Abstract
The broomrape family (Orobanchaceae) is an excellent model system for comparative evolutionary studies that focus on various genomic aspects associated with or being the result of the transition to heterotrophy. This chapter provides a family-wide summary of our current knowledge of the extraordinarily dynamic genomic evolution in Orobanchaceae. Several candidate genes that have been newly recruited in parasite-specific pathways have been identified by transcriptome sequencing. While little information is available on the evolution of mitochondrial genomes, studies of plastid genes and genomes of members of Orobanchaceae bring to light the first insights into the complex and differential patterns of reductive evolution of plastid chromosomes following the loss of photosynthesis. The chapter also discusses the need for large-scale transcriptome and genome sequencing to determine basic parasite-specific genetics and genome dynamics that may have potential for the development of novel strategies to control weedy Orobanchaceae.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The broomrape family, Orobanchaceae, is widely recognized as the model group to study genomic evolution in parasitic plants, especially because it is the only parasitic family to include the entire range of evolutionary transitional stages, from a fully autotrophic via semi-heterotrophic to completely holo-heterotrophic lifestyle (Westwood et al. 2010). Orobanchaceae, encompassing an estimated number of 2,000 species,Footnote 1 are confidently placed in the large and diverse group of Lamiales, which contains a great number of species with highly specialized life forms including desiccation tolerance, carnivory, and parasitism (Schäferhoff et al. 2010). The parasitic lifestyle has brought about numerous morphological and developmental changes. Substantial progress has been made during the past few years in uncovering basic genetic reconfigurations and signalling pathways necessary in establishing a haustorial connection to another plant. Nevertheless, little is known about the evolution of nuclear and mitochondrial genes and genomes in Orobanchaceae, even 20 years after the first plastid genome of a non-photosynthetic member of the family has been sequenced. This is especially astonishing given the great advance in molecular biological methods and sequencing technologies over the past 5–10 years. Reasons therefore are manifold—as usual. Owing to the great diversity of the family, evolutionary studies based upon molecular data are restricted to only a few members of either species-rich and commonly distributed genera or to members of considerable ecological importance such as Orobanche, Phelipanche, and Striga. Additionally, in vitro cultivation of obligate parasites is often difficult or requires permits in some countries, hampering genetic and reverse genetic approaches. Finally, the trend for rather large genomes renders Orobanchaceae challenging objects for genomic surveys.
This chapter summarizes the current knowledge of the genomic evolution in Orobanchaceae. The following sections provide overviews about nuclear, mitochondrial, and plastid genomics and about horizontal DNA transfer. Subsequently, a short concluding paragraph outlines some prospects on where genomics in the broomrape family may be headed in the next few years (see also Sect. 4.5 for the evolution of parasite-specific functions).
2 The Nuclear Genome
2.1 Nuclear Genes
Nothing is known about the evolution of nuclear coding regions in the Orobanchaceae, and thus very little is known about the molecular basis of parasite-specific life stages. Recently, a hydroxylase (PRCYP707A1) functioning in the abscisic acid (ABA) catabolic pathway in Phelipanche ramosa has been identified as playing a major role during germination of seeds after exposure to germination stimulants (Lechat et al. 2012). The same study also identified two heat shock proteins and a few more transcripts associated with ABA cascades. Even though those transcripts have not been characterized in detail, preliminary lines of evidence link those transcripts to proteins active during seed germination in Arabidopsis thaliana. Experiments with the holoparasitic Orobanche minor have shown that the involvement of phytochromes (PHY) during germination, shoot elongation, and anthocyanin content differs from that observed in photosynthetic plants, suggesting reconfigured regulatory cascades involving at least PHY proteins A and B (Takagi et al. 2009). Relative to Arabidopsis, 26 amino acids are substituted in PHYA of O. minor (Trakulnaleamsai et al. 2005). Of these, some substitutions perhaps hold the potential to alter protein function, thereby contributing to unusual light responses in the holoparasite compared to autotrophic plants (Trakulnaleamsai et al. 2005; Takagi et al. 2009). Given that expression patterns as well as cellular localizations of PHYA in O. minor are comparable to autotrophic plants and that the chromatophore-binding site in PHYA is highly conserved, the reported amino acid changes may also represent results of coevolution with rapidly evolving PHYA-interacting photosynthesis genes.
In Striga hermonthica, Yoshida et al. (2010) found 589 assembled fragments of expressed genes (unigenes) that are not similar to known plant genes, implying that at least some of these may be specific to parasitism. Sequencing of cDNAs from the facultative hemiparasite Triphysaria versicolor revealed an up-regulation of more than a hundred unigenes during early haustorium initiation. These fragments were assigned similarity to proteins functioning in quinone detoxification, transcription and regulatory processes, membrane transport, and the citric acid cycle (Matvienko et al. 2001). Two of these transcripts, a quinone oxidoreductase (QR1) and a protein associated to plant signalling pathways (TvPirin), have been further characterized as essential for haustorium initiation after contact to host roots or exposure to haustorium-inducing chemicals (Bandaranayake et al. 2010; see also Sects. 4.4 and 4.5). However, not much is known about the molecular evolution of those genes, and future studies will have to show whether genes relevant for the development of lateral haustoria of Triphysaria are also essential for the induction of terminal haustoria (see Sect. 4.4). Deep sequencing of ultrathin slices of host-parasite interface tissue of T. versicolor furthermore revealed the differential expression of a β-expansin gene (TvEXPB1) when the parasite was grown in the presence of different hosts (Honaas et al. 2013). However, it still needs to be elucidated whether cell wall modifying proteins and their differential expression are common among other Orobanchaceae.
Using three Orobanchaceae species differing in their extent of heterotrophy, the ongoing large-scale transcriptome-sequencing approach of the Parasitic Plant Genome Project (PPGP, Westwood et al. 2010, 2012) aims, among other aspects, at discovering and studying genes that are exclusive to specific ontogenetic stages. In a first study, Wickett et al. (2011) found that expression of nuclear-encoded photosynthesis subunits in aboveground tissue is considerably reduced in the obligate hemiparasite S. hermonthica compared to the facultative hemiparasite T. versicolor. No expression of nuclear-encoded photosynthesis genes was detected in P. aegyptiaca, where these genes might have become pseudogenes or have already been deleted from the genome. In contrast, genes for chlorophyll synthesis were still expressed in Phelipanche (Wickett et al. 2011), corroborating results of the detection of trace amounts of chlorophyll a in some holoparasites of Orobanchaceae (Epifagus, Myzorrhiza cooperi [syn. Orobanche cooperi], Aphyllon uniflora [syn. O. uniflora]) and other families (Cummings and Welschmeyer 1998).
A survey of the evolution of the small ribosomal RNA subunit (SSU) found that parasitic plants possess significantly elevated nucleotide substitution rates (Nickrent and Duff 1996). However, comparative studies across parasitic and myco-heterotrophic plants (see Sect. 1.8) did not show a significant acceleration in the SSU evolution in several Orobanchaceae holoparasites, and the pattern of rate acceleration across lineages remains widely elusive (Lemaire et al. 2011).
2.2 Chromosome Numbers
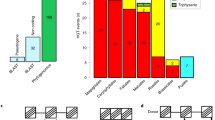
Chromosome numbers have been the focus of several studies on hemiparasitic Orobanchaceae, although the vast majority of these reports lacked an explicit evolutionary context. Chromosome numbers and ploidy are highly variable in the family and apparently do not correlate with genome size. The current knowledge of chromosome numbers and genome sizes in Orobanchaceae is graphically summarized in Fig. 15.1. Lindenbergia and Schwalbea, members of the first branching lineages, have n = 16 (Hjertson 1995) and n = 18 (Kondo et al. 1981), respectively. Being mostly diploid with the exception of one tetraploid species, the closely related sister group of Orobanchaceae, Rehmannia, harbours n = 14 chromosomes (Albach et al. 2007). In the light of current data, there seems to be a slight trend towards higher chromosome numbers in the exclusively holoparasitic Orobanche clade (see Sect. 14.2.2.3), although polyploidy appears to be common in some of the hemiparasites as well (e.g. Castilleja, Striga, Euphrasia; Tank et al. 2009; Kondo et al. 1981; Barker et al. 1988; Iwo et al. 1993). Except for Phelipanche with n = 12, most genera of the holoparasite clade have n = 19 or more chromosomes (Fig. 15.1; Schneeweiss et al. 2004). Plots of synonymous substitutions of selected expressed genes revealed unambiguously that Phelipanche must have undergone at least one whole-genome duplication after the split from hemiparasitic ancestors (Wickett et al. 2011), corroborating previous hypotheses of ancient duplication events in the Orobanche clade (Schneeweiss et al. 2004). Differences in chromosome number indicate that lineages within this clade might have undergone at least one or more rounds of polyploidization. Inconsistent chromosome morphologies imply that these events might have even occurred independently (Schneeweiss et al. 2004). It will be interesting to see whether independent changes and ancient polyploidization also occurred in other holoparasitic groups, especially in the light of the severe ecological changes accompanying the transition to the non-photosynthetic lifestyle. A change of ploidy level in O. transcaucasica apparently coincides with a shifted host range, suggesting that, at least in this particular case, genome duplication favours ecological differentiation from its progenitors (Schneeweiss et al. 2004).
It is unclear whether several independent rounds of polyploidization and dysploidization (i.e. reduction of ploidy/chromosome numbers) in the hemiparasites of the Old and New World led to the great diversity of chromosome number, which ranges from n = 8 to n = 20 (Fig. 15.1). Changes of ploidy level are a substantial basis for speciation among angiosperms, allowing offsprings to settle into new niches (Wood et al. 2009). Niching due to host range shifts as a result of polyploidization or dysploidization may thus be a more significant aspect for speciation in parasitic lineages. At this point, such putative correlations between the degrees of parasitism and chromosomal evolution including ploidy are, however, hypothetical at best. Caryological studies in most of the tropical and/or Asian hemi- and holoparasitic lineages (i.e. nearly 50 % of all Orobanchaceae genera) are still lacking.
Evolution of chromosome number and genome size in Orobanchaceae. Arrow, the origin of parasitism. Thin branches, autotrophic and photosynthetic heterotrophs; thick branches, non-photosynthetic heterotrophs; dashed branches, uncertain placement. Tree topology after Schneeweiss (see Chap. 14). Question mark indicates uncertain chromosome counts. Chromosome data: IPCN (Goldblatt and Johnson 1979), Fedorov (1969), and Moore (1982). Genome size data: 1Yoshida et al. (2010); Estep et al. (2012); 2Castro et al. 2012; 3Hanson et al. (2002); 4Zonneveld et al. (2005); Nagl and Fusenig (1979); 5Weiss-Schneeweiss et al. (2006); 6Piednoël et al. (2012).
2.3 Genome Size
In line with the rampant, even though uncorrelated occurrence of polyploidy, genome sizes vary greatly within Orobanchaceae. Lindenbergia (1C = 0.45 Gbp), Schwalbea (1C = 0.56 Gbp; Piednoël et al. 2012), and Odontites (1C = 0.55–0.56 Gbp; Hanson et al. 2002) possess small genomes, the sizes of which are comparable to those of poplar and rice. Other photosynthetic Orobanchaceae such as Rhinanthus and Melampyrum have considerably larger genomes, with that of Melampyrum reaching almost three times the size of the human genome (Fig. 15.1; Hanson et al. 2002). The smallest genome of the holoparasitic Orobanche clade is found in O. cumana (Weiss-Schneeweiss et al. 2006). The Orobanche genus contains some polyploids (e.g. O. transcaucasica, O. gracilis) that may exceed 1C = 5.5 Gbp. Among diploids, O. crenata with 1C = 2.8 Gbp ranks among the biggest according to currently available measurements (Fig. 15.1; Weiss-Schneeweiss et al. 2006). In contrast to Orobanche, Phelipanche has larger genomes on average (Weiss-Schneeweiss et al. 2005), but the largest genomes in Orobanchaceae have been described so far for species of Cistanche with 1C = 8.7Gbp in C. phelypaea (Weiss-Schneeweiss et al. 2006). Even larger ones may occur in other Cistanche species (N. Ataei, D. Quandt, and H. Weiss-Schneeweiss, unpublished data). Nevertheless, compared to the (cryptically) photosynthetic heterotrophs of other parasitic angiosperm families such as Cuscuta (1C = 0.57–32.1 Gbp, McNeal et al. 2007a) or members of Santalales (1C = 0.3–80.2 Gbp, Martin 1983; Hanson et al. 2001; Zonneveld 2010), the range of genome sizes is rather moderate in Orobanchaceae, a fact that contributes to its status as the ‘model family’ among parasitic plants.
As in most plant genomes, the abundance of repetitive DNA contributes substantially to genome size differences in Orobanchaceae. The five economically most important species of Striga show considerable genomic variation with respect to the 14 largest genus-specific repeat families residing in the genomes with more than a few hundred copies (Estep et al. 2012). Those repetitive DNAs account for 10–19 % of the nuclear genomes of Striga species, but they are not strictly correlated with genome size. They belong to classes commonly found in angiosperm genomes with transposable elements being the most abundant. The analyses of repeat classes point towards a ploidy series in the genus Striga (Estep et al. 2012). Interestingly, the variability among different populations of single species of Striga, e.g. S. asiatica or S. gesnerioides, is moderate or even low, respectively (Botanga et al. 2002; Botanga and Timko 2005, 2006; see Sect. 19.2).
The genomes of seven holoparasitic broomrapes and two photosynthetic Orobanchaceae were characterized employing a whole-genome shotgun pyrosequencing approach (Piednoël et al. 2012). The proportion of repeat DNA sequences is low in the small-sized genomes of the nonparasite Lindenbergia and the hemiparasite Schwalbea with repetitive elements accounting for no more than 30 % of the genomes. As implied by chromosomal and genome size data, divergent dynamics of genome evolution exist in the sister groups Orobanche and Phelipanche. This hypothesis is corroborated by differing quantities of genus-specific clusters of transposable elements (Piednoël et al. 2012). The proportion of long and short interspersed nuclear elements (LINE and SINE, respectively) seems to be generally lower in Phelipanche than in Orobanche. LINEs contribute to the increase in genome size (as do many retrotransposons) in that they autonomously copy themselves. Phelipanche spp. may have evolved a more sophisticated machinery for silencing transposable elements, which results in a more stable genomic and chromosomal evolution. Control and regulatory mechanisms for transposable elements are lineage-specific and contribute widely to genome stability (e.g. He et al. 2012). It will be interesting to see whether genome size evolution is related to host range and/or to the degree of parasitism. For instance, in some plants, nutrient limitation leaves behind genomic signatures (Acquisti et al. 2009a, b), but obligate parasites may not be affected by those limitations in the same way because of the host-provided nutrient supply.
Polyploid Orobanchaceae tend to a reduction of the monoploid genome size (1Cx value) after events of polyploidization, which is in congruence with several nonparasitic polyploid angiosperm lineages (Leitch and Bennett 2004). In most cases, 1Cx values from polyploids are smaller than those of diploid relatives (Weiss-Schneeweiss et al. 2005). Although the genetic mechanisms are still poorly understood, genome-size reduction may be selected for because of, e.g. biophysical (e.g. chromosome pairing in meiosis and mitosis) and biochemical reasons (‘biochemical economy’) (Leitch and Bennett 2004; Leitch and Leitch 2012). Perhaps there is a trade-off between genomic plasticity that comes with genome size and nutritional constraints. An obligate parasitic way of life might favour moderately to large-sized genomes irrespective of the ability to carry out photosynthesis, enhancing chances of sub- or neofunctionalization of duplicated genes that contribute to host specificity and host adaptation, leading eventually to speciation within parasite lineages.
Several other Orobanchaceae groups may have had comparable scenarios of frequent increase and decrease of chromosome number and genome size like those observed in the Orobanche clade. Independent events of polyploidization have also been hypothesized for some other lineages (e.g. Euphrasia, Lathraea) based upon duplications of the phytochrome A gene (Bennett and Mathews 2006).
3 The Plastid Genome
The plastid chromosome (plastome) is the best understood cellular genome in angiosperms. The plastome normally has a highly conserved structure with a large and a small single-copy region (LSC and SSC, respectively) that are separated from each other by two large and virtually identical inverted repeats. Plastomes encode a large set of subunits for the photosynthesis apparatus including genes for photosystems I and II, the cytochrome complex, an ATP synthase, and an NAD(P)H complex as well as few genes involved in photosynthetic energy gain (rbcL, ccsA, cemA) or lipid synthesis (accD). Several proteins for the genetic apparatus are solely plastid encoded including several ribosomal protein genes, a plastid-encoded polymerase complex, as well as few others involved in either transcript maturation (matK) or protein turnover (infA, clpP, photosystem assembly factors ycf3, ycf4). The essential function of the two largest plastid genes (ycf1, ycf2) is as yet unknown, but both reading frames are conserved among photosynthetic and non-photosynthetic land plants (Wicke et al. 2011). Based on protein-domain comparisons, both proteins probably function in housekeeping processes rather than having a metabolic function (Wolfe 1994; Boudreau et al. 1997; Drescher et al. 2000). The plastid genome normally also harbours two sets of four ribosomal RNA genes as well as 30 tRNA genes, the latter of which enable the delivery of all codons due to (extended) wobbling and superwobbling (Lagerkvist 1978; Rogalski et al. 2008; Alkatib et al. 2012).
Due to its compact nature and its prime role in photosynthesis, the evolution of the plastid genome of non-photosynthetic plants has received attention early on. Already in 1990, dePamphilis and Palmer reported the loss of all genes for the plastid NAD(P)H dehydrogenase complex from the plastome of the holoparasite Epifagus virginiana. Soon the complete plastid genome sequence of E. virginiana was described (Wolfe et al. 1992b). Massive gene loss led to an extraordinary structure of the plastome which is reduced to less than half the size of that of photosynthetic relatives. Nevertheless, the relative order of genes in LSC, SSC, and the inverted repeats remains largely colinear to photosynthetic plants (Fig. 15.2). Besides ndh genes, most genes involved in light and dark reaction of photosynthesis are completely absent from the plastome; only a few photosynthesis-related genes reside in the plastome as pseudogenes (e.g. ΨrbcL, ΨatpA). Furthermore, several genes encoding proteins of the genetic apparatus are (functionally) lost including tRNA genes, the plastid-encoded polymerase complex, and some ribosomal protein genes (Morden et al. 1991; Wolfe et al. 1992b). Comparable dramatic reductions of plastid DNAs occur in a variety of parasitic plants, including Cuscuta species (Funk et al. 2007; McNeal et al. 2007b), mistletoes (Nickrent and García 2009), green algae (Knauf and Hachtel 2002; de Koning and Keeling 2006) as well as myco-heterotrophic plant lineages, including non-photosynthetic orchids (Logacheva et al. 2011; Delannoy et al. 2011) and achlorophyllous Ericaceae (Braukmann and Stefanović 2012). As in some of the other parasites, residing plastid genes of the translation apparatus in Epifagus evolved significantly faster than those of nonparasitic relatives (Wolfe et al. 1992a). Nevertheless, the retained plastid genes of E. virginiana are transcribed, mature, and are translated into functional RNAs and proteins (Morden et al. 1991; Wolfe et al. 1992a; Ems et al. 1995; Lohan and Wolfe 1998; also see Wimpee et al. 1991, 1992).
Comparison of plastid genome structure of Epifagus virginiana (inner circle) and the nonparasitic plant Nicotiana tabacum (outer circle). The large inverted repeats are indicated as thickened chromosomal segments relative to the large and the small single-copy regions. Genes are coloured according to their function with the name of genes depicted on the outer circle. Pseudogenes are coloured in grey. Thin lines from the Nicotiana to the Epifagus genome indicate structural reorganizations due to massive gene loss. genes No line between Nicotiana and Epifagus indicates that the region has been lost in the latter; a dashed connection indicates pseudogenization in Epifagus. Genome maps were drawn using OGDRAW (Lohse et al. 2007) based upon Shinozaki et al. (1986) and Wolfe et al. (1992b)
In terms of gene losses, other holoparasitic Orobanchaceae lineages possess considerably different plastomes than E. virginiana, indicating that reductive evolution of plastid DNA is a highly lineage-specific process within Orobanchaceae (and presumably within other parasitic plant lineages as well). Extensive restriction-mapping experiments suggested that Conopholis americana has an even smaller plastid genome (ca. 42kb) than the closely related E. virginiana, mainly due to the loss of one large inverted repeat (Downie and Palmer 1992; Colwell 1994). Other large deletions are comparable to those of Epifagus, implying that functional reduction is similar in both species (Colwell 1994). Conversely, restriction mapping and PCR screens suggest that the plastid genome of the holoparasite Lathraea clandestina is ca. 100–110 kb in size with gene synteny mostly colinear to Epifagus and most photosynthetic plants (Delavault et al. 1996). Reductive evolution of the plastid genome in Lathraea has obviously not proceeded as far as in E. virginiana. Still, some losses have also affected the plastid genome of Lathraea: most prominently all SSC-located ndh genes have been deleted (Delavault et al. 1996). This is in line with data from other holoparasitic and myco-heterotrophic lineages with minimally reduced plastomes (Funk et al. 2007; McNeal et al. 2007b; Wickett et al. 2008; Delannoy et al. 2011; Logacheva et al. 2011).
Further support for the hypothesis of a highly lineage-specific reductive plastome evolution comes from studies using a broad taxon sampling, but focusing only on a few plastome regions. Because of its prime function during photosynthetic carbon fixation, most data is available for the plastid gene rbcL, which encodes the large subunit of RuBisCO. Some non-photosynthetic lineages (e.g. Lathraea, Harveya, Myzorrhiza [syn. Orobanche sect. M.]) preserve an intact reading frame for rbcL (Delavault et al. 1995, 1996; Wolfe and dePamphilis 1997, 1998; Randle and Wolfe 2005). In Lathraea, however, rbcL is transcribed by a nuclear-encoded polymerase rather than by the normally used plastome-encoded polymerase, which also transcribes most of the other photosynthesis genes (Lusson et al. 1998). Accordingly, it is not surprising that plastome-encoded polymerase-specific promoter regions are lost in Epifagus (Morden et al. 1991) and also in some other parasites (Krause et al. 2003; Berg et al. 2004). Several non-photosynthetic species harbour only a pseudogene copy (e.g. Aphyllon [syn. Orobanche sect. Gymnocaulis], Hyobanche, most Orobanche s. str.; Wolfe and dePamphilis 1997; Delavault and Thalouarn 2002; Manen et al. 2004; Young and dePamphilis 2005), and several lines of evidence indicate that the rbcL-gene region is deleted from the plastomes of Phelipanche (Manen et al. 2004; Park et al. 2007a; Wicke et al. in prep.). Low levels of rbcL expression have been detected in holoparasitic Harveya and Lathraea (Lusson et al. 1998; Randle and Wolfe 2005). Myzorrhiza corymbosa maintains functional upstream and downstream untranslated regulatory elements, which is indicative of maintained transcription activity (Wolfe and dePamphilis 1997); expression data is, however, lacking. The function of the translated polypeptide transcribed from rbcL has not been investigated in holoparasites. A function of RuBisCO that is unrelated to photosynthesis has been speculated (e.g. Wolfe and dePamphilis 1997; Leebens-Mack and dePamphilis 2002), corroborated by findings that link RuBisCO to amino acid synthesis and a glycolysis-bypassing pathway (Tolbert 1997; Schwender et al. 2004). The transcript of an rbcL pseudogene has been detected in Hyobanche (Randle and Wolfe 2005), implying that regulation of the largely nuclear-encoded transcription (and transcript processing) machineries lacks behind plastid DNA evolution, at least in this particular case.
Evolutionary analyses of rps2 and matK show low rates of nucleotide substitution of these genes in rbcL-preserving lineages (dePamphilis et al. 1997; Young and dePamphilis 2005), suggesting that rps2 and matK are under purifying selection and, thus, still functional. Furthermore, Orobanche minor retains most plastid tRNAs, although some only as pseudogenes (Lohan and Wolfe 1998), and it retains several DNA fragments that are deleted from the plastome of Epifagus and Conopholis. Taken together, this and the previously mentioned studies point towards less-reduced plastid genomes in Harveya, Hyobanche, Myzorrhiza, and Orobanche. The reasons for these lineage-specific reductions are not yet understood, but the time since transition to holoparasitism seems to play a most relevant role. In general, older holoparasitic Orobanchaceae lineages, such as Epifagus, have greater reductions than younger ones, such as Lathraea and the Harveya/Hyobanche lineages (Leebens-Mack and dePamphilis 2002; also consider phylogenetic relationships depicted in Fig. 15.1).
Besides time, factors influencing the rate of gene loss and physical reduction of the plastome are still widely elusive. However, there seems to be a strong correlation between the deletion of a plastomic gene region and its physical proximity to indispensable genes of housekeeping or metabolic function (Wicke et al. submitted). The localization of a gene that has become dispensable after the loss of photosynthesis is apparently also protected by its location within an operon that encodes genes of various functional complexes (Wicke et al. submitted). Given the high gene density of plastid chromosomes, both effects are not mutually exclusive for the protection of physical gene deletion. A complex interaction of species-specific repair and recombination rates may further be elucidated as important factors in “regulating” how plastid genome reduction proceeds in holoparasites (Wicke et al. in prep.).
Structural reorganization of the plastid DNAs (e.g. inversions) in parasites occurs to a considerably smaller extent (if at all), compared with the amount of segmental DNA losses. The only reports of large-scale structural changes come from Colwell (1994) and Downie and Palmer (1992), who revealed the independent loss of one of the inverted repeats in and its Conopholis and S. asiatica, respectively. Outside Orobanchaceae, the highly reduced plastome of the underground orchid Rhizanthella has severe alterations around the inverted repeats (Delannoy et al. 2011), whereas the less dramatically reduced genome of the closely related bird’s-nest orchid Neottia nidus-avis shows no genomic rearrangements (Logacheva et al. 2011). Small inversions were reported in plastomes of some species of Cuscuta, but no large-scale plastomic reconfigurations were found (Krause 2011). Thus, the generally high degree of structural conservation reported for the majority of angiosperm plastomes (Wicke et al. 2011) appears to be upheld in Orobanchaceae holoparasites for a long duration after the loss of photosynthesis. In contrast, ongoing research suggests that the relaxation of functional constraints and subsequent gene loss rapidly commence after the transition to a (obligate) heterotrophic way of life (Wicke et al. submitted).
4 The Mitochondrial Genome
Unlike plastomes, mitochondrial genomes (chondriomes) are highly susceptible to the incorporation of both horizontally transferred DNA and DNA from other cellular genomes (Won and Renner 2003; Bergthorsson et al. 2004; Davis et al. 2005; Knoop et al. 1996, 2011). Additionally, the high variability of size and gene content of plant chondriomes makes them difficult targets for phylogenetic and comparative evolutionary studies (Knoop et al. 2011).
As of this writing, no complete sequence of a chondriome of a parasitic flowering plant is available. Only few studies focused on the molecular evolution of mitochondrial genes, although several genes (e.g. matR, atp1, and coxI) have been employed to infer the placement of parasitic plant lineages in the tree of flowering plants. Despite the fact that DNA evolves normally more slowly than nuclear and plastid DNAs (Wolfe et al. 1987), some holoparasitic lineages (e.g. Apodanthaceae, Rafflesiaceae) exhibit elevated nucleotide substitution rates in the mitochondrial small ribosomal RNA (mtSSU) as well as in the mitochondrial genes coxI, atp1, matR, and in exons B and C of nad1 (Duff and Nickrent 1997; Nickrent et al. 2004; Barkman et al. 2004, 2007; Filipowicz and Renner 2010). However, no rate acceleration has been found in holoparasitic Orobanchaceae genera such as Epifagus, Orobanche, and Boschniakia (supposedly Kopsiopsis; Mower et al. 2004; Barkman et al. 2007). Hemiparasitic Orobanchaceae (e.g. Lamourouxia, Agalinis, Pedicularis, Hedbergia, Parentucellia, Bartsia, Buchnera) also appear to evolve at similar evolutionary rates as Lindenbergia (Mower et al. 2004).
Little is known about the evolution of macro- and microstructural changes, such as (small) insertions, deletions, and inversions in coding and non-coding mitochondrial DNAs of parasitic plants. Duff and Nickrent (1997) reported a slight increase of indel events in the mtSSU of non-asterid parasite lineages. In contrast, mtSSU in Epifagus shows only an insignificant length variation (2 nt) compared to photosynthetic relatives. Remarkably, parasitic plants frequently possess an intron in the coxI gene. The coxI intron is found in ten of the at least 12 independently evolved angiosperm lineages with a parasitic lifestyle (Barkman et al. 2007). The source of the intron remains largely unclear. While an initial acquisition of the coxI intron via horizontal homing from a fungus seems likely for some angiosperms (Vaughn et al. 1995; Adams et al. 1998; Cho and Palmer 1999; Seif et al. 2005, cp. Cusimano et al. 2008), most gains in parasites seem to have occurred by horizontal plant-to-plant transfers (Sanchez-Puerta et al. 2008; see below). The close interaction between a parasitic plant and its host further supports the hypothesis of a plant donor of parasite coxI introns.
5 Horizontal DNA Transfer
The identification of true horizontal DNA transfers from one plant to another and its origin can be very problematic (critically reviewed in Renner and Bellot 2012). Parasite–host systems appear to be especially prone to horizontal gene/DNA transfer (HGT) (see Sect. 6.5.2). In plants, the uptake and incorporation of DNA from another species occur more frequently in mitochondrial DNA, although comparative data of functional and non-functional HGT is still widely lacking for the nuclear genome. Prominent cases of HGT involving mitochondrial genes concern the atp1 region of parasitic plants of the Rafflesiaceae and Apodanthaceae (Davis and Wurdack 2004; Nickrent et al. 2004). A chimeric copy consisting of parasite-specific and horizontally acquired genic parts of atp1 was found in the mitochondria in Pilostyles thurberi (Apodanthaceae; Barkman et al. 2007). Copies of host atp1 appear to have also been independently transferred to species of the Bartsia clade of Orobanchaceae, and to Cuscuta (Mower et al. 2004, 2010). Other mitochondrial genes involved in HGT have not yet been identified in Orobanchaceae.
The transfer of macromolecules such as RNAs predominantly from the host into the parasite was reported for Triphysaria versicolor (Tomilov et al. 2008) and Phelipanche aegyptiaca (Aly et al. 2009; see Sect. 6.5.1). Phelipanche seems to also take up a host protein (Aly et al. 2011). A similar phenomenon was found in Cuscuta (reviewed by LeBlanc et al. 2012); comparable data from other parasitic plant families are currently lacking. Although the cellular components involved in RNA trafficking in host–parasite systems supposedly differ according to haustorial anatomy (see Sect. 3.9.3), current data suggest that parasites may have access to a great variety of host RNAs, including those encoding proteins that are functionally located in host plastids (e.g. rbcS, LeBlanc et al. 2012). Mobile RNAs may eventually also be incorporated into the nuclear genome of the parasite. Such cases have already been reported for an expressed nuclear gene of unknown function in Striga hermonthica (Yoshida et al. 2011) and also for Rafflesia (Xi et al. 2012).
Another case of HGT involves fragments of the plastid regions rbcL, rps2, and trnL-F, which appear to have been transmitted from Orobanche into some species of Phelipanche (Park et al. 2007a). Current data on plastid genome evolution in both genera suggest, however, that the horizontally acquired fragments from at least two out of the five originally studied Phelipanche species do not reside in the plastid genome (Wicke et al. submitted). Those fragments may be located in either the nuclear or the mitochondrial genome, as hypothesized earlier (Park et al. 2007a). Regardless of the location of the horizontally acquired copies, putative HGT remains highly interesting as it might involve transmission via a host plant as the vector, even though a vertical transmission may, however, also be considered (Park et al. 2007b).
6 Conclusions
Orobanchaceae possess highly dynamic genomes, which is in part due to the rampant occurrence of polyploidy as implied by genome-size data and chromosomal evolution. Transcript-profiling and transcriptome-sequencing projects have already identified several genes that are candidates for being newly recruited in parasite-specific developmental pathways, and large-scale sequencing in combination with basic genetic work has revealed complex patterns of macromolecule trafficking and signalling in selected host–parasite systems. Studies of plastid genes and genomes of members of Orobanchaceae have additionally brought to light the first insights into the complexity and differential dynamics in the process of plastid genome reduction after the loss of photosynthesis.
Ongoing and future research and research networks will allow the stepwise elucidation of the physiological evolution of the Orobanchaceae from the autotrophic to holo-heterotrophic lifestyle.
Despite the great progress that has already been achieved, our understanding of genomic evolution in Orobanchaceae is still hampered by a substantial lack of data on, for instance, chromosome numbers, genome sizes, gene content and organization, gene expression, and epigenetic variation (see Chap. 13). Further basic and comparative research are needed, including large-scale transcriptome and genome sequencing, to determine basic parasite-specific genetics and to elucidate the complex (co-)evolution of the parasites and their hosts.
Orobanchaceae, the largest and most diverse family of parasitic angiosperms has already proven to be highly suitable for studying the functional basis of a parasitic lifestyle in higher plants. Several projects are currently underway that will shed further light on genomic evolution as well as on the extent of horizontal gene transfer. Besides crucial physiological and ultrastructural works, genome surveys utilizing the rapidly developing sequencing technologies and large-scale proteomic approaches should be a key element in understanding the evolution of parasitism and all adaptations that come with the parasitic way of life, e.g. haustorium formation, host recognition, and nutrient acquisition. The sequencing of reference genomes of Orobanchaceae species is inevitable (though challenged by its genome size) and will eventually be an important step towards finding effective ways for control of the weedy species (see Chaps. 21 and 24).
The availability of reverse genetic approaches is another key point that will bring parasitic plant research to another level. Agrobacterium-mediated transformation systems have already been established successfully for the hemiparasites Triphysaria versicolor (Tomilov et al. 2007), Phtheirospermum japonicum (Ishida et al. (2011), and for the holoparasite Phelipanche aegyptiaca (Fernandez-Aparicio et al. 2011). Thus, the broomrape family provides a unique framework to experimentally test the function of putative parasite-specific genetic elements and to study physiological evolution across close relatives with differing degrees of heterotrophy.
Notes
- 1.
This chapter uses the most recent taxonomic changes outlined in Chap. 14.
References
Acquisti C, Elser JJ, Kumar S (2009a) Ecological nitrogen limitation shapes the DNA composition of plant genomes. Mol Biol Evol 26:953–956
Acquisti C, Kumar S, Elser JJ (2009b) Signatures of nitrogen limitation in the elemental composition of the proteins involved in the metabolic apparatus. Proc R Soc B 276:2605–2610
Adams KL, Clements MJ, Vaughn JC (1998) The Peperomia mitochondrial coxI group I intron: timing of horizontal transfer and subsequent evolution of the intron. J Mol Evol 46:689–696
Albach DC, Li H-Q, Zhao N, Jensen SR (2007) Molecular systematics and phytochemistry of Rehmannia (Scrophulariaceae). Biochem Syst Ecol 35:293–300
Alkatib S, Fleischmann TT, Scharff LB, Bock R (2012) Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res 40:6713–6724
Aly R, Cholakh H, Joel DM, Leibman D, Steinitz B, Zelcer A, Naglis A, Yarden O, Gal-On A (2009) Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol J 7:487–498
Aly R, Hamamouch N, Abu-Nassar J, Wolf S, Joel DM, Eizenberg H, Kaisler E, Cramer C, Gal-On A, Westwood JH (2011) Movement of protein and macromolecules between host plants and the parasitic weed Phelipanche aegyptiaca Pers. Plant Cell Rep 30:2233–2241
Bandaranayake PCG, Filappova T, Tomilov AA, Tomilova NB, Jamison-McClung D, Ngo Q, Inoue K, Yoder JI (2010) A single-electron reducing quinone oxidoreductase Is necessary to induce haustorium development in the root parasitic plant Triphysaria. Plant Cell 22:1404–1419
Barker WR, Kiehn M, Vitek E (1988) Chromosome numbers in Australian Euphrasia (Scrophulariaceae). Plant Syst Evol 158:161–164
Barkman TJ, Lim S-H, Salleh KM, Nais J (2004) Mitochondrial DNA sequences reveal the photosynthetic relatives of Rafflesia, the world’s largest flower. Proc Natl Acad Sci USA 101:787–792
Barkman TJ, McNeal JR, Lim SH, Coat G, Croom HB, Young ND, dePamphilis CW (2007) Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol 7:248
Bennett JR, Mathews S (2006) Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am J Bot 93:1039–1051
Berg S, Krause K, Krupinska K (2004) The rbcL genes of two Cuscuta species, C. gronovii and C. subinclusa, are transcribed by the nuclear-encoded plastid RNA polymerase (NEP). Planta 219:541–546
Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD (2004) Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA 101:17747–17752
Botanga CJ, Timko MP (2005) Genetic structure and analysis of host and non-host interactions of Striga gesnerioides. Phytopathology 95:1166–1173
Botanga CJ, Timko MP (2006) Phenetic relationships among different races of Striga gesnerioides (Willd.) Vatke from West Africa. Genome 49:1351–1365
Botanga CJ, Kling JG, Berner DK, Timko MP (2002) Genetic variability of Striga asiatica (L.) Kuntz based on AFLP analysis and host-parasite interaction. Euphytica 128:375–388
Boudreau E, Turmel M, Goldschmidt-Clermont M, Rochaix J-D, Sivan S, Michaels A, Leu S (1997) A large open reading frame (orf1995) in the chloroplast DNA of Chlamydomonas reinhardtii encodes an essential protein. Mol Gen Genet 253:649–653
Braukmann T, Stefanović S (2012) Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol Biol 79:5–20
Castro M, Castro S, Loureiro J (2012) Genome size variation and incidence of polyploidy in Scrophulariaceae sensu lato from the Iberian Peninsula. AoB Plants 2012: pls037. doi:10.1093/aobpla/pls037
Cho KY, Palmer JD (1999) Multiple acquisitions via horizontal transfer of a group I intron in the mitochondrial cox1 gene during evolution of the Araceae family. Mol Biol Evol 16:1155–1165
Colwell AE (1994) Genome evolution in a non-photosynthetic plant, Conopholis americana. Ph.D. Thesis, Washington University. Division of Biology and Biomedical Sciences, St. Louis, WA, USA
Cummings MP, Welschmeyer NA (1998) Pigment composition of putatively achlorophyllous angiosperms. Plant Syst Evol 210:105–111
Cusimano N, Zhang L-B, Renner SS (2008) Reevaluation of the cox1 group I intron in Araceae and angiosperms indicates a history dominated by loss rather than horizontal transfer. Mol Biol Evol 25:265–276
Davis C, Wurdack KJ (2004) Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science 305:676–678
Davis CC, Anderson WR, Wurdack KJ (2005) Gene transfer from a parasitic flowering plant to a fern. Proc R Soc B 272:2237–2242
de Koning AP, Keeling PJ (2006) The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol 4:12
Delannoy E, Fujii S, des Francs CC, Brundrett M, Small I (2011) Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol 28:2077–2086
Delavault PM, Thalouarn P (2002) The obligate root parasite Orobanche cumana exhibits several rbcL sequences. Gene 297:85–92
Delavault PM, Sakanyan V, Thalouarn P (1995) Divergent evolution of two plastid genes, rbcL and atpB, in a non-photosynthetic parasitic plant. Plant Mol Biol 29:1071–1079
Delavault PM, Russo NM, Lusson NA, Thalouarn P (1996) Organization of the reduced plastid genome of Lathraea clandestina, an achlorophyllous parasitic plant. Physiol Plant 96:674–682
dePamphilis CW, Palmer JD (1990) Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 348:337–339
dePamphilis CW, Young ND, Wolfe AD (1997) Evolution of plastid gene rps2 in a lineage of hemiparasitic and holoparasitic plants: many losses of photosynthesis and complex patterns of rate variation. Proc Natl Acad Sci USA 94:7367–7372
Downie SR, Palmer JD (1992) Restriction site mapping of the chloroplast DNA inverted repeat – a molecular phylogeny of the Asteridae. Ann Mo Bot Gard 79:266–283
Drescher A, Ruf S, Calsa T, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22:97–104
Duff RJ, Nickrent DL (1997) Characterization of mitochondrial small-subunit ribosomal RNAs from holoparasitic plants. J Mol Evol 45:631–639
Ems SC, Morden CW, Dixon CK, Wolfe KH, dePamphilis CW, Palmer JD (1995) Transcription, splicing and editing of plastid RNAs in the nonphotosynthetic plant Epifagus virginiana. Plant Mol Biol 29:721–733
Estep MC, Gowda BS, Huang K, Timko MP, Bennetzen JL (2012) Genomic characterization for parasitic weeds of the genus Striga by sample sequence analysis. Plant Genome 5:30–41
Fedorov AA (ed) (1969) Khromosomnye chisla tsetkovykh rasteny (Chromosome numbers of flowering plants). Izdatel’stvo Nauka, Leningrad
Fernandez-Aparicio M, Rubiales D, Bandaranayake PCG, Yoder J, Westwood J (2011) Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca. Plant Methods 7:36
Filipowicz N, Renner SS (2010) The worldwide holoparasitic Apodanthaceae confidently placed in the Cucurbitales by nuclear and mitochondrial gene trees. BMC Evol Biol 10:219
Funk H, Berg S, Krupinska K, Maier U, Krause K (2007) Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol 7:45
Goldblatt P, Johnson DE (1979) Index to plant chromosome numbers. Missouri Botanical Garden, St. Louis. http://www.tropicos.org/Project/IPCN. Accessed August 2012
Hanson L, McMahon KA, Johnson MAT, Bennett MD (2001) First nuclear DNA C-values for another 25 angiosperm families. Ann Bot 88:851–858
Hanson L, Leitch IJ, Bennett MD (2002) Unpublished data from the Jodrell Laboratory, Royal Botanic Gardens, Kew. Accessed via the Kew C-Value Database at data.kew.org/cvalues/ in August 2012
He F, Zhang X, Hu J-Y, Turck F, Dong X, Goebel U, Borevitz JO, de Meaux J (2012) Widespread interspecific divergence in cis-regulation of transposable elements in the Arabidopsis genus. Mol Biol Evol 29:1081–1091
Hjertson ML (1995) Taxonomy, phylogeny and biogeography of Lindenbergia (Scrophulariaceae). Bot J Linn Soc 119:265–321
Honaas L, Wafula E, Yang Z, Der J, Wickett N, Altman N, Taylor C, Yoder J, Timko M, Westwood J, dePamphilis (2013) Functional genomics of a generalist parasitic plant: Laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol 13:9
Ishida JK, Yoshida S, Ito M, Namba S, Shirasu K (2011) Agrobacterium rhizogenes-mediated transformation of the parasitic plant Phtheirospermum japonicum. PLoS One 6(10):e25802. doi:10.1371/journal.pone.0025802
Iwo GA, Husaini SWH, Olaniyan GO (1993) Cytological observations and distribution of Striga species in central part of Nigeria. Feddes Repertorium 104:497–501
Knauf U, Hachtel W (2002) The genes encoding subunits of ATP synthase are conserved in the reduced plastid genome of the heterotrophic alga Prototheca wickerhamii. Mol Genet Genomics 267:492–497
Knoop V, Unseld M, Marienfeld J, Brandt P, Sunkel S, Ullrich H, Brennicke A (1996) Copia-, gypsy- and LINE-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics 142:579–585
Knoop V, Volkmar U, Hecht J, Grewe F (2011) Mitochondrial genome evolution in the plant lineage. In: Kempten F (ed) Advances in plant biology – mitochondrial genomes. Springer Science & Business, New Yorlk, pp 3–29
Kondo K, Segawa M, Musselman LJ, Mann WF (1981) Comparative ecological study of the chromosome races in certain root parasitic plants of the southeastern U.S.A. Bol Soc Broteriana 53:793–807
Krause K (2011) Piecing together the puzzle of parasitic plant plastome evolution. Planta 234:647–656
Krause K, Berg S, Krupinska K (2003) Plastid transcription in the holoparasitic plant genus Cuscuta: parallel loss of the rrn16 PEP-promoter and of the rpoA and rpoB genes coding for the plastid-encoded RNA polymerase. Planta 216:815–823
Lagerkvist U (1978) “Two out of three”: an alternative method for codon reading. Proc Natl Acad Sci USA 75:1759–1762
LeBlanc M, Kim G, Westwood JH (2012) RNA trafficking in parasitic plant systems. Front Plant Sci 3:203
Lechat M-M, Pouvreau J-B, Péron T et al (2012) PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J Exp Bot 63:5311–5322
Leebens-Mack JH, dePamphilis CW (2002) Power analysis of tests for loss of selective constraint in cave crayfish and nonphotosynthetic plant lineages. Mol Biol Evol 19:1292–1302
Leitch IJ, Bennett MD (2004) Genome downsizing in polyploid plants. Biol J Linn Soc Lond 82:651–663
Leitch AR, Leitch IJ (2012) Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol 194:629–646
Lemaire B, Huysmans S, Smets E, Merckx V (2011) Rate accelerations in nuclear 18S rDNA of mycoheterotrophic and parasitic angiosperms. J Plant Res 124:561–576
Logacheva MD, Schelkunov MI, Penin AA (2011) Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol 3:1296–1303
Lohan AJ, Wolfe KH (1998) A subset of conserved tRNA genes in plastid DNA of nongreen plants. Genetics 150:425–433
Lohse M, Drechsel O, Bock R (2007) Organellar Genome DRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52:267–274
Lusson NA, Delavault PM, Thalouarn P (1998) The rbcL gene from the non-photosynthetic parasite Lathraea clandestina is not transcribed by a plastid-encoded RNA polymerase. Curr Genet 34:212–215
Manen J-F, Habashi C, Jeanmonod D, Park J-M, Schneeweiss GM (2004) Phylogeny and intraspecific variability of holoparasitic Orobanche (Orobanchaceae) inferred from plastid rbcL sequences. Mol Phylogenet Evol 33:482–500
Martin NJ (1983) Nuclear DNA variation in the Australasian Loranthaceae. In: Calder M, Berhnhardt P (eds) Biology of mistletoes. Academic Press, New York, pp 277–293
Matvienko M, Torres MJ, Yoder JI (2001) Transcriptional responses in the hemiparasitic plant Triphysaria versicolor to host plant signals. Plant Physiol 127:272–282
McNeal JR, Arumugunathan K, Kuehl J, Boore J, dePamphilis C (2007a) Systematics and plastid genome evolution of the cryptically photosynthetic parasitic plant genus Cuscuta (Convolvulaceae). BMC Biol 5:55
McNeal JR, Kuehl J, Boore J, de Pamphilis C (2007b) Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol 7:57
Moore DM (1982) Flora Europaea Check-List and chromosome index. Cambridge University Press, Cambridge UK
Morden CW, Wolfe KH, dePamphilis CW, Palmer JD (1991) Plastid translation and transcription genes in a nonphotosynthetic plant – Intact, missing and pseudogenes. EMBO J 10:3281–3288
Mower JP, Stefanovic S, Young GJ, Palmer JD (2004) Gene transfer from parasitic to host plants. Nature 432:165–166
Mower JP, Stefanovic S, Hao W, Gummow J, Jain K, Ahmed D, Palmer J (2010) Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol 8:150
Nagl W, Fusenig HP (1979) Types of chromatin organization in plant nuclei. Plant Syst Evol Suppl 2:221–223
Nickrent DL, Duff RJ (1996) Molecular studies or parasitic plants using ribosomal RNA. In: Moreno MT, Cubero JI, Berner D, Joel D, Musselman LJ, Parker C (eds) Advances in parasitic plant research. Junta de Andalucia, Dirección General de Investigación Agraria, Cordoba, Spain, pp 28–52
Nickrent DL, García M (2009) On the brink of holoparasitism: plastome evolution in dwarf mistletoes (Arceuthobium, Viscaceae). J Mol Evol 68:603–615
Nickrent DL, Blarer A, Qiu Y-L, Vidal-Russell R, Anderson FE (2004) Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evol Biol 4:40
Park J-M, Manen J-F, Schneeweiss GM (2007a) Horizontal gene transfer of a plastid gene in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae). Mol Phyl Evol 43:974–985
Park J-M, Schneeweiss GM, Weiss-Schneeweiss H (2007b) Diversity and evolution of Ty1-copia and Ty3-gypsy retroelements in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae). Gene 387:75–86
Piednoël M, Aberer AJ, Schneeweiss GM, Macas J, Novak P, Gundlach H, Temsch EM, Renner SS (2012) Next-generation sequencing reveals the impact of repetitive DNA across phylogenetically closely related genomes of Orobanchaceae. Mol Biol Evol 29:3601–3611
Randle CP, Wolfe AD (2005) The evolution and expression of RBCL in holoparasitic sister-genera Harveya and Hyobanche (Orobanchaceae). Am J Bot 92:1575–1585
Renner SS, Bellot S (2012) Horizontal gene transfer in eukaryotes: fungi-to-plant and plant-to-plant transfers of organellar DNA. In: Bock R, Knoop V (eds) Genomics of chloroplasts and mitochondria. Springer, Heidelberg, pp 223–235
Rogalski M, Karcher D, Bock R (2008) Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol 15:192–198
Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD (2008) Frequent, phylogenetically local horizontal transfer of the cox1 group I intron in flowering plant mitochondria. Mol Biol Evol 25:1762–1777
Schäferhoff B, Fleischmann A, Fischer E, Albach D, Borsch T, Heubl G, Müller KF (2010) Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences. BMC Evol Biol 10:352
Schneeweiss GM, Palomeque T, Colwell AE, Weiss-Schneeweiss H (2004) Chromosome numbers and karyotype evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am J Bot 91:439–448
Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y (2004) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432:779–782
Seif E, Leigh J, Liu Y, Roewer I, Forget L, Lang BF (2005) Comparative mitochondrial genomics in zygomycetes: bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Res 33:734–744
Shinozaki K, Ohme M, Tanaka M et al (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5:2043–2049
Takagi K, Okazawa A, Wada Y, Mongkolchaiyaphruek A, Fukusaki E, Yoneyama K, Takeuchi Y, Kobayashi A (2009) Unique phytochrome responses of the holoparasitic plant Orobanche minor. New Phytol 182:965–974
Tank DC, Egger JM, Olmstead RG (2009) Phylogenetic classification of subtribe Castillejinae (Orobanchaceae). Syst Bot 34:182–197
Tolbert NE (1997) The C2 oxidative photosynthetic carbon cycle. Annu Rev Plant Physiol Plant Mol Biol 48:1–25
Tomilov AA, Tomilova NB, Yoder JI (2007) Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta 225:1059–1071
Tomilov AA, Tomilova NB, Wroblewski T, Michelmore R, Yoder JI (2008) Trans-specific gene silencing between host and parasitic plants. Plant J 56:389–397
Trakulnaleamsai C, Okazawa A, An C-I, Kajiyama S, Fukusaki E, Yoneyama K, Takeuchi Y, Kobayashi A (2005) Isolation and characterization of a cDNA encoding phytochrome A in the non-photosynthetic parasitic plant, Orobanche minor Sm. Biosci Biotechnol Biochem 69:71–78
Vaughn JC, Mason MT, Sper-Whitis GL, Kuhlman P, Palmer JD (1995) Fungal origin by horizontal transfer of a plant mitochondrial group I intron in the chimeric coxI gene of Peperomia. J Mol Evol 41:563–572
Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM (2006) Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am J Bot 93:148–156
Westwood JH, Yoder JI, Timko MP, dePamphilis CW (2010) The evolution of parasitism in plants. Trends Plant Sci 15:227–235
Westwood JH, dePamphilis CW, Das M, Fernández-Aparicio M, Honaas LA, Timko MP, Wafula EK, Wickett NJ, Yoder JI (2012) The Parasitic Plant Genome Project: new tools for understanding the biology of Orobanche and Striga. Weed Sci 60:295–306
Wickett NJ, Zhang Y, Hansen SK, Roper JM, Kuehl JV, Plock SA, Wolf PG, dePamphilis CW, Boore JL, Goffinet B (2008) Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol 25:393–401
Wickett NJ, Honaas LA, Wafula EK, Das M, Huang K, Wu B, Landherr L, Timko MP, Yoder J, Westwood JH, dePamphilis CW (2011) Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr Biol 21:2098–2104
Wimpee CF, Wrobel R, Garvin D (1991) A divergent plastid genome in Conopholis americana, an achlorophyllous parasitic plant. Plant Mol Biol 17(166):161
Wimpee CF, Morgan R, Wrobel RL (1992) Loss of transfer RNA genes from the plastid 16S–23S ribosomal RNA gene spacer in a parasitic plant. Curr Genet 21:417–422
Wolfe KH (1994) Similarity between putative ATP-binding sites in land plant plastid ORF2280 proteins and the FtsH/CDC48 family of ATPases. Curr Genet 25:379–383
Wolfe AD, dePamphilis CW (1997) Alternate paths of evolution for the photosynthetic gene rbcL in four nonphotosynthetic species of Orobanche. Plant Mol Biol 33:965–977
Wolfe AD, dePamphilis CW (1998) The effect of relaxed functional constraints on the photosynthetic gene rbcL in photosynthetic and nonphotosynthetic parasitic plants. Mol Biol Evol 15:1243–1258
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Wolfe KH, Morden CW, Ems SC, Palmer JD (1992a) Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol 35:304–317
Wolfe KH, Morden CW, Palmer JD (1992b) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA 89:10648–10652
Won H, Renner SS (2003) Horizontal gene transfer from flowering plants to Gnetum. Proc Natl Acad Sci USA 100:10824–10829
Wood TE, Takebayashic N, Abrahamsen MS, Mayrose I, Greenspoond PB, Rieseberg LH (2009) The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA 106:13875–13879
Xi Z, Bradley R, Wurdack K, Wong KM, Sugumaran M, Bomblies K, Rest J, Davis C (2012) Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics 13:227
Yoshida S, Ishida JK, Kamal N, Ali A, Namba S, Shirasu K (2010) A full-length enriched cDNA library and expressed sequence tag analysis of the parasitic weed, Striga hermonthica. BMC Plant Biol 10:55
Young ND, dePamphilis CW (2005) Rate variation in parasitic plants: correlated and uncorrelated patterns among plastid genes of different function. BMC Evol Biol 5:16
Zonneveld BJM (2010) New record holders for maximum genome size in Eudicots and Monocots. J Bot 2010:4 pages
Zonneveld BJM, Leitch IJ, Bennett MD (2005) First nuclear DNA amounts in more than 300 angiosperms. Ann Bot 96:229–244
Acknowledgments
I thank Gerald Schneeweiss for the helpful comments on this manuscript and I am grateful to Kai Müller for critical discussion. I also thank Najibeh Ataei, Dietmar Quandt, and Hanna Weiss-Schneeweiss for sharing their unpublished data. Support from the Austrian Science Fund (FWF grant P19404 to G. Schneeweiss), the University of Vienna, and the German Academic Exchange Service (DAAD) for own research is highly appreciated.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Wicke, S. (2013). Genomic Evolution in Orobanchaceae. In: Joel, D., Gressel, J., Musselman, L. (eds) Parasitic Orobanchaceae. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-38146-1_15
Download citation
DOI: https://doi.org/10.1007/978-3-642-38146-1_15
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-38145-4
Online ISBN: 978-3-642-38146-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)