonella-induced dysbiosis in chickens (Atterbury et al. 2011). This follows older attempts made to use BALOs in animal models: Nakamura (1972) effectively treated Shigella flexneri-induced keratoconjunctivitis in rabbit with Bdellovibrio, but BALO use against pathogens in the intestinal tract of rabbits was unsuccessful (Westergaard and Kramer 1977). Very few studies explored the potential of BALOs against phytopathogens: BALOs efficiently eradicated Xanthomonas oryzae from rice paddy field water and caused a rapid decline in populations of Pectobacterium carotovorum subp. carotovorum in soil (Uematsu 1980). BALO isolates from the rhizosphere of soybean were used to control bacterial blight caused by Pseudomonas glycinea (Scherff 1973). Reduction in disease severity and systemic symptoms was significant.
BALOs can also be employed as theoretical models for understanding the evolution of the eukaryotic cell (Davidov and Jurkevitch 2009; Guerrero et al. 1986); they also are convenient empirical models for testing hypotheses pertaining to ecological and evolutionary theories (Gallet et al. 2007, 2009; Wilkinson 2007).
Isolation
BALOs are isolated as bacteriophages are, using a prey as “bait.” The sample, or serial dilutions of it, is mixed with a potential prey bacterium in melted soft agar and poured onto an agar plate containing a diluted growth medium. The bacterial predator forms lytic, transparent plaques that have to be differentiated from those formed by protozoa or bacteriophages. A drawback is that only BALOs able to prey on the proposed bacterium can be retrieved. As BALOs vary in host range, no single bacterial species can potentially support the growth of all isolates. However, Vibrio parahaemolyticus was shown to be an effective host for the retrieval of Bacteriovoracaceae from marine environments (Pineiro et al. 2008). Another limitation of the method stems from the presence of much higher levels of non-predatory bacteria in the sample that can grow on the plate and blur the detection of plaques.
Direct Isolation of Bdellovibrio from Environmental Samples
The most common approach is based on the use of one or more filtration steps with or without differential centrifugation of the sample analyzed.
Based on Stolp (1981), a water sample or 50-g soil in 500 mL in sterile buffer is shaken vigorously for 1 h and then centrifuged for 5 min at 2,000 g to remove gross particles. The supernatant is passed through a series of membrane filters of decreasing pore size (3.0, 1.2, 0.8, and 0.45 μm). Filtrates from the last two steps are serially diluted, and 100-μL aliquots are mixed with approximately 109 cells of the prey bacterium in molten soft top low-nutrient or buffer agar. The mixture is poured onto a low-nutrient or buffer agar plate and incubated at 28–32 °C. Rapidly developing lytic regions visible after 24 h are the result of bacteriophage multiplication. These plaques are usually small and do not grow further. They should be marked to differentiate them from the slower-growing BALOs. Plaques becoming visible within 2–3 days and showing further expansion for a few more days are potentially BALOs (Fig. 1.4 ). Small and highly motile BALO cells can be detected by examination of crushed plaque material in wet mounts with a phase contrast microscope.
Media. DNA: (diluted nutrient agar (Seidler and Starr 1969b)); 0.08 % Difco Nutrient Broth is supplemented with CaCl2 · 2H2O, 2 mM, and MgCl2 · 6H2O, 3 mM after autoclaving, and pH is adjusted to 7.2 with 0.1 N NaOH.
Bottom agar: 1.2–1.5 %. Top agar: 0.6 %.
Plating: Aliquots of 4-mL molten top agar are kept at 42 °C in a water bath prior to mixing with prey and sample suspensions.
Marine BALOs require salts to grow. Therefore, the medium used for isolating marine strains should contain at least 25 % sea water or appropriate salts (Marbach and Shilo 1978). A common method used to isolate marine BALOs is that of (Schoeffield and Williams 1990): 5 mL of the sample is added to 3.3 mL of molten top agar having 0.7 mL of the prey suspension for a final agar concentration of 0.65 %. The mixture is then poured onto large Petri dishes and incubated at 25 °C.
Media. Polypeptone (Pp 20) medium (Williams et al. 1982): Filtered ocean water, 1 L; Polypeptone, 1 g; agar, 15 g for bottom agar and 6.5 g for top agar; pH 7.7–7.8.
Synthetic marine salt solution (Marbach and Shilo 1978): NaCl, 500 mM; KCl, 10 mM; MgSO4, 25 mM; MgCl2, 25 mM; CaCl2, 10 mM.
Specific Enrichment for Bdellovibrio
Stolp (1968) devised a method yielding large numbers of BALOs, thereby greatly facilitating the isolation of predators on a specific host bacterium whenever quantification is not needed. This approach has been applied to obtain BALOs lytic to Rhizobium and Agrobacterium (Parker and Grove 1970), Legionella (Richardson 1990), and Azospirillum brasilense from 2-year-stored air-dried soils (Germida 1987).
Based on Stolp, modified by Ruby (1991): One-hundred milliliter aliquots of a dense suspension of the prospective prey bacterium (1010 cells mL−1) are prepared in DN medium or HM buffer in Erlenmeyer flasks. The sample (soil, 100 mg; sewage, 0.5 mL; river water, 1 mL) is added and the flasks are incubated on a rotary shaker. The suspension is examined daily over 2–4 days for lysis (reduction in optical density) and for the presence of small, highly motile presumptive BALO cells or bdelloplasts by phase contrast microscopy. If no BALOs are apparent, the incubation can be prolonged or a 1-ml aliquot can be transferred into a fresh suspension of substrate bacterium. When BALOs are detected, the enrichment culture is centrifuged for 5 min at 2,000 g (250 g) (Germida 1987)and the supernatant filtered through a 0.45-μm membrane. Serial dilutions are plated on the prospective prey bacterium to obtain plaques. Developing plaques are checked microscopically for small, highly motile cells.
HM buffer: Hydroxyethyl piperazine-N′-2-ethanesulfonic acid, 25 mM adjusted to pH 7.6 with NaOH and supplemented with CaCl2 · 2H2O, 1 mM and MgCl2 · 6H2O, 1 mM.
Isolation and Cultivation of H-I Mutants
H-I BALOs are isolated by introducing WT attack phase cells in a rich medium such as PYE (g L−1-peptone, 10; yeast extract, 3; MgCl2, 3 mM; CaCl2, 2 mM; pH 7.4–7.6) without prey, selecting for growing isolates. A drawback of this approach is the spurious growth of residual prey cells in the medium. To overcome this shortcoming, lytic suspensions are filtered through a 0.45 μm membrane, efficiently separating BALO cells from prey bacteria but resulting in low recovery rates (Shilo and Bruff 1965). An efficient approach utilizes streptomycin-resistant (Smr) host-dependent isolates. After cultivation with a streptomycin-sensitive prey, a lytic suspension containing Smr AP cells is inoculated onto a complete medium amended with streptomycin, allowing the growth of H-I mutants but restricting that of the Sm-sensitive prey (Seidler and Starr 1969b). Recently, Dashiff and Kadouri (2009) used prey auxotrophic to diaminopimelic acid (DAP) that could not grow in a complete medium devoid of DAP. They obtained H-I mutants without filtration or selection for antibiotic resistance at a very high frequency (up to 10−2) (Dashiff and Kadouri 2009).
Mutants can grow as a suspension or form yellowish colonies on a solid medium. Saprophytic (type I) H-I mutants are essentially cultured with buffered heat-killed prey bacteria (109 CFU mL−1, MgCl2, 3 mM; CaCl2, 2 mM, HEPES or DNB, pH 7.4–7.6) or on PYE medium amended with cell extract (usually derived from prey bacteria, 0.3 mg mL−1 protein). Both media are suitable for liquid as well as for agar-based growth. Type I H-I mutants cultivated on heat-killed prey have to be laid over a semisolid medium using the double-layer agar plating (Schwudke et al. 2001). Colonies are then surrounded by clear halos (Roschanski et al. 2011).
Growth Requirements
With axenic mutants at hand, the minimal nutritional requirements of BALOs can be addressed. Ishiguro (1974) successfully developed a chemically defined medium for axenic B. bacteriovorus consisting of (g L−1): Na2HPO4, 7; KH2PO4, 3; NaCl, 0.5; MgSO4, 0.2; NH4Cl. Mutants were capable of assimilating ammonium and required no additional vitamins. Various amino acids, organic acids, and glycerol – but no sugars – were utilized to different efficiencies.
Conclusions
The field of predatory interactions between bacteria, as represented by studies of the Bdellovibrio and like organisms, has been revived in the past decade by the interest of new research groups and the introduction of modern molecular biology, genetics, and genomics technologies. These have enable to gain a much more detailed understanding of the taxonomy and phylogeny of this diverse group, to appreciate the intricate molecular mechanisms at play during the cell cycle of these fascinating bacteria as well as of their genomic particularities. Based on these significant advances, novel applications of predatory bacteria in medicine, agriculture, and environmental sciences may now become a reality.
References
Abram D, CastroeMelo J, Chou D (1974) Penetration of Bdellovibrio bacteriovorus into host cells. J Bacteriol 118:663–680
Araki Y, Ruby EG (1988) A soluble enzyme activity that attaches free diaminopimelic acid to bdelloplast peptidoglycan. Biochemistry 27:2624–2629
Atterbury RJ, Hobley L, Till R, Lambert C, Capeness MJ, Lerner TR, Fenton AK, Barrow P, Sockett RE (2011) Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl Environ Microbiol 77:5794–5803
Baer ML, Ravel J, Chun J, Hill RT, Williams HN (2000) A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int J Syst Evol Microbiol 50(Pt 1):219–224
Baer ML, Ravel J, Pineiro SA, Guether-Borg D, Williams HN (2004) Reclassification of salt-water Bdellovibrio sp. as Bacteriovorax marinus sp. nov. and Bacteriovorax litoralis sp. nov. Int J Syst Evol Microbiol 54:1011–1016
Barabote RD, Rendulic S, Schuster SC, Saier MH Jr (2007) Comprehensive analysis of transport proteins encoded within the genome of Bdellovibrio bacteriovorus. Genomics 90:424–446
Barel G, Sirota A, Volpin H, Jurkevitch E (2005) Fate of predator and prey proteins during browth of Bdellovibrio bacteriovorus on Escherichia coli and Pseudomonas syringae prey. J Bacteriol 187:329–335
Beck S, Schwudke D, Strauch E, Appel B, Linscheid M (2004) Bdellovibrio bacteriovorus strains produce a novel major outer membrane protein during predacious growth in the periplasm of prey bacteria. J Bacteriol 186:2766–2773
Borgnia MJ, Subramaniam S, Milne JLS (2008) Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J Bacteriol 190:2588–2596
Butan C, Hartnell LM, Fenton AK, Bliss D, Sockett RE, Subramaniam S, Milne JLS (2011) Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J Bacteriol 193:1341–1350
Chang CY, Hobley L, Till R, Capeness M, Kanna M, Burtt W, Jagtap P, Aizawa S, Sockett RE (2011) The Bdellovibrio bacteriovorus twin-arginine transport system has roles in predatory and prey-independent growth. Microbiology 157:3079–3093
Chauhan A, Cherrier J, Williams HN (2009) Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. Proc Natl Acad of Sci USA 106(11):4301–4306. doi:10.1073/pnas.0809671106
Chen H, Athar R, Zheng G, Williams HN (2011) Prey bacteria shape the community structure of their predators. ISME J 5(8):1314–1322
Chen H, Young S, Berhane T-K, Williams HN (2012) Predatory Bacteriovorax communities ordered by various prey species. PLoS One 7:e34174
Chu WH, Zhu W (2009) Isolation of Bdellovibrio as biological therapeutic agents used for the treatment of Aeromonas hydrophila infection in fish. Zoonoses and Public Health 57(4):258–64. doi:10.1111/j.1863-2378.2008.01224.x
Cotter TW, Thomashow MF (1992a) A conjugation procedure for Bdellovibrio bacteriovorus and its use to identify DNA sequences that enhance the plaque-forming ability of a spontaneous host-independent mutant. J Bacteriol 174:6011–6017
Cotter TW, Thomashow MF (1992b) Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J Bacteriol 174:6018–6024
Cover WH, Rittenberg SC (1984) Change in the surface hydrophobicity of substrate cells during bdelloplast formation by Bdellovibrio bacteriovorus 109J. J Bacteriol 157:391–397.
Dashiff A, Kadouri DE (2009) A new method for isolating host-independent variants of Bdellovibrio bacteriovorus using E. coli auxotrophs. Open Microbiol J 3:87–91
Dashiff A, Kadouri DE (2011) Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Mol Oral Microbiol 26:19–34
Davidov Y, Jurkevitch E (2004) Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax–Peredibacter clade as Bacteriovoracaceae fam. nov. Int J Syst Evol Microbiol 54:1439–1452
Davidov Y, Jurkevitch E (2009) Predation between prokaryotes and the origin of eukaryotes. Bioessays 31:748–757
Davidov Y, Friedjung A, Jurkevitch E (2006a) Structure analysis of a soil community of predatory bacteria using culture-dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and-like organisms. Environ Microbiol 8:1667–1673
Davidov Y, Huchon D, Koval SF, Jurkevitch E (2006b) A new α-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol 8:2179–2188
Diedrich DL, Denny CF, Hashimoto T, Conti SF (1970) Facultatively parasitic strain of Bdellovibrio bacteriovorus. J Bacteriol 101:989–996
Diedrich DL, Portnoy Duran CA, Conti SF (1984) Acquisition of Escherichia coli outer membrane proteins by Bdellovibrio sp. strain 109D. J Bacteriol 159:329–334
Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E (2008) Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl Environ Microbiol 74:7152–7162
Doskina TV (1973) Isolation of parasitic and saprophytic strains of Bdellovibrio bacteriovorus from natural waters. Gig Sanit 38:84–85
Evans KJ, Lambert C, Sockett RE (2007) Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J Bacteriol 189:4850–4859
Fenton AK, Hobley L, Butan C, Subramaniam S, Sockett RE (2010a) A coiled-coil-repeat protein “Ccrp”in Bdellovibrio bacteriovorus prevents cellular indentation, but is not essential for vibroid cell morphology. FEMS Microbiol Lett 313:89–95
Fenton AK, Kanna M, Woods RD, Aizawa SI, Sockett RE (2010b) Shadowing the actions of a predator: Backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol 192:6329–6335
Fenton AK, Lambert C, Wagstaff PC, Sockett RE (2010c) Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J Bacteriol 192:1299–1311
Framatico PM, Cooke PH (1996) Isolation of bdellovibrios that prey on Escherichia coli 0157:H7 and Salmonella species and application for removal of prey from stainless steel surfaces. J Food Safety 16:161–173
Friedberg D (1977) Effect of light on Bdellovibrio bacteriovorus. J Bacteriol 131:399–404
Fry J, Staples DG (1976) Distribution of Bdellovibrio bacteriovorus in sewage works, river water, and sediments. Appl Environ Microbiol 31:469–474
Gadkari D, Stolp H (1975) Energy metabolism of Bdellovibrio bacteriovorus. I. Energy production, ATP pool, energy charge. Arch Microbiol 102(3):179–185
Gadkari D, Stolp H (1976) Energy metabolism of Bdellovibrio bacteriovorus. II. P/O ratio and ATP pool turnover rate. Arch Microbiol 108:125–132
Gallet R, Alizon S, Comte P-A, Gutierrez A, Depaulis F, van Baalen M, Michel E, Muller-Graf CDM (2007) Predation and disturbance Interact to shape prey species diversity. Am Nat 170:143–154
Gallet R, Tully T, Evans MKE (2009) Ecological conditions affect evolutionary trajectory in a predator–prey system. Evolution 63:641–651
Germida JJ (1987) Isolation of Bdellovibrio spp. that prey on Azospirillum brasilense in soil. Can J Microbiol 33:459–461
Gordon RF, Stein MA, Diedrich DL (1993) Heat shock-induced axenic growth of Bdellovibrio bacteriovorus. J Bacteriol 175:2157–2161
Gray KM, Ruby EG (1990) Prey-derived signals regulating duration of the developmental growth phase of Bdellovibrio bacteriovorus. J Bacteriol 172:4002–4007
Gray KM, Ruby EG (1991) Intercellular signalling in the bdellovibrio developmental growth cycle. Microbial Cell-Cell Interactions ASM, Washington, DC
Guerrero R, Pedros-Alio C, Esteve I, Mas J, Chase D, Margulis L (1986) Predatory prokaryotes: predation and primary consumption evolved in bacteria. Proc Natl Acad Sci U S A 83:2138–2142
Hespell RB, Odelson DA (1978) Metabolism of RNA-ribose by Bdellovibrio bacteriovorus during intraperiplasmic growth on Escherichia coli. J Bacteriol 136:936–946
Hespell RB, Rosson RA, Thomashow MF, Rittenberg SC (1973) Respiration of Bdellovibrio bacteriovorus strain 109J and its energy substrates for intraperiplasmic growth. J Bacteriol 113:1280–1288
Hespell R, Thomashow MF, Rittenberg SC (1974) Changes in cell composition and viability of Bdellovibrio bacteriovorus during starvation. Arch Microbiol 97:313–327
Hobley L, Fung RK, Lambert C, Harris MA, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S, Gomelsky M, Sockett RE (2012a) Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog 8:e1002493
Hobley L, Lerner TR, Williams LE, Lambert C, Till R, Milner DS, Basford SM, Capeness MJ, Fenton AK, Atterbury RJ, Harris MA, Sockett RE (2012b) Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genomics 13:670
Iida Y, Hobley L, Lambert C, Fenton AK, Sockett RE, Aizawa S (2009) Roles of multiple flagellins in flagellar formation and flagellar growth post bdelloplast lysis in Bdellovibrio bacteriovorus. J Mol Biol 394:1011–1021
Ishiguro EE (1974) Minimum nutritional requirements for growth of host-independent derivatives of Bdellovibrio bacteriovorus strain 109 Davis. Can J Microbiol 20:263–264
Jurkevitch E, Minz D, Ramati B, Barel G (2000) Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl Environ Microbiol 66:2365–2371
Kadouri D, O'Toole GA (2005) Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol 71:4044–4051
Kadouri D, Venzon NC, O'Toole GA (2007) Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol 73:605–614
Karunker I, Rotem O, Dori-Bachash M, Jurkevitch E, Sorek R (2013) A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PLoS One 8:e61850
Kelley J, Turng B, Williams H, Baer M (1997) Effects of temperature, salinity, and substrate on the colonization of surfaces in situ by aquatic bdellovibrios. Appl Environ Microbiol 63:84–90
Kessel M, Shilo M (1976) Relationship of Bdellovibrio elongation and fission to host cell size. J Bacteriol 128:477–480
Kikuchi Y, Bomar L, Graf J (2009) Stratified bacterial community in the bladder of the medicinal leech, Hirudo verbana. Environ Microbiol 11:2758–2770
Koval SF, Hynes SH, Flannagan RS, Pasternak Z, Davidov Y, Jurkevitch E (2012) Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int J Syst Evol Microbiol 63(Pt 1):146–151
Kuenen JG, Rittenberg SC (1975) Incorporation of long-chain fatty acids of the substrate organism by Bdellovibrio bacteriovorus during intraperiplasmic growth. J Bacteriol 121:1145–1157
LaMarre AG, Straley SC, Conti SF (1977) Chemotaxis toward amino acids by Bdellovibrio bacteriovorus. J Bacteriol 131:201–207
Lambert C, Smith MC, Sockett RE (2003) A novel assay to monitor predator–prey interactions for Bdellovibrio bacteriovorus 109J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ Microbiol 5:127–132
Lambert C, Evans KJ, Till R, Hobley L, Capeness M, Rendulic S, Schuster SC, Aizawa S, Sockett RE (2006) Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol Microbiol 60:274–286
Lambert C, Chang CY, Capeness MJ, Sockett RE (2010a) The first bite–profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5:e8599
Lambert C, Ivanov P, Sockett RE (2010b) A transcriptional “Scream” early response of E. coli prey to predatory invasion by Bdellovibrio. Curr Microbiol 60:419–427
Lambert C, Fenton AK, Hobley L, Sockett RE (2011) Predatory Bdellovibrio bacteria use gliding motility to scout for prey on surfaces. J Bacteriol 193:3139–3141
Lambert C, Till R, Hobley L, Sockett RE (2012) Mutagenesis of RpoE-like sigma factor genes in Bdellovibrio reveals differential control of groEL and two groES genes. BMC Microbiol 12:99
Lambina VA, Afinogenova AV, Romai Penabad S, Konovalona SM, Pushkareva AP (1982) Micavibrio admirandus gen. et sp. nov. Mikrobiologiya 51:114–117
Lerner TR, Lovering AL, Bui NK, Uchida K, Aizawa S-I, Vollmer W, Sockett RE (2012) Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog 8:e1002524
Mahmoud KK, Koval SF (2010) Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio. Microbiology 156:1040–1051
Martinez V, Jurkevitch E, Garcia JL, Prieto MA (2012) Reward for bdellovibrio bacteriovorus for preying on a polyhydroxyalkanoate producer. Environ Microbiol
Marbach A, Shilo M (1978) Dependence of marine bdellovibrios on potassium, calcium, and magnesium ions. Appl Environ Microbiol 36:169–177
McCann MP, Solimeo HT, Cusick F Jr, Panunti B, McCullen C (1998) Developmentally regulated protein synthesis during intraperiplasmic growth of Bdellovibrio bacteriovorus 109J. Can J Microbiol 44:50–55
Medina AA, Kadouri DE (2009) Biofilm formation of Bdellovibrio bacteriovorus host-independent derivatives. Res Microbiol 160:224–231
Medina AA, Shanks RM, Kadouri DE (2008) Development of a novel system for isolating genes involved in predator-prey interactions using host independent derivatives of Bdellovibrio bacteriovorus 109J. BMC Microbiol 8:33
Mills E, Pultz IS, Kulasekara HD, Miller SI (2011) The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol 13:1122–1129
Monnappa AF, Dwidar M, Mitchell RJ (2013) Application of bacterial predation to mitigate recombinant bacterial populations and their DNA. Soil Biol Biochem 57:427–435
Morehouse KA, Hobley L, Capeness M, Sockett RE (2011) Three motAB stator gene products in Bdellovibrio bacteriovorus contribute to motility of a single flagellum during predatory and prey-independent growth. J Bacteriol 193:932–943
Muller FD, Beck S, Strauch E, Linscheid MW (2011) Bacterial predators possess unique membrane lipid structures. Lipids 46:1129–1140
Nakamura M (1972) Alteration of Shigella pathogenicity by other bacteria. Am J Clin Nutr 25:1441–1451
Nguyen NA, Sallans L, Kaneshiro ES (2008) The major glycerophospholipids of the predatory and parasitic bacterium Bdellovibrio bacteriovorus HID5. Lipids 43:1053–1063
Parker CA, Grove PL (1970) Bdellovibrio bacteriovorus parasitizing Rhizobium in Western Australia. J Appl Microbiol 33:253–255
Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, Jurkevitch E (2012) By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J 7(4):756–769
Pineiro S, Stine CO, Chauhan A, Steyert SR, Smith R, Williams HN (2007) Global survey of diversity among environmental saltwater Bacteriovoracaceae. Environ Microbiol 9:2441–2450
Pineiro SA, Williams HN, Stine OC (2008) Phylogenetic relationships amongst the saltwater members of the genus Bacteriovorax using rpoB sequences and reclassification of Bacteriovorax stolpii as Bacteriolyticum stolpii gen. nov., comb. nov. Int J Syst Evol Microbiol 58:1203–1209
Qi Z, Zhang X-H, Boon N, Bossier P (2009) Probiotics in aquaculture of China – Current state, problems and prospect. Aquaculture 290:15–21
Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett ER, Schuster S (2004) A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692
Rice TD, Williams HN, Turng BF (1998) Susceptibility of bacteria in estuarine environments to autochthonous bdellovibrios. Microb Ecol 35:256–264
Richardson IR (1990) The incidence of Bdellovibrio spp. in man-made water systems: coexistence with legionellas. J Appl Bacteriol 69:134–140
Rittenberg SC, Hespell RB (1975) Energy efficiency of intraperiplasmic growth of Bdellovibrio bacteriovorus. J Bacteriol 121:1158–1165
Rittenberg SC, Shilo M (1970) Early host damage in the infection cycle of Bdellovibrio bacteriovorus. J Bacteriol 102:149–160
Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E (2011) Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J Bacteriol 193:1745–1756
Rosson RT, Rittenberg SC (1979) Regulated breakdown of Escherichia coli deoxyribonucleic acid during intraperiplasmic growth of Bdellovibrio bacteriovorus 109J. J Bacteriol 140:620–633
Ruby EG (1991) The genus Bdellovibrio. In: Balows HGTA, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes. Springer, New York, pp 3400–3415
Ruby EG, McCabe JB (1986) An ATP transport system in the intracellular bacterium, Bdellovibrio bacteriovorus 109J. J Bacteriol 167:1066–1070
Ruby EG, Rittenberg SC (1983) Differentiation after premature release of intraperiplasmically growing Bdellovibrio bacteriovorous. J Bacteriol 154:32–40
Ruby EG, Rittenberg SC (1984) Attachment of diaminopimelic acid to bdelloplast peptidoglycan during intraperiplasmic growth of Bdellovibrio bacteriovorus 109J. J Bacteriol 158:597–602
Ruby EG, McCabe JB, Barke JI (1985) Uptake of intact nucleoside monophosphates by Bdellovibrio bacteriovorus 109J. J Bacteriol 163:1087–1094
Sanchez-Amet A, Torrella F (1990) Formation of stable bdelloplasts as a starvation-survival strategy of marine bdellovibrios. Appl Environ Microbiol 56(9):2717–2725
Schelling M, Conti S (1986) Host receptor sites involved in the attachment of Bdellovibrio bacteriovorus and Bdellovibrio stolpii. FEMS Microbial Lett 36:319–323
Scherff RH (1973) Control of bacterial blight of soybean by Bdellovibrio bacteriovorus. Phytopathology 63:400–402
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schoeffield AJ, Williams HN (1990) Efficiencies of recovery of bdellovibrios from brackish-water environments by using various bacterial species as prey. Appl Environ Microbiol 56:230–236
Schwudke D, Strauch E, Krueger M, Appel B (2001) Taxonomic studies of predatory bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Syst Appl Microbiol 24:385–394
Schwudke D, Linscheid M, Strauch E, Appel B, Zahringer U, Moll H, Muller M, Brecker L, Gronow S, Lindner B (2003) The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid A containing alpha-D-Mannoses that replace phosphate residues: similarities and differences between the lipid As and the lipopolysaccharides of the wild type strain B. bacteriovorus HD100 and its host-independent derivative HI100. J Biol Chem 278:27502–27512
Seidler RJ, Starr MP (1968) Structure of the flagellum of Bdellovibrio bacteriovorus. J Bacteriol 95:1952–1955
Seidler R, Starr MMP (1969a) Factors affecting the intracellular parasitic growth of Bdellovibrio bacteriovorus developing within Escherichia coli. J Bacteriol 97:912–923
Seidler RJ, Starr MP (1969b) Isolation and characterization of host-independent bdellovibrios. J Bacteriol 100:769–785
Shemesh Y, Jurkevitch E (2004) Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ Microbiol 6:12–18
Shemesh Y, Yaacov D, Susan K, Jurkevitch E (2003) Small eats big: ecology and diversity of Bdellovibrio and like organisms, and their dynamics in predator–prey interactions. Agronomie 23:433–439
Shilo M, Bruff B (1965) Lysis of gram-negative bacteria by host-independent ectoparasitic Bdellovibrio bacteriovorus isolates. J Gen Microbiol 40:317–328
Snyder AR, Williams HN, Baer ML, Walker SOC (2002) 16S rDNA sequence analysis of environmental Bdellovibrio-and-like organisms (BALO) reveals extensive diversity. Int J Syst Evol Microbiol 52:2089–2094
Sockett ER, Lambert C (2004) Bdellovibrio as therapeutic agents: a predatory renaissance? Nat Rev Microbiol 2:669–675
Sourjik V, Wingreen NS (2012) Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268
Starr MP (1975) Bdellovibrio as a symbiont: the association of bdellovibrios with other bacteria interpreted in terms of a generalized scheme for classifying organismic associations. Symp Soc Exp Biol 29:93–124
Stein MA, McAllister SA, Torian BE, Diedrich DL (1992) Acquisition of apparently intact and unmodified lipopolysaccharides from Escherichia coli by Bdellovibrio bacteriovorus. J Bacteriol 174:2858–2864
Stolp H (1967) Lysis von bacterien durch den parasiten Bdellovibrio bacteriovorus, Film C972. I.W.F., Gottingen
Stolp H (1968) Ein rauberischer backterienparasit. Die Naturwissenschaften 55:57–63
Stolp H (1981) The genus Bdellovibrio. In: Starr MP, Stolp H, Troper HG, Balows A, Schegel HG (eds) The prokaryotes. Springer, Berlin, pp 618–629
Stolp H, Petzold H (1962) Untersuchungen uber einen obligat parasitischen Mikroorganismus mit lytischer Aktivitiit fur Pseudomonas Bakterien. Phytopathol Z 45:364–370
Stolp H, Starr MP (1963) Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29:217–248
Straley SC, Conti SF (1977) Chemotaxis by Bdellovibrio bacteriovorus toward prey. J Bacteriol 132:628–640
Straley SC, LaMarre AG, Lawrence LJ, Conti SF (1979) Chemotaxis of Bdellovibrio bacteriovorus toward pure compounds. J Bacteriol 140:634–642
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Thomashow MF, Cotter TW (1992) Bdellovibrio host dependence: the search for signal molecules and genes that regulate the intraperiplasmic growth cycle. J Bacteriol 174:5767–5771
Thomashow MF, Rittenberg SC (1978a) Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: solubilization of Escherichia coli peptidoglycan. J Bacteriol 135:998–1007
Thomashow MF, Rittenberg SC (1978b) Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: N-deacetylation of Escherichia coli peptidoglycan amino sugars. J Bacteriol 135:1008–1014
Thomashow MF, Rittenberg SC (1978c) Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: attachment of long-chain fatty acids to Escherichia coli peptidoglycan. J Bacteriol 135:1015–1023
Thomashow LS, Rittenberg SC (1985) Isolation and composition of sheathed flagella from Bdellovibrio bacteriovorus 109J. J Bacteriol 163:1047–1054
Tudor JJ, Conti SF (1977) Characterization of bdellocysts of Bdellovibrio sp. J Bacteriol 131(1):314–322
Tudor J, Conti SF (1978) Characterization of germination and activation of Bdellovibrio bdellocysts. J Bacteriol 133:130–138
Tudor JJ, McCann MP, Acrich IA (1990) A new model for the penetration of prey cells by bdellovibrios. J Bacteriol 172:2421–2426
Uematsu T (1980) Ecology of Bdellovibrio parasitic to rice bacterial leaf blight pathogen, Xanthomonas oryzae. Rev Plant Protect Res 13:12–26
Van Essche M, Quirynen M, Sliepen I, Van Eldere J, Teughels W (2009a) Bdellovibrio bacteriovorus attacks Aggregatibacter actinomycetemcomitans. J Dent Res 88:182–186
Van Essche M, Sliepen I, Loozen G, Van Eldere J, Quirynen M, Davidov Y, Jurkevitch E, Boon N, Teughels W (2009b) Development and performance of a quantitative PCR for the enumeration of Bdellovibrionaceae. Environ Microbiol Rep 1:228–233
Varon M (1979) Selection of predation-resistant bacteria in a continuous culture. Nature 277:386–388
Varon M, Seijffers J (1975) Symbiosis-independent and symbiosis-incompetent mutants of Bdellovibrio bacteriovorus 109J. J Bacteriol 124:1191–1197
Varon M, Shilo M (1968) Interaction of Bdellovibrio bacteriovorus and host bacteria: I Kinetic studies of attachment and invasion of Escherichia coli B by Bdellovibrio bacteriovorus. J Bacteriol 95:744–753
Varon M, Shilo M (1969) Attachment of Bdellovibrio bacteriovorus to cell wall mutants of Salmonella spp. and Escherichia coli. J Bacteriol 97:977–979
Wang Z, Kadouri D, Wu M (2011) Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics 12:453
Westergaard JM, Kramer TT (1977) Bdellovibrio and the intestinal flora of vertebrates. Appl Environ Microbiol 34:506–511
Wilkinson MHF (2007) Mathematical modelling of predatory prokaryotes. In: Jurkevitch E (ed) Predatory prokaryotes, vol 4. Springer, Berlin/Heidelberg, pp 93–130
Williams HN (1988) A study of the distribution of bdellovibrios in estuarine sediment over an annual cycle. Microb Ecol 15:9–20
Williams H, Falkler W, Shay D (1982) Seasonal distribution of bdellovibrios at the mouth of the Patuxent river in the Chesapeake bay. Can J Microbiol 28:111–116
Williams HN, Schoeffied AJ, Guether D, Kelley J, Shah D, Falkler WA (1995) Recovery of bdellovibrios from submerged surfaces and other aquatic habitats. Microb Ecol 29:39–48
Wurtzel O, Dori-Bachash M, Pietrokovski S, Jurkevitch E, Sorek R (2010) Mutation detection with next-generation resequencing through a mediator genome. PLoS One 5:e15628
Zheng G, Wang C, Williams HN, Pineiro SA (2008) Development and evaluation of a quantitative real-time PCR assay for the detection of saltwater Bacteriovorax. Environ Microbiol 10:2515–2526
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this entry
Cite this entry
Rotem, O., Pasternak, Z., Jurkevitch, E. (2014). Bdellovibrio and Like Organisms. In: Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-39044-9_379
Download citation
DOI: https://doi.org/10.1007/978-3-642-39044-9_379
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-39043-2
Online ISBN: 978-3-642-39044-9
eBook Packages: Biomedical and Life SciencesReference Module Biomedical and Life Sciences