Abstract
Phylogenetic studies of immunity have revealed that about 500 million years ago, two types of recombinatorial adaptive immune systems (AIS) arose in vertebrates. Jawed vertebrates diversify the repertoire of immunoglobulin-domain-based B and T cell antigen receptors mainly through the rearrangement of gene segments and somatic hypermutation, whereas an alternative AIS of jawless vertebrates is based on variable lymphocyte receptors (VLRs) which diversify through recombinatorial usage of leucine-rich repeat (LRR) units. None of the fundamental recognition elements of jawed vertebrates AIS have been found in jawless vertebrates. Despite differences in molecular architecture, the parallel ‘two-arms’ of the AIS evolved within the context of preexisting innate immunity and maintained over a long period of time in jawed and jawless vertebrates, respectively, as a consequence of powerful and enduring evolutionary selection pressure by pathogens and other factors.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
In order to survive in the competitive ‘struggle for existence’ in an environment, organisms must be able to protect themselves from pathogens. This requirement of self-defense in the ongoing struggle for survival led to the evolution of complex immune systems. Biologists have found that simplest multicellular life forms like sponges (phylum Porifera) have many of the elements used by vertebrates for immune response and pathogen defense. These ancient defense strategies defend against infection by potential pathogens in a relatively non-specific manner, collectively known as innate immunity, which can be found in representative species at almost every level of the evolutionary tree of life. In addition to the innate immunity, another layer of complexity in immune defenses emerged during animal evolution with the appearance of adaptive immunity in vertebrates around 500 million years ago (Boehm et al. 2012; Cooper and Alder 2006). The adaptive immune system (AIS) allows specific recognition and mounting of a protective response against numerous pathogens. Adaptive immunity is mediated through various genetic and cellular processes that create appropriate somatic variants of antigen-binding receptors under evolutionary pressure by pathogens and other factors (Flajnik and Kasahara 2010). The core elements of AIS are now mechanistically understood, such as compartmental differentiation of lymphocytes, the generation of immune recognition diversity and the supporting cellular complexity that selects and expands cell populations expressing suitable antigen-binding receptor variants and immunological memory. Immunologists and evolutionary biologists long believed that these general features of the adaptive immunity are exclusive to the jawed vertebrates. However, the recent discovery of a lymphoid cell-based system of adaptive immunity in jawless vertebrates (lamprey and hagfish) was surprisingly similar to the AIS in jawed vertebrates (Pancer et al. 2004, 2005; Rogozin et al. 2007; Guo et al. 2009). We are within reach of important breakthroughs in our understanding of how AIS evolved in the context of well-developed innate immunity and how these molecularly disparate systems are related to the evolutionary acquisition of immunological complexity.
2 Immune Response Molecules in Invertebrates
Common elements deployed for innate immune defense in invertebrates may provide insight into how and when our complex AIS evolved. Two protein families, which contain either the leucine-rich-repeat (LRR) motifs or the immunoglobulin superfamily (IgSF) domains, are widely involved for immune defense. Leucine-rich repeat containing proteins are consisted of multiples of 20–30 amino acid units to form horseshoe-like solenoid structures in which the concave surface is formed by parallel β sheets and the convex surface by an array of helices (Buchanan and Gay 1996). The Toll-like receptors (TLRs) are well-defined examples of LRR-containing proteins which function as pattern-recognition receptors (PRRs) that constitute key components of innate immune systems throughout the animal kingdom (Hoffmann et al. 1999).
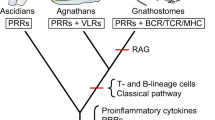
Members of the IgSF also serve important immune defense functions in invertebrates. IgSF members with important roles in innate immunity include the fibrinogen-related proteins (FREPs) in snails (Zhang et al. 2004), down syndrome cell adhesion molecule (Dscam) in insects (Watson et al. 2005), variable region-containing chitin-binding proteins (VCBPs) in amphioxus and sea squirt (Cannon et al. 2002). These molecules undergo repertoire diversification by alternative splicing or even somatic mutation to generate potential antigen recognition capacity. Both LRRs motifs and IgSF domains were readily available for co-option to provide the molecular basis for use in the somatic diversification of variable lymphocyte receptor (VLR) in jawless vertebrates or immunoglobulin (Ig)/T cell receptor (TCR) in jawed vertebrates. The hypothetical evolutionary scheme of innate and AIS is shown in Fig. 17.1.
Overview of the evolution of innate and adaptive immune systems. The stages at which different families of immune molecules emerged are shown in the species phylogeny. The figure is modified from Hirano et al. (2011)
3 Brief Overview of the AIS of Jawed Vertebrates
The most genetically distant organisms in which the Ig-domain-based AIS, as characterized in humans, is found are the cartilaginous fish. The well-defined mechanisms of the Ig-based AIS are mentioned briefly here for comparative purposes. The two major lineages of clonally diverse lymphocytes that specifically recognize and respond to antigens are named T and B lymphocytes, because they are generated in the thymus and the bone marrow (or the avian bursa of Fabricius), respectively (Cooper et al. 1965; Greaves et al. 1968). During their early developmental stages, the T and B lymphocyte progenitors rearrange different sets of variable (V), diversity (D), and/or joining (J) gene segments to generate the antigen-binding regions of the TCRs and B cell receptors (BCRs) (Tonegawa 1983). The recombination activating genes (RAG1/RAG2) encode enzymes that mediate V(D)J rearrangement (Schatz et al. 1989). The antigen-binding regions of the different V(D)J combinations are diversified further through splicing variability and the enzymatic addition of nucleotides in the joints created during V(D)J segment assembly (Dudley et al. 2005). This random nature of diversification inevitably results in the generation of Ig/TCR repertoire expansion. The membrane bound and secreted antibodies made by B lineage cells typically recognize exposed determinants (epitopes) of intact molecules, including surface protein and carbohydrate moieties of invasive microbes (Klein and Horejsi 1997). In contrast, the TCRs recognize peptide fragments of antigens presented by accessory cells within cell surface molecules encoded by the major histocompatibility complex (MHC) class I and class II genes (Klein and Horejsi 1997). Therefore, TCRs typically recognize antigens that have been partially digested within the antigen presenting cells, primarily dendritic cells and phagocytic cells.
The acquisition of a mechanism for gene rearrangement to produce clonally diverse Igs and TCRs was critical for adaptive immunity in jawed vertebrates. Discovery of multiple V, D, and J gene segments with specific recombination signal sequences (RSSs) provided insight into the recombinatorial system employed in Ig and TCR loci to generate clonal diversity (Hedrick et al. 1984; Tonegawa 1983; Yanagi et al. 1984). The RAG1/RAG2 proteins were found to recognize the RSSs flanking the V(D)J gene segments to initiate the double-stranded DNA breaks and the recruitment of other proteins required for recombination (Jung and Alt 2004; Oettinger et al. 1990). The RAG1 and RAG2 proteins form a transposase that can excise DNA containing the RSSs and reinsert it elsewhere, thus supporting the theory that RAG1/RAG2 genes were once components of a transposable element (Agrawal et al. 1998; Hiom and Gellert 1997).
4 VLR-Based Alternative Adaptive Immune System in Jawless Vertebrates
An alternative AIS that uses Variable lymphocyte receptor as antigen receptors has been discovered in extant jawless vertebrates (lampreys and hagfish) only recently (Pancer et al. 2004, 2005; Rogozin et al. 2007). Jawless vertebrates mount specific responses to pathogens, elicit allograft recognition, and display other general immune-type responses that are characteristic of cellular immunity, but they do not use a V(D)J recombination-mediated form of adaptive immunity as found jawed vertebrates (Hirano et al. 2011; McCurley et al. 2012). The VLR-based alternative AIS of jawless vertebrates displays an anticipatory receptor repertoire complexity comparable to that of the Ig-domain-based AIS of jawed vertebrates.
The thymus-derived T lymphocytes and bone marrow-derived B lymphocytes are the cellular pillars of adaptive immunity in the jawed vertebrates. Cells with similar morphological features and molecular machinery of lymphocytes in jawed vertebrates are also found in lampreys and hagfish (Mayer et al. 2002; Nagata et al. 2002; Najakshin et al. 1999; Uinuk-Ool et al. 2002). These findings along with earlier observations that lampreys and hagfish produce specific agglutinins following immunization with bacteria and foreign red blood cells suggested that jawless vertebrates could have an AIS. However, from transcriptome analysis no MHC, TCR, BCR, and RAG orthologues genes were found in jawless vertebrates, and this failure added to skepticism about the presence of adaptive immunity in agnathans. This view was dramatically changed when the VLR genes were identified as the key elements for AIS in lampreys and hagfish (Pancer et al. 2004, 2005).
4.1 VLR Discovery and Diversity Generation in Jawless Vertebrates
Since the transcriptome analysis of lymphocyte-like cells of naïve lamprey did not reveal evidence for equivalent AIS of jawed vertebrates, lamprey larvae were stimulated by an antigen and mitogen mixture to survey the transcriptome of activated lamprey lymphocytes. The objective was to catch the lamprey lymphocytes in the act of an immune response. Large lymphoblastoid cells in blood were then sorted by their light scatter characteristics and used for the construction of a cDNA library (Pancer et al. 2004). Still no orthologs of Ig, TCR, and MHC genes were detected, but this experiment revealed a large number of transcripts for uniquely diverse Leucine-rich repeat proteins, which were named VLRs because of their lymphocyte-restricted expression and sequence diversity. Each VLR transcript was found to encode a conserved signal peptide (SP) followed by highly variable LRR modules: a 27–38 residue N-terminal LRR (LRRNT), the first 18-residue LRR (LRR1), several 24-residue variable LRRs (LRRV), one 13-residue connecting peptide LRR (LRRCP), and a 48–65 residue C-terminal LRR (LRRCT).
After the discovery of the first lamprey VLR gene (now known as VLRB), two hagfish VLR genes, VLRA and VLRB, were identified in an expressed sequence tags (EST) database of hagfish leukocyte transcripts (Pancer et al. 2005). The VLRA gene in lamprey was identified in a subsequent search of the draft sequence database of the sea lamprey genome (Rogozin et al. 2007). The sequence homology indicates that the hagfish VLRA and VLRB genes are homologous to the lamprey VLRA and VLRB genes, respectively. Another VLR gene, designated VLRC, has been identified recently through an analysis of the sea lamprey EST database (Kasamatsu et al. 2010). The predicted VLRC structure is very similar to that of lamprey VLRA and VLRB, except that VLRC lacks the thumb-like protrusion encoded in the LRRCT inserts of VLRA and VLRB that are important for antigen recognition. Phylogenetically, the VLRC gene is close to VLRA genes of lamprey and hagfish (Kasamatsu et al. 2010). The discovery of VLRC raises interesting questions about the function of VLRC-expressing lymphocytes, their antigen-binding potential, and their role in pathogen responses. Whether or not hagfish possess a VLRC homolog is presently unknown.
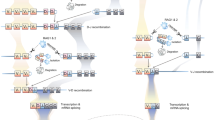
All of the germline VLR genes are incomplete, in that they have coding sequences only for the leader sequence, incomplete amino- and carboxy-terminal LRR subunits (LRRNT and LRRCT), and for the stalk region (Fig. 17.2) (Hirano et al. 2011). There are two exons; the first exon encodes only a portion of the 5′ untranslated region while the second exon encodes the rest of the 5′ untranslated region, a sig, a 5′ portion of the LRRNT, a 3′ portion of the LRRCT, and the stalk region. For hagfish VLRA and VLRB and for lamprey VLRA, the 5′ LRRNT sequence is separated from the 3′ LRRCT sequence by a short noncoding intervening sequence that does not contain canonical splice donor and acceptor sites. The lamprey VLRB gene is more complex in that it has a 5′ LRRNT coding sequence present between two intervening sequences (Pancer et al. 2004). Each germline VLR gene is flanked by hundreds of different LRR module-encoding genomic donor cassettes, which are used as randomly selected templates to add the LRR cassettes needed for production of a mature VLR gene (Fig. 17.3). From a genomic point of view, the incomplete germline VLR gene resembles the constant gene segment, whereas the clusters of different types of donor genomic cassettes represent the functional correlates of the clusters of variable, diversity, and joining gene segments of Ig or TCR loci in jawed vertebrates (Das et al. 2013).
Organization of VLR genes and assembled VLR protein. Germline VLR genes in lamprey and hagfish are shown in upper panel. The incomplete germline VLR genes contain regions encoding for portions of the LRRNT and LRRCT separated by noncoding intervening sequences and for the invariant stalk region. Assembled VLR protein. Mature VLR protein consist of a signal peptide (SP), an LRRNT, an LRR1, up to eight LRRV modules, a connecting peptide (CP), an LRRCT, and an invariant stalk region. The figure is modified from Hirano et al. (2011)
Mechanism of VLR gene assembly. The germline VLR genes are flanked by hundreds of donor LRR cassettes. The noncoding intervening sequence between portions of the LRRNT and LRRCT is replaced by the step-wise incorporation of genomic donor cassettes based on a short region of sequence homology (~6–30 nt. The figure is modified from Hirano et al. (2011)
A gene conversion-like mechanism has been postulated for the complex VLR gene assembly process (Cooper and Alder 2006; Nagawa et al. 2007) in which the intervening sequence is replaced in a stepwise manner by random selection of flanking LRR cassettes to serve as templates for adding the necessary sequences to complete a VLR gene (Fig. 17.3). However, recent study indicates preferential usage of certain donor cassettes during development of the VLR repertoire (Das et al. 2013). The assembly process can be initiated at either the 5′ LRRNT or the 3′ LRRCT ends (Alder et al. 2005; Nagawa et al. 2007; Das et al. 2013) using short stretches of nucleotide homology (6–30 bp) between donor sequences and acceptor sequences (Nagawa et al. 2007; Das et al. 2013). In silico analysis of the diversity of VLRB gene sequences suggests a potential repertoire of >1,014 distinct receptors, i.e., a magnitude comparable to that of the theoretical repertoire diversity of our AIS (Alder et al. 2005).
5 Ancient T-like and B-like Lymphocyte Populations in Jawless Vertebrates
The lymphocytes expressing the VLRA or VLRB in lamprey are remarkably similar to T and B lymphocytes in jawed vertebrates (Guo et al. 2009). The lamprey VLRB-expressing lymphocytes resemble B cells of jawed vertebrates in that they respond to immunization with pathogens or foreign erythrocytes with proliferation, lymphoblastoid transformation and differentiation into plasmacytes that secrete VLRB antibodies specific for protein or carbohydrate epitopes (Alder et al. 2008; Herrin et al. 2008). On the other hand, the VLRA-expressing lymphocytes also respond to immunization, but they do not secrete the VLRA proteins either before or after immunization (Hirano et al. 2011). VLRB cells can bind to native antigens directly; however, it is still not clear whether VLRA cells could recognize native or processed antigens.
A limited transcriptome analysis indicates that VLRA and VLRB cells have different gene expression profiles. VLRB+ lymphocytes preferentially express orthologues of genes that are preferentially expressed by B cells in jawed vertebrates (Guo et al. 2009): the hematopoietic progenitor homing receptor CXCR4, two components of the BCR-mediated signaling cascades, Syk and the B cell adaptor protein (BCAP), the chemotactic inflammatory cytokine IL-8, the IL-17 receptor, and the TLR orthologues TLR2abc, TLR7, and TLR10, the ligation of which is important for B cell activation. By contrast, VLRA+ lymphocytes preferentially express genes orthologues to those typically expressed by the T cells in jawed vertebrates: GATA1/2, c-Rel, aryl hydrocarbon receptor (AHR), and BCL11b transcriptional factors used for T cell differentiation, the CCR9 chemokine receptor involved in homing of lymphocyte progenitors to the thymus, the Notch1 T cell fate-determining molecule, the CD45 tyrosine phosphatase receptor protein that is essential for T cell development, and proinflammatory cytokine genes interleukin-17 (IL-17) (Guo et al. 2009). All these evidence indicate that characteristics of VLRA+ and VLRB+ lymphocytes resemble those of mammalian T and B cells, respectively. Therefore, it can be postulated that the T-like and B-like cells were evolved before the divergence of jawless and jawed vertebrates (Fig. 17.4).
6 Conclusion
The discovery of VLR-type antigen receptors in lamprey and hagfish has provided unparalleled insight into the origins of adaptive immunity at the early stages of vertebrate evolution. Both VLRs in jawless vertebrates and BCRs/TCRs in jawed vertebrates rely on combinatorial diversity to generate a vast repertoire of immune receptors; however, the genes of Variable lymphocyte receptors show no structural similarity to those of BCRs or TCRs. VLR genes probably exist only in jawless vertebrates, whereas Ig, TCR, and MHC genes are possibly restricted in jawed vertebrates. Recent studies on the immune system of jawless vertebrates indicate that the preliminary separation of T and B lineage cells occurred earlier than the emergence of an anticipatory receptor system in the common ancestor of jawed and jawless vertebrates. Considering the monophyletic relationship of lamprey and hagfish (Takezaki et al. 2003; Heimberg et al. 2010), it is possible that the VLR-based antigen receptors in jawless vertebrates and Ig-domain-based antigen receptors in jawed vertebrates developed independently. In an alternative scenario, it is also possible that if the common ancestor possessed both VLR- and Ig-domain-based anticipatory receptors; after separation, the jawed and jawless vertebrates lost their VLR-based and Ig-domain-based anticipatory receptors, respectively. Taking into account the striking similarities in cellular and molecular basis of immune function, studies of the alternative AIS in jawless vertebrates may yield insight into several aspects of adaptive immunity in jawed vertebrates that remain elusive. Furthermore, the parallelism of lymphocyte lineages in jawed and jawless vertebrates may also guide in future studies on the origins of the different hematopoietic lineages in non-vertebrate species.
References
Agrawal A, Eastman QM, Schatz DG (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744–751
Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z (2005) Diversity and function of adaptive immune receptors in a jawless vertebrate. Science 310:1970–1973
Alder MN, Herrin BR, Sadlonova A, Stockard CR, Grizzle WE, Gartland LA, Gartland GL, Boydston JA, Turnbough CL Jr, Cooper MD (2008) Antibody responses of variable lymphocyte receptors in the lamprey. Nat Immunol 9:319–327
Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD (2012) VLR-based adaptive immunity. Annu Rev Immunol 30:203–220
Buchanan SG, Gay NJ (1996) Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol 65:1–44
Cannon JP, Haire RN, Litman GW (2002) Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol 3:1200–1207
Cooper MD, Alder MN (2006) The evolution of adaptive immune systems. Cell 124:815–822
Cooper MD, Peterson RD, Good RA (1965) Delineation of the thymic and bursal lymphoid systems in the chicken. Nature 205:143–146
Das S, Hirano M, Aghaallaei N, Bajoghli B, Boehm T, Cooper MD (2013) Organization of lamprey variable lymphocyte receptor C locus and repertoire development. Proc Natl Acad Sci USA 110:6043–6048
Dudley DD, Chaudhuri J, Bassing CH, Alt FW (2005) Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol 86:43–112
Flajnik MF, Kasahara M (2010) Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11:47–59
Greaves MF, Roitt IM, Rose ME (1968) Effect of bursectomy and thymectomy on the responses of chicken peripheral blood lymphocytes to phytohaemagglutinin. Nature 220:293–295
Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD (2009) Dual nature of the adaptive immune system in lampreys. Nature 459:796–801
Hedrick SM, Cohen DI, Nielsen EA, Davis MM (1984) Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature 308:149–153
Heimberg AM, Cowper-Sal-lari R, Semon M, Donoghue PC, Peterson KJ (2010) microRNAs reveal the interrelationships of hagfish, lampreys, and gnathostomes and the nature of the ancestral vertebrate. Proc Natl Acad Sci USA 107:19379–19383
Herrin BR, Alder MN, Roux KH, Sina C, Ehrhardt GR, Boydston JA, Turnbough CL Jr, Cooper MD (2008) Structure and specificity of lamprey monoclonal antibodies. Proc Natl Acad Sci USA 105:2040–2045
Hiom K, Gellert M (1997) A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell 88:65–72
Hirano M, Das S, Guo P, Cooper MD (2011) The evolution of adaptive immunity in vertebrates. Adv Immunol 109:125–157
Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA (1999) Phylogenetic perspectives in innate immunity. Science 284:1313–1318
Jung D, Alt FW (2004) Unraveling V(D)J recombination; insights into gene regulation. Cell 116:299–311
Kasamatsu J, Sutoh Y, Fugo K, Otsuka N, Iwabuchi K, Kasahara M (2010) Identification of a third variable lymphocyte receptor in the lamprey. Proc Natl Acad Sci USA 107:14304–14308
Klein J, Horejsi V (1997) Immunology. Blackwell Science, Oxford
Mayer WE, Uinuk-Ool T, Tichy H, Gartland LA, Klein J, Cooper MD (2002) Isolation and characterization of lymphocyte-like cells from a lamprey. Proc Natl Acad Sci USA 99:14350–14355
McCurley N, Hirano M, Das S, Cooper MD (2012) Immune related genes underpin the evolution of adaptive immunity in jawless vertebrates. Curr Genomics 13:86–94
Nagata T, Suzuki T, Ohta Y, Flajnik MF, Kasahara M (2002) The leukocyte common antigen (CD45) of the Pacific hagfish, Eptatretus stoutii: implications for the primordial function of CD45. Immunogenetics 54:286–291
Nagawa F, Kishishita N, Shimizu K, Hirose S, Miyoshi M, Nezu J, Nishimura T, Nishizumi H, Takahashi Y, Hashimoto S et al (2007) Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol 8:206–213
Najakshin AM, Mechetina LV, Alabyev BY, Taranin AV (1999) Identification of an IL-8 homolog in lamprey (Lampetra fluviatilis): early evolutionary divergence of chemokines. Eur J Immunol 29:375–382
Oettinger MA, Schatz DG, Gorka C, Baltimore D (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248:1517–1523
Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD (2004) Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430:174–180
Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, Kasahara M, Cooper MD (2005) Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA 102:9224–9229
Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z (2007) Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol 8:647–656
Schatz DG, Oettinger MA, Baltimore D (1989) The V(D)J recombination activating gene, RAG-1. Cell 59:1035–1048
Takezaki N, Figueroa F, Zaleska-Rutczynska Z, Klein J (2003) Molecular phylogeny of early vertebrates: monophyly of the agnathans as revealed by sequences of 35 genes. Mol Biol Evol 20:287–292
Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302:575–581
Uinuk-Ool T, Mayer WE, Sato A, Dongak R, Cooper MD, Klein J (2002) Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc Natl Acad Sci USA 99:14356–14361
Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D (2005) Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309:1874–1878
Yanagi Y, Yoshikai Y, Leggett K, Clark SP, Aleksander I, Mak TW (1984) A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature 308:145–149
Zhang SM, Adema CM, Kepler TB, Loker ES (2004) Diversification of Ig superfamily genes in an invertebrate. Science 305:251–254
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Das, S., Hirano, M. (2013). Variable Lymphocyte Receptors in Jawless Vertebrates: Illuminating the Origin and Early Evolution of Adaptive Immunity. In: Pontarotti, P. (eds) Evolutionary Biology: Exobiology and Evolutionary Mechanisms. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-38212-3_17
Download citation
DOI: https://doi.org/10.1007/978-3-642-38212-3_17
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-38211-6
Online ISBN: 978-3-642-38212-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)