Abstract
Image registration plays an important role within pulmonary image analysis. The task of registration is to find the spatial mapping that brings two images into alignment. Registration algorithms designed for matching 4D lung scans or two 3D scans acquired at different inflation levels can catch the temporal changes in position and shape of the region of interest. Accurate registration is critical to post-analysis of lung mechanics and motion estimation. In this chapter, we discuss lung-specific adaptations of intensity-based registration methods for 3D/4D lung images and review approaches for assessing registration accuracy. Then we introduce methods for estimating tissue motion and studying lung mechanics. Finally, we discuss methods for assessing and quantifying specific volume change, specific ventilation, strain/ stretch information and lobar sliding.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Image Registration
- Registration Method
- Registration Algorithm
- Registration Accuracy
- Maximal Principal Strain
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Image registration can be used to determine a spatial mapping that matches images collected at different time points, or using different imaging modalities. It has been widely used in radiotherapy for various applications, such as in motion studies, dose accumulation/composite, dose response evaluation, adaptive planning, auto contouring etc. for various radiotherapy treatment sites such as brain, lung, head neck, prostate, and cervix. In motion studies, image registration has been used to evaluate the internal target volume (ITV) which encompasses the clinical target volume (CTV) and internal margin (IM) to account for the variation in size, shape and position, e.g. filling of bladder and movements of respiration [54]. In auto contouring, the contours drawn by physicians on the initial data set can be propagated to the data set of interest after image registration is performed to define the gross tumor volume (GTV) or the organs at risk (OAR) [95]. Such image pair can be intra-modality images such as 4D CT images or inter-modality images such as the images of PET-CT image and planning CT image. In addition to the primary contours drawn by the physicians, the OARs can also be defined as atlas from population so that they can be propagated to any new patient data set when the contour task is required. The auto contour greatly reduced the total treatment planning time and allows for the potential fast same day simulation, planning and treatment clinical work flow [34]. Similarly, in the dose accumulation/composite, instead of propagating the contours, the planning dose from previous treatments can be propagated to the later treatment scans so that the total planning dose can be accumulated for further dose response evaluation or adaptive planning [100].
In lung cancer radiotherapy, image registration is a key tool as one seeks to link images across modalities, across time, or between lung volumes in the use of pulmonary investigations [39, 46, 58, 60], due to the inherent motion of the respiratory system. For example, registration can be used to determine the spatial locations of corresponding voxels in a sequence of pulmonary scans, as discussed in Chap. 6. The computed correspondences immediately yield the displacement fields corresponding with the motion of the lung between a pair of images. Using the image registration, the auto contouring, dose accumulation/composite, adaptive planning tools etc. as introduced above can be built upon. Furthermore, because of the lung tissue’s mechanical property during expansion and contraction is highly related to its ventilation function, the image registration has also been extended to the functional lung imaging as discussed in Chap. 13.
Chapters 2 and 3 introduced different methods for 4D thoracic CT image acquisition. Imaging allows non-invasive study of lung behavior and image registration has been used to examine lung mechanics and pulmonary functions [31, 43, 74]. Some groups have utilized non-invasive imaging and image registration techniques to examine the linkage between estimates of regional lung expansion and local lung ventilation [26, 32, 42–44, 74, 88, 89, 102]. Guerrero et al. used two CT images, acquired at different lung inflations, and optical flow image registration to estimate regional ventilation to identify functioning versus non-functioning lung tissue for radiotherapy treatment planning [43, 44]. Sundaram and Gee used serial magnetic resonance imaging to quantify lung kinematics in statically acquired sagittal cross-sections of the lung at different inflations [88, 89]. Using non-linear image registration, they estimated a dense displacement field from one image to the other, and from the displacement field they computed regional lung strain. Christensen et al. used consistent image registration to match images across cine-CT sequences, and estimate rates of local tissue expansion and contraction [26]. Their measurements matched well with spirometry data. Reinhardt et al. used image registration to match lung CT volumes across different levels of inflation [32, 74]. They calculated local specific volume changes as an index of regional ventilation, and compared specific volume change to xenon-CT based estimates of regional ventilation in sheep. Gorbunova et al. developed a weight preserving image registration method for monitoring disease progression [42]. Yin et al. proposed a new similarity cost preserving the lung tissue volume, and compared the new cost function driven registration method with SSD driven registration in the estimation of regional lung function [102].
Local lung expansion can be estimated by using registration to match images acquired at different levels of inflation. Tissue expansion (and thus, specific volume change) can be estimated by calculating the Jacobian determinant of the transformation field derived by image registration [74]. The tissue strain tensor can also be calculated from the transformation field. Since both the Jacobian matrix and the strain tensor are formed using partial derivatives of the transformation field, it is important that the underlying registration transformation model be well-behaved with respect to these derivatives if the functional and mechanical results obtained are to be useful.
2 Intensity-Based Registration
The goal of registration is to find the spatial mapping that will bring the moving image into alignment with the fixed image. Many image registration algorithms have been proposed and various features such as landmarks, contours, surfaces and volumes have been utilized to manually, semi-automatically or automatically define correspondences between two images [39, 59, 60]. Chapters 5 and 6 introduced feature-based and intensity-based registration techniques. The input data to the registration process is usually two images; one is defined as the moving or template image\(I_1\), and the other is defined as the fixed or target image\(I_2\). The transform defines how points from the moving image \(I_1\) are mapping to their corresponding points in the fixed image \(I_2\). In three dimensional space, let \({\varvec{x}}=(x, y, z)^T\) define a voxel coordinate in the image domain of the fixed image \(I_2\). The transformation \({\varvec{T}}\) is a \((3 \times 1)\) vector-valued function defined on the voxel lattice of fixed image, and \({\varvec{T}}({\varvec{x}})\) gives the corresponding location in moving image to the point \({\varvec{x}}\). The cost function represents the similarity measure of how well the fixed image is matched by a warped moving image. The optimizer is used to optimize the similarity criterion over search space defined by transformation parameters.
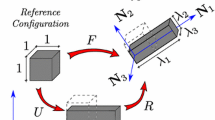
4D image registrations have been developed for spatial and temporal motion estimation in a 4D image sequence. 4D CT images are given as a sequence of 3D images representing different respiratory phases in the breathing cycle as discussed in Chaps. 2 and 3. Typical values are \(10\) phases from 0 to 90 % phase at 10 % intervals, where 0 % represents maximum inhale and 50 % approximately maximum exhalation. Most registration approaches use a pairwise registration paradigm, including the reference-strategy which registers each phase to a chosen reference phase (e.g. the end of expiration), and the consecutive-strategy which describes deformations respect to the neighboring time point. Figure 7.1 depicts the reference-strategy with the 50 % image as reference. For a voxel localization \({\varvec{x}}\) in the reference phase, the corresponding localization in the rest of the phases is given by the computed transformation, and in this way, the trajectory of a point is easily defined. Using the consecutive strategy, transformations have to be concatenated to compute a trajectory, whereby interpolation errors may be introduced.
Registration scheme for motion field extraction of a 4D CT data set using the reference-strategy. Each phase of the 4D CT image is registered with a chosen reference phase (e.g. the end of expiration) (figure adapted from [57])
In addition to pair-wise methods, groupwise strategies have been developed which simultaneously align multiple phases in a 4D image sequence. In these approaches, all the phase images in a 4D data set are input into the registration algorithm without assigning fixed or moving image [16, 62, 93, 99]. In this chapter, we introduce lung motion estimation from pair-wise registration between images acquired at different inflation levels.
2.1 Preprocessing
Many registration techniques for lung CT images require image preprocessing steps. Common preprocessing operations include downsampling large datasets, extracting region of interest (ROI), and initial pre-aligning images. As the CT imaging techniques improve, it is now possible to image lung structures with high spatial resolution, and thus produce large CT datasets. In many cases, downsampling is needed to resize the original data and to improve the registration efficiency and robustness. A further common preprocessing step is an initial alignment to roughly catch the translation, rotation, and scaling between two images, i.e. by an affine pre-registration.
One of the major challenges for registration methods in lung motion estimation is the occurrence of sliding motion between the visceral and parietal pleura (see Sect. 4.2.1) and between the individual lung lobes [4, 12, 33]. Sliding motion contradicts common regularization models applied to avoid discontinuities like gaps or foldings in the computed transformation. To cope with sliding motion, many registration methods utilize a lung segmentation mask to restrict registration to the inside of the lung. In the inter-institutional study Evaluation of Methods for Pulmonary Image Registration (EMPIRE10) [66], 16 out of 20 participating methods applied masking in at least on step of the algorithm. Due to the large density difference between the air-filled lungs and surrounding tissue, robust automatic algorithms exist for lung segmentation in CT images [52]. Figure 7.2 gives an illustration of pulmonary CT images with renderings of the lung segmentations. Image pair was acquired during breath-holds near functional residual capacity (maximum exhale) and total lung capacity (maximum inhale). Although first approaches for automatic segmentation of lung lobes exist [76, 92], lobe segmentation remains challenging because of imperceptible fissures in 4D CT images. Performing registration in the region of interest may help improve registration accuracy and efficiency, but registration is limited to the object (lung or lobe) and provides no information about other image regions. Recently, novel regularization approaches has been presented to explicitly model the sliding motion along organ boundaries (see Sect. 7.2.3.3).
2.2 Similarity Criteria
Intensity-based registration utilizes the intensity information of two images to measure how well they are aligned. It takes advantage of the strong contrast between the lung parenchyma and the chest wall, and between the parenchyma and the blood vessels and larger airways. To solve the intensity-based image registration problem, it is usually assumed that intensities of corresponding voxels are related to each other in some way. Many criteria to construct the intensity relationship between corresponding points have been suggested for aligning two images, as discussed in Chap. 6. Cost functions such as mean square difference (MSD), correlation coefficient, mutual information, pattern intensity, and gradient correlation are routinely used for image registration [50, 72].
2.2.1 Sum of Squared Difference (SSD)
A simple and common similarity function is the sum of squared difference (SSD), which measures the intensity difference at corresponding points between two images. Mathematically, it is defined by
where \(I_1\) and \(I_2\) are the moving and fixed images, respectively. \(\varOmega \) denotes the union of lung regions in fixed image and deformed moving image. The underlying assumption of SSD is that the image intensity at corresponding points between two images should be similar. This is true when registering images of the same modality. In such cases, if the images are perfectly mapped, the corresponding intensities should be identical, which means the same underlying structure has the same intensity value in the two images.
Illustration of histogram matching before SSD registration between breath-hold maximum exhale and maximum inhale images from a human subject. a A sagittal slice from maximum exhale. b A sagittal slice from maximum inhale. c The sagittal slice of b after histogram modification so that its grayscale range is similar to that of a
However, considering the change in CT intensity as air inspired and expired during the respiratory cycle, the grayscale range are different within the lung region in two CT images acquired at different inflation levels. To balance this grayscale range difference, normalization of the intensities are needed. For example, a histogram matching procedure [98] can be used before SSD registration to modify the histogram of moving image so that it is similar to that of fixed image. Figure 7.3 gives an illustration of histogram matching before SSD registration between a pair of images acquired at breath-hold maximum exhale and maximum inhale from a human subject. Alternative approaches use a-priori knowledge about density changes in the lungs to preprocess images before the registration [81]. Please note that intensity normalization is only needed for SSD similarity function discussed here. It is not necessary for the similarity functions discussed below.
2.2.2 Similarity Functions for Multi-Modal Registration
As mentioned above, CT intensity is a measure of tissue density and therefore changes as the tissue density changes during inflation and deflation. The registration problem under this circumstance is similar to the multi-modality image registration, where similarity functions based on correlation coefficients (CC) [7, 48] or on mutual information (MI) [27, 61, 91, 94] are well suited and widely used. In the EMPIRE10 study for lung registration [66], 10 out of 20 participants used multi-modal similarity metrics based on CC or MI.
The local normalized cross-correlation (NCC) between two images \(I_1\) and \(I_2\) assumes a linear dependency between the image intensities and is defined by
where \({\varvec{x}}_i\in \mathcal{N }({\varvec{x}})\) iterates through a neighborhood of voxel \({\varvec{x}}\) and \(\mu \) is the mean value within this neighborhood.
Mutual information expresses the amount of information that one image contains about the other one. Analogous to the Kullback-Leibler measure, the negative mutual information cost of two images is defined as [61, 91]
where \(p(i,j)\) is the joint intensity distribution of transformed moving image \(I_1\circ {\varvec{T}}\) and fixed image \(I_2\); \(p_{I_1\circ {\varvec{T}}}(i)\) and \(p_{I_2}(j)\) are their marginal distributions, respectively. The histogram bins of \(I_1\circ {\varvec{T}}\) and \(I_2\) are indexed by \(i\) and \(j\). Misregistration results in a decrease in the mutual information, and thus, increases the similarity cost \(C_{\mathrm{MI}}\). Note that the MI metric does not assume a linear relationship between the intensities of the two images (see Sect. 6.1.4.5 too). Multi-modal metrics omit the need of intensity normalization in lung registration, but the computational effort of these measures is increased compared to SSD.
2.2.3 Sum of Squared Tissue Volume Difference (SSTVD)
Beside the widely used standard similarity functions for mono- or multi-modal registration, a number of lung-specific similarity metrics have been developed recently.
A recently developed similarity function, the sum of squared tissue volume difference (SSTVD) [42, 102], accounts for the variation of intensity in the lung CT images during respiration. This similarity criterion minimizes the local difference of tissue volume inside the lungs scanned at different pressure levels. This criterion is based on the assumption that the tissue volume is constant between inhale and exhale.Footnote 1
Assume that lung is a mixture of two materials: air and tissue/blood (non-air). Then the Hounsfield units (HU) in lung CT images is a function of tissue and air content. From the HU of CT lung images, the regional tissue volume and air volume can be estimate following the air-tissue mixture model by Hoffman et al. [49]. Let \(v({\varvec{x}})\) be the volume element at location \({\varvec{x}}\). Then the tissue volume\(V({\varvec{x}})\) within the volume element can be estimated as
where we assume that \(HU_{air}=-1000\) and \(HU_{tissue}=0\). Let \(v_1({\varvec{T}}({\varvec{x}}))\) and \(v_2({\varvec{x}})\) be the anatomically corresponding volume elements, and \(V_1({\varvec{T}}({\varvec{x}}))\) and \(V_2({\varvec{x}})\) be the tissue volumes within anatomically corresponding volume elements, respectively. Then the intensity similarity function SSTVD is defined as [101, 102]
The Jacobian determinant (often simply called the Jacobian) of a transformation \(J({\varvec{T}})\) estimates the local volume changes resulted from mapping an image through the deformation [10, 21, 25]. In 3D space, let \({\varvec{T}}(x,y,z) = [T_x(x,y,z), T_y(x,y,z), T_z(x,y,z)]^T\) be the vector-valued transformation which deforms the moving image \(I_1\) to the fixed image \(I_2\). The Jacobian of the transformation \(J({\varvec{T}}(x,y,z))\) at location \((x, y, z)^T\) is defined as
Thus, the tissue volumes in image \(I_1\) and \(I_2\) are related by \(v_1({\varvec{T}}({\varvec{x}}))=v_2({\varvec{x}})\cdot J({\varvec{T}}({\varvec{x}}))\), and Eq. (7.5) can be rewritten as
An alternative approach based on the assumption that the total mass (tissue volume) of the lung is conserved was proposed by Castillo et al. [17], named “combined compressible local-global optical flow” (CCLG). The main idea of this approach is to substitute the conservation of mass equation for the constant intensity assumption in the classical optical flow formulation of Horn and Schunk [51].
2.2.4 Sum of Squared Vesselness Measure Difference (SSVMD)
Feature information extracted from the intensity image is important to help guide the image registration process. During the respiration cycle, blood vessels keep their tubular shapes and tree structures. Therefore, the spatial and shape information of blood vessels can be utilized to help improve the registration accuracy. Blood vessels have larger HU values than that of parenchyma tissues. The intensity difference between parenchyma and blood vessels can effectively help intensity-based registration. However, as the blood vessel branches, the diameter of vessel becomes smaller and smaller. The small blood vessels are difficult to see because of their low intensity contrast. Therefore, grayscale information of the small vessels give almost no contribution to similarity functions directly based on intensity. In order to better utilize the information of blood vessel locations, the vesselness measure (VM) computed from intensity image can be used.
The vesselness measure is based on analyses of eigenvalues of the Hessian matrix of the image intensity. These eigenvalues can be used to indicate the shape of the underlying object. In 3D lung CT images, tubular structures such as blood vessels (bright) are associated with one negligible eigenvalue and two similar non-zero negative eigenvalues [40]. Ordering eigenvalues of a Hessian matrix by magnitude \(|\lambda _1|\le |\lambda _2|\le |\lambda _3|\), the Frangi’s vesselness function [40] is defined as
with
where \(R_A\) distinguishes between plate-like and tubular structures, \(R_B\) accounts for the deviation from a blob-like structure, and \(S\) differentiates between tubular structure and noise. \(\alpha \), \(\beta \), \(\gamma \) control the sensitivity of the vesselness measure. In lung CT images, an example parameter setting is \(\alpha =0.5\), \(\beta =0.5\), and \(\gamma =5\).
The Hessian matrix is computed by convolving the intensity image with second and cross derivatives of the Gaussian function. For a multiscale analysis, the response of the vesselness filter will achieve the maximum at a scale which approximately matches the size of vessels to detect. Therefore, the final vesselness measure is estimated by computing Eq. (7.8) for a range of scales and selecting the maximum response: \(F=\max _{\sigma _{min}\le \sigma \le \sigma _{max}}F(\lambda )\). Here \(\sigma \) is the standard deviation of the Gaussian function [36].
The vesselness image is rescaled to [0, 1] and can be considered as a probability-like estimate of vesselness features. Larger vesselness value indicates the underlying object is more likely to be a vessel structure, as shown in Fig. 7.4. The sum of squared vesselness measure difference (SSVMD) is designed to match similar vesselness patterns in two images. Given \(F_1({\varvec{x}})\) and \(F_2({\varvec{x}})\) as the vesselness measures of images \(I_1\) and \(I_2\) at location \({\varvec{x}}\) respectively, the vesselness cost function is formed as [13, 14]
Mismatch from vessel to tissue structures will result a larger SSVMD cost. This similarity metric can be used together with any other intensity-based volumetric metrics to help guide the registration process and improve matching accuracy.
2.3 Transformation Constraints
Enforcing constraints on the transformation helps to generate more meaningful registration results. In general, the computed transformation should be smooth and maintain the topology of the transformed images. Furthermore, the registration method should be symmetric, and should produce the same correspondences between two images independent of the choice of fixed and moving images.
2.3.1 Regularization Constraints
Continuum mechanical models such as linear elasticity [22, 23] and viscous fluid [22, 24] can be used to regularize the transformations. For example, linear-elastic constraint of the form
can be used to regularize the displacement fields where
The linear elasticity operator \(L\) has the form of \(L {\varvec{u}}({\varvec{x}}) = - \alpha \nabla ^2 {\varvec{u}}({\varvec{x}}) - \beta \nabla ( \nabla \cdot {\varvec{u}}({\varvec{x}})) + \gamma {\varvec{u}}({\varvec{x}})\) where \(\nabla = \left[ \frac{\partial }{\partial x}, \frac{\partial }{\partial y}, \frac{\partial }{\partial z} \right] \) and \(\nabla ^2 = \nabla \cdot \nabla = \left[ \frac{\partial ^2}{\partial x^2} +\frac{\partial ^2}{\partial y^2} + \frac{\partial ^2}{\partial z^2} \right] \). In general, \(L\) can be any nonsingular linear differential operator [63], e.g. a Laplacian regularization constrain is given by
Further regularization terms are discussed in Sect. 6.1.5.
The purpose of the regularization constraints are to constrain the transformation to obey the laws of continuum mechanics and ensure it maintains the topology of two images. Using a linear differential operator as defined in Eq. (7.11) can help to smooth the transformation, and to eliminate abrupt changes in the displacement fields. However, it can not prevent the transformation from folding onto itself, i.e. destroying the topology of the images under transformation [25].
To help maintain desirable properties of the moving and fixed images during deformation, another regularization example can be a constraint that prevents the Jacobian of transformations from going to zero or infinity. The Jacobian is a measurement to estimate the pointwise expansion and contraction during the deformation (see Sect. 7.2.2.3). A constraint that penalizes small and large Jacobian is given by [21]
Further examples of regularization constraints that penalize large and small Jacobians can be found in Sect. 6.1.5, and Ashburner et al. [6].
2.3.2 Inverse Consistency Constraint
In order to find better correspondence mapping and reduce pairwise registration error, one method is to jointly estimate the forward and reverse transformations between two images while minimizing the inverse consistent error.
Define forward transformation \({\varvec{T}}\) deforms \(I_1\) to \(I_2\) and the reverse transformation \({\varvec{G}}\) deform \(I_2\) to \(I_1\). A meaningful map between two anatomical images should be one-to-one, i.e. each point in image \(I_1\) is mapped to only one point in image \(I_2\) and vice versa. However, many unidirectional image registration techniques have the problem that their similarity cost function does not uniquely determine the correspondence between two images. The reason is that the local minima of similarity cost functions cause the estimated forward mapping \({\varvec{T}}\) to be different from the inverse of the estimated reverse mapping \({\varvec{G}}^{-1}\). To overcome correspondence ambiguities, transformations \({\varvec{T}}\) and \({\varvec{G}}\) can be jointly estimated. Ideally, \({\varvec{T}}\) and \({\varvec{G}}\) should be inverses of one another, i.e. \({\varvec{T}}={\varvec{G}}^{-1}\). In order to couple the estimation of \({\varvec{T}}\) and \({\varvec{G}}\) together, an inverse consistency constraint (ICC) [21] is imposed as
The constraint is minimized and the corresponding transformations are said to be inverse-consistent if \({\varvec{T}}= {\varvec{G}}^{-1}\).
2.3.3 Sliding Preserving Regularization
The physiological characteristics of the lung motion imply discontinuities between the motion of lung and rib cage contradicting common regularization schemes. As discussed in Sect. 7.2.1, most lung-specific registration algorithms address this problem using lung segmentation masks.
Recently, novel regularization approaches were presented to explicitly model the sliding motion along organ boundaries. Ruan et al. [78] uses a regularization that preserves large shear values to allow for sliding motion. Schmidt-Richberg et al. [83] addressed sliding motion by a direction-dependent regularization at organ boundaries extending the common diffusion registration by distinguishing between normal- and tangential-directed motion. The idea of direction-dependent regularization was adopted in other publications, too [30, 71].
2.4 Parameterization, Optimization and Multi-Resolution Scheme
2.4.1 Transformation Parameterization
The transformation model defines how one image can be deformed to match another. It can be a simple rigid or affine transformation, or a non-linear transformation such as the spline-based registrations [90], elastic models [9], fluid models [19], finite element (FE) models [38], etc. The lung is composed of non-homogenous, soft tissue, interlaced by branching networks of airways, arteries, and veins. Lung tissue expansion varies within in the lung depending on body orientation, the direction of gravitation forces, the pattern of airway and vessel branching, disease conditions, and other factors. Since lung expansion is non-uniform, non-linear transformation models are needed to track tissue expansion across changes in lung volume.
To represent the locally varying geometric distortions, the transformation can be represented through different forms. There are three common parameterizations used for intensity-based registration methods of lung CT. The first type of transformation is based on B-splines, as introduced in Sect. 6.2.1. B-splines [79] are well suited for shape modeling and are efficient to capture the local nonrigid motion between two images. Considering the computational efficiency and accuracy requirement, the cubic B-Spline based parameterization is commonly chosen to represent the transformation. In the EMPIRE10 study 10 out of 20 algorithms used a B-Spline parametrization as transformation model [66]. Other basis functions such as thin-plate splines(TPS) [8], Fourier series [5], elastic body spline(EBS) [29] can also be used to parameterize the deformation.
The second type of transformation is dense deformable vector field (DVF), which is introduced in Sect. 6.2.2. The DVF is a non-parametric model representing the deformation by displacement vectors \({\varvec{u}}({\varvec{x}})\) for each voxel location. The transformation is then given by \({\varvec{T}}({\varvec{x}})={\varvec{x}}+{\varvec{u}}({\varvec{x}})\).
In the third type, the transformation is represented by a velocity field\({\varvec{v}}:{\varOmega }\rightarrow \mathbb R ^{d}\) in order to ensure that the transformation is diffeomorphic. Diffeomorphisms define a globally one-to-one smooth and continuous mapping, and therefore, preserve topology and are suitable for the study of pulmonary kinematics. Given a time-dependent velocity field \({\varvec{v}}:{\varOmega }\times [0,1]\rightarrow \mathbb R ^{d}\), one defines the ordinary differential equation (ODE): \(\partial _t {\varvec{\phi }}({\varvec{x}},t)={\varvec{v}}({\varvec{\phi }}({\varvec{x}},t),t)\), with \({\varvec{\phi }}({\varvec{x}},0)={\varvec{x}}\). For sufficient smooth \({\varvec{v}}\) and fixed \(t\), e.g. \(t=1\), the solution \({\varvec{T}}({\varvec{x}})={\varvec{\phi }}({\varvec{x}},1)\) of this ODE is known to be a diffeomorphism on \({\varOmega }\) [103]. Diffeomorphic registration algorithms are discussed in more detail in Sect. 10.4.2. Several approaches use diffeomorphic registration methods to model lung motions [28, 35, 41, 82, 86].
2.4.2 Optimization
Most registration algorithms employ standard optimization techniques to the optimal transformation, as discussed in Chap. 6. There are several existing methods in numerical analysis such as the partial differential equation (PDE) solvers to solve the elastic and fluid transformation, gradient descent, conjugate gradient method, Newton, Quasi-Newton, LBFGS, etc [56, 68, 69, 73]. One efficient optimization method is a limited-memory, quasi-Newton minimization method with bounds (L-BFGS-B) [11, 104]. It is well suited for optimization with a high dimensional parameter space. In addition, this algorithm allows bound constraints on the independent variables. For example, if the transformation are represented using B-splines, then the bound constraints can be applied on B-spline coefficients so that it is sufficient to guarantee the local injectivity (one-to-one property) of transformation [18], i.e. the transformation maintains the topology of two images. According to the analysis from Choi and Lee [18], the displacement fields are locally injective all over the domain if B-Spline coefficients satisfy the condition that \(c_x \le \delta _x/K, c_y \le \delta _y/K, c_z \le \delta _z/K\), where \(c_x, c_y, c_z\) are B-Spline coefficients, \(\delta _x, \delta _y, \delta _z\) are B-Spline grid sizes along each direction, and K is a constant approximately equal to 2.479772335.
2.4.3 Multi-Resolution Scheme
In order to improve speed, accuracy and robustness of registration algorithms, a spatial multiresolution procedure from coarse to fine is often used. The basic idea of multiresolution is that registration is first performed at a coarse scale where the images have much fewer pixels, which is fast and can help eliminate local minima. The resulting spatial mapping from the coarse scale is then used to initialize registration at the next finer scale. This process is repeated until registration is performed at the finest scale.
3 Registration Accuracy Assessment
Validation and evaluation of image registration accuracy is an important task to quantify the performance of registration algorithms. Due to the absence of a ‘gold standard’ to judge a registration algorithm, various evaluation methods are needed to validate the performance of image registration with respect to different properties of transformations. Focusing on lung image registration, the following image features and information can be used to measure registration accuracy:
-
Features extracted from image, such as landmarks, airway and vessel trees, fissures, and object volumes;
-
Transformation properties, such as zero singularities and inverse consistency properties;
-
Additional information that is not used in the registration data, such as ventilation map estimated from Xenon-CT image;
All the features and image information mentioned above provide different perspectives for registration accuracy measurement. Additional validation methods to measure registration accuracy can be found in Chap. 8.
3.1 Evaluation Methods Used in the EMPIRE10 Challenge
Researchers usually test their algorithms on their own data, which varies widely. To make a fair comparison of different methodologies, EMPIRE10 challenge (Evaluation of Registration Methods on Thoracic CT) was designed to evaluate the performance of different lung registration algorithms on the same set of 30 thoracic CT pairs.
All the registration results were evaluated in four aspects: (1) alignment of the lung boundaries, (2) alignment of the major fissures, (3) alignment of correspondence of annotated point pairs, (4) analysis of singularities in the deformation field. We will shortly discuss these validation methods, full details can be found in [66].
3.1.1 Landmark Matching Accuracy
Landmarks are point features of an object. Anatomical landmarks have biological meaning. Intra-subject images of the lung contain identifiable landmarks such as airway-tree and vascular-tree branch points. An example of the landmark distribution is shown in Fig. 7.5 with the assistance of a semi-automatic system [67]. The Euclidean distance between registration-predicted landmark position and its true position is defined as landmark error.
In the EMPIRE10 study, landmarks were generated semi-automatically by three independent experts.
3.1.2 Fissure Alignment Distance
The human lungs are divided into five independent compartments which are called lobes. Lobar fissures are the division between adjacent lung lobes. The lobes can be segmented [92] and the fissures represent important physical boundaries within the lungs. Therefore, alignment of fissures is an important evaluation category. The statistics of fissure positioning error (FPE) can be used to evaluate the fissure alignment. The FPE is defined as the minimum distance between a point on the deformed fissure and the closest point on the corresponding target fissure. Mathematically, this metric can be stated as
for a given point \({\varvec{x}}\) in \(F_1\), where \(F_1\) (\(F_2\), resp.) is the set of all points in the fissure in image \(I_1\) (\(I_2\), resp.) and \(d(\cdot )\) defines the Euclidean distance.
3.1.3 Relative Overlap of Lung Segmentations
The relative overlap (RO) is used to measure how well the corresponding parenchymal regions of interest agree with each other. The RO metric is given by
where \(S_1\) and \(S_2\) are corresponding regions of interest in images \(I_1\) and \(I_2\), respectively. \(S_1\circ {\varvec{T}}\) corresponds to a segmentation transformed from image \(I_1\) to \(I_2\). The relative overlap of segmentations can be evaluated on the whole parenchyma region, or subvolumes of left lung, right lung, or even on the lobe level whenever the segmentations are available.
3.1.4 Validation on Transformation Properties
Good matching accuracy on the feature locations does not guarantee that regions away from the features are correctly aligned. In order to reveal how well a transformation preserves the topology between two images during the deformation, the Jacobian determinant of the transformation field derived by image registration can be used for singularity assessment.
The Jacobian determinant \(J\) at a given point gives important information about the behavior of transformation \({\varvec{T}}\) at that point. If the Jacobian value at \({\varvec{x}}\) is zero, then \({\varvec{T}}\) is not invertible near \({\varvec{x}}\). A negative Jacobian indicates \({\varvec{T}}\) reverses orientation, which folds the domain [20, 25]. A positive Jacobian means the transformation \({\varvec{T}}\) preserves orientation near \({\varvec{x}}\). These indications of Jacobian are listed in Eq. (7.18).
The Jacobian evaluates the quality of transformations by measuring how well the transformation preserves topology. For lung registration, a meaningful and physically plausible deformation within a continuous region should be diffeomorphic, and thus contains no singularities (\(J \le 0\)).
Other measures of transformation properties can be defined, e.g. the inverse consistency error (see Sect. 8.1). It measures the consistency between a forward transformation and a reverse transformation between two images. Ideally, composing the forward and reverse transformations together should produce the identity map when there is no inverse consistency error.
3.1.5 Registration Algorithms for Lung Motion Estimation
The EMPIRE10 competition included a wide variety of registration algorithms (transformation models, similarity measures, etc.). Most algorithms used lung-specific preprocessing (masking, histogram-matching, etc.), but beside this, only a selection of algorithms were tailored towards thoracic CT applications. A conclusion of the EMPIRE10 study was “[..] generic algorithms which were not tailored for this data performed extremely well and many different approaches to registration were shown to be successful.” [66]. Accurate registration of lung CT images is possible, as shown by an average TRE of less than 1 mm for the top six algorithms in this study. Another important result was that landmark matching accuracy provided the most useful reference for distinguishing between registration algorithm results.
In the following, we review some of the registration algorithms that participated in this study: Song et al. proposed the algorithm which includes affine transformation and different diffeomorphic transformation by maximizing cross correlation between images [86]. Han proposed a hybrid feature-constrained deformable registration method. The features are detected based on robust 3D SIFT descriptors, and then a mutual information based registration is used to match those features. The feature correspondences are used to guide an intensity-based deformable image registration which maximizes the mutual information and minimize the normalized sum of squared differences between images [47]. Ruhaak et al. presented a variational approach which drives the registration towards exact lung boundary matching. They used the normalized gradient field to measure the distance focusing on image edges instead of intensities, and a curvature regularizer is applied to penalize the second order derivatives and yields smooth solutions [80]. Muenzing et al. developed a novel regularization model incorporates information from two different knowledge sources: anatomical aspect and statistical aspect in which the regularization is considered as a machine learning problem. Predominant anatomical structures are extracted and used to model the ROI as composition of anatomical objects. And finally, a link function is formulated to combine the two pieces of information [65]. Modat et al. proposed the NiftyReg package which contains a global and a local registration algorithm [64]. A block-matching technique is used in the global registration, as proposed by Ourselin et al. [70]. The Free-Form Deformation (FFD) algorithm is used in the local registration stage to maximize the normalized mutual information. Staring et al. proposed the algorithm including a combination of an affine as well as non-rigid transformations by maximizing the normalized correlation coefficient, which is implemented in elastix [87]. Schmidt-Richberg et al. developed a robust registration approach which consists of an affine alignment, a shape-based adjustment of lung surfaces and an intensity-based diffeomorphic image registration. The algorithm has been optimized for the registration of lung CT data [82]. Cao et al. developed a nonrigid registration method to preserve both parenchymal tissue volume and vesselness measure. The transformation is regularized and its local injectivity is guaranteed [15]. Kabus et al. proposed a fast elastic registration algorithm that can be used in a multi-resolution setting. It is initialized by an affine pre-registration of the lungs followed by simultaneously minimizing of the similarity measure calculated as the sum of squared differences as well as the regularizing term based on the Navier-Lame equation [55]. The elastic regularizer assumes the lung tissue can be characterized as an elastic and compressible material. All these methods achieved good results in the evaluation challenge.
3.2 Validation Using Additional Information
3.2.1 Correlation Between Lung Expansion and Xe-CT Estimates of Specific Ventilation
Anatomical reference can usually provide features such as landmarks at the regions with high contrast which can be recognized by either human observer or computer algorithms. However, they are not be able to assess registration accuracy at the regions where no high contrast landmarks are available. Registration results in estimate of the regional specific ventilation (regional lung function). By comparing it with functional information, such as the xenon-CT based estimate of the regional specific ventilation, it is possible to assess the registration accuracy using lung function.
Previous studies have shown that the degree of regional lung expansion is directly related to specific ventilation (sV) [74]. A good registration should produce a deformation map (Jacobian map) which has high correlation with Xe-CT estimates of ventilation map.
An example of Jacobian map derived from registering CT images between positive end-expiratory pressure (PEEP) 10 cm \(\mathrm{H}_2\mathrm{O}\) and 25 cm \(\mathrm{H}_2\mathrm{O}\) for one sheep subject, and its sV measures resulted from Xe-CT image analysis [45, 85] are shown on a transverse slice in Fig. 7.6a, b. The correlation between ventilation map and Jacobian map can be utilized to validate the registration results. A higher correlation indicates a better registration. As shown in Fig. 7.6, both sV and Jacobian maps show a similar ventral to dorsal gradient. High specific ventilation should correspond with large tissue expansion.
Color-coded maps overlaid on a transverse slice showing (a) the Jacobian of transformation between PEEP 10 cm \(\mathrm{H}_2\mathrm{O}\) and 25 cm \(\mathrm{H}_2\mathrm{O}\) for one sheep subject, and (b) the corresponding specific ventilation (1/min) from the Xe-CT analysis. Blue and purple regions show larger deformation in a and higher ventilation in b, while red and orange regions show smaller deformation in a and lower ventilation in b
4 Application: Assessment of Lung Biomechanics
CT imaging of the lungs provides a new opportunity for assessment of lung function by non-rigid image registration of a pair of scans at different inflation levels. After finding out the transforms and correspondence for each voxel between two images, we are ready for motion estimation and mechanical analysis in a regional level. Post-analysis of the registration results reflects the mechanical properties of local lung tissue. The computed measurements from dense displacement fields can be used to estimate regional tissue motion, analyze regional pulmonary function and help diagnose lung diseases.
Measurements resulting from image registration reveal details of local tissue deformation patterns. Tissue motion can be quantified by parameters and index maps from:
-
Displacement field analysis, quantifying the magnitude and direction of local tissue movement;
-
Specific volume change and ventilation analysis, quantifying specific volume and specific air volume change through the deformation;
-
Strain analysis, quantifying the major deformation magnitude and direction;
-
Anisotropic deformation analysis, quantifying regional deformation patterns;
-
Shear stretch analysis, quantifying the lobar sliding.
4.1 Displacement Vector Field
The displacement fields can be directly used to assess the magnitude and direction of local volume movement. Figure 7.7 shows a 3D displacement filed computed from two 3D CT data sets, acquired at breath-hold maximum exhale and breath-hold maximum inhale. The two 3D CT images are registered using a Laplacian regularization and a combination of the squared tissue volume difference (SSTVD) and the squared vesselness measure difference (SSVMD) as distance measure (see Sects. 7.2.2.3, 7.2.2.4, and 7.2.3.1). The vector field gives the direction of tissue motion, and the length of vectors reflects the motion magnitude. Notice that regions near the diaphragm have larger tissue motions, and are moving downwards.
Figure 7.8 demonstrates how tissue movement may be tracked during respiratory cycles using 4DCT data. The 4DCT data sets were acquired using a Siemens Biograph 40-slice CT scanner operating in helical mode, and reconstructed at 0 %, 20 %, 40 %, 60 %, 80 %, 100 % during inspiration (noted as 0IN, 20IN, 40IN, 60IN, 80IN, 100IN), and 80 %, 60 %, 40 %, 20 % during expiration (noted as 80EX, 60EX, 40EX, 20EX). Again, a Laplacian regularization and \(C_{ SSTVD }\) combined with \(C_{ SSVMD }\) were used for the registration of the images acquired at different inflation levels. The tissue ROI is noted in Fig. 7.8a, and its motion is shown in the Y (dorsal-to-ventral) direction and Z (base-to-apex) direction in Fig. 7.8b. The motion of the ROI mainly occurred in the Z direction, and moved around 15 mm from 0IN to 100IN. During inspiration, its movement along Z direction in each interval was quite uniform (around 3 mm in each of the five intervals). During expiration, its backwards movement was much faster from 100IN to 80EX, and from 80EX to 60EX intervals (totally around 11 mm in the two intervals), while in the rest three intervals it only moved around 4 mm to its original position.
4.2 Specific Volume Change
Different measures can be defined to quantify lung tissue deformation patterns. Specific volume change (\({ sVol }\)) measures the volume change of local structure under deformation, and regions of local expansion or contraction can be identified. The most intuitive way to calculate \({ sVol }\) is derived through the Jacobian determinant of the transformation \(J({\varvec{T}})\). Using a Lagrangian reference frame, a Jacobian value of one corresponds to zero expansion or contraction. Local tissue expansion corresponds to a Jacobian greater than one and local tissue contraction corresponds to a Jacobian less than one. These indications of Jacobian are listed in Eq. (7.18).
As derived in Sect. 13.2 the specific volume change can be expressed as
Note that \({ sVol }\) is linearly related to the Jacobian and therefore it is possible to use the Jacobian measurement to reflect the specific volume change.
In Fig. 7.9, the specific volume change maps reflected by Jacobian show local volume deformation during exhalation stage. Since the CT images were acquired with subjects in the supine orientation, the more dependent region of lung is the dorsal region since it is closest to the direction of the force of gravity. Thus, there is more ventilation in the dorsal region. The maps also reflect the fact that vessels have little deformation during respiratory cycles while lung tissues and airways deform a lot.
4.3 Specific Ventilation
Specific ventilation measures the specific air volume change, and can be calculated from the intensity information of corresponding regions. More details about specific ventilation can be found in Chap. 13. The lung density is represented by CT grayscale in Hounsfield units (HU), which is defined such that the HU of water and air are 0 and -1000, respectively. Since the lung density decreases as it inflates with air, changes in the lung CT density during inflation can also be used to quantify regional mechanical properties. Therefore, given CT images of a lung region at two different pressures, it is possible to estimate the regional air volume change based on the HU values at corresponding regions. Assuming that the fraction of air in a region located at \({\varvec{x}}\) is given by \(F_{ air }({\varvec{x}}) = - I({\varvec{x}})/1000\), where \(I\) is the intensity function of a image, the specific ventilation sV can be calculated by [84]
A derivation of this equation and more details about specific ventilation can be found in Chap. 13.
Figure 7.10 shows an example specific ventilation map computed from images pair acquired at positive end-expiratory pressure (PEEP) of 10 cm \(\mathrm{H}_2\mathrm{O}\) and 15 cm \(\mathrm{H}_2\mathrm{O}\) for one sheep subject. During this experiment, the sheep was anesthetized, mechanically ventilated, and CT scans covering the thorax were acquired at 0, 5, 10, 15, 20, and 25 cm \(\mathrm{H}_2\mathrm{O}\) airway pressures. The consistent image registration method [21] was used to register the sheep data with different airway pressures. An obvious dorsal to ventral gradient is noticed from the \({ sV }\) map. This agrees with well known physiology that subjects positioned in the supine posture have more ventilation in the dorsal region.
4.4 Strain Tensors
In classical mechanics, deformation of structures is characterized by the regional distribution of a strain or stretch tensor. Some of the most common strain tensors, such as Linear strain tensor, Green-St. Venant strain tensor, and Eulerian-Almansi strain tensor are discussed in Chap. 4.
These strain tensors can be computed from the deformation field and used to analyze the stress caused by the geometrical deformation of the lung. In there most general form, the strain tensors are real symmetric matrices. Through singular value decomposition (SVD), they can be represented as a set of orthogonal eigenvectors, along which there is no shear, but only expansion or compression. These eigenvalues and eigenvectors are denoted as principal strains and principal directions. The maximal eigenvalue for each tensor is defined as maximal principal strain, and its corresponding eigenvector is called maximal principal direction. The principal strains and directions provide valuable information on preferential directionalities in deformation.
Figure 7.11 illustrates the maximal principal direction, maximal principal strain, and Jacobian together on coronal slice for one sheep subject when deforming images from PEEP 0 cm \(\mathrm{H}_2\mathrm{O}\) to PEEP 10 cm \(\mathrm{H}_2\mathrm{O}\). Strain tensors provide complementary information to the Jacobian for analyzing lung tissue expansion. The regions near diaphragm have larger maximal principal strain. The maximal principal strain on the lung boundaries are towards the chest wall.
Figure 7.12 shows maps of the maximal principal strain from linear strain, Green strain, and Eulerian strain tensors. Although they are of different ranges, regional patterns are similar among all three strain measures. The regions near lung boundaries and near the heart has larger maximal principal strains, while the dorsal regions have less maximal principal strains.
4.5 Anisotropy Analysis
Regional deformation of the lung during inspiration and expiration is more than just volume change. Volume change may also have orientational preference anisotropy of deformation [77, 97]. For instance, regions closest to the diaphragm are likely to experience more volume change in the vertical orientation. And regions closest to the heart may be more constrained from expanding normal to the heart. Volume change and deformation anisotropy are independent quantities as a region may undergo no volume change but still have deformed significantly, such as the case that the lengthening in one orientation is compensated by contraction along another orientation. Therefore, without orientational preference, regional volume change alone is not enough to characterize lung deformation.
4.5.1 Regional Stretch
In continuum mechanics, the deformation gradient tensor \(\mathbf{F }\) is the same as the Jacobian matrix \(\mathbf{J }\) of the transformation, which describes the continuum deformation from point-wise displacements, and it can be decomposed into stretch and rotation components:
where the \(\mathbf{U }\) is the right stretch tensor and \(\mathbf{R }\) is an orthogonal rotation tensor.
The Cauchy-Green deformation tensor is defined as
In order to obtain stretch information \(\mathbf{U }\) from this equation, it is first necessary to evaluate the principal directions of \(\mathbf{C }\), denoted here by the eigenvector \(N_1\), \(N_2\) and \(N_3\) and their corresponding eigenvalues \(\lambda _1^2\), \(\lambda _2^2\) and \(\lambda _3^2\). Therefore, after eigendecomposition and taking the square root of the eigenvalues of \(\mathbf{C }\), we can get the eigenvalues of \(\mathbf{U }\): \(\lambda _1\), \(\lambda _2\) and \(\lambda _3\) (ordered as \(\lambda _1 > \lambda _2 > \lambda _3\)). The eigenvalues of \(\mathbf{U }\) are defined as principal stretches and calculated as
4.5.2 Distortion Index (DI)
The ratio of the length in the direction of maximal stretch over the length in the direction of minimal stretch is defined as distortion index (DI)
The DI value is always larger or equal to 1. A big DI value indicates an anisotropic expansion, while a DI values approximately 1 represents an isotropic expansion.
Figure 7.13 gives an example of maps showing Jacobian, principal linear strain and distortion index from PEEP 0 cm \(\mathrm{H}_2\mathrm{O}\) to PEEP 10 cm \(\mathrm{H}_2\mathrm{O}\) registration for one sheep subject. Comparison between Jacobian and principal strain together with the distortion index (DI) map can reflect more lung tissue deformation information. For the region near the aorta in the left lung (black rectangular region), the Jacobian is large while the principal strain is relatively small. This illustrates that this region experienced an isotropic expansion, shown as red in the DI map (small DI value approximately 1) . For the region near the heart (red rectangular region), the Jacobian is near 1 while its principal strain is large. This illustrates that region experienced an anisotropic expansion, shown as purple in the DI map (larger DI value approximately 2). This anisotropic expansion may be caused by the blocking of the heart. Regional deformation is significantly anisotropic at the posterior end of lungs, but more isotropic at the anterior end.
Amelon et al. [3] has proposed another method to quantify the magnitude of anisotropy by defining the anisotropy deformation index (ADI)
It takes the three stretch factors into consideration, and thus has better discriminability among different anisotropy deformation patterns. This ADI calculation ranges from 0 to \(\infty \) where 0 indicates perfectly isotropic deformation.
4.6 Quantification of Lobar Sliding
The lung is divided into five lobes, three in the right lung and two in the left lung. It is thought that the lobes slide with respect to each other during breathing to reduce parenchymal distortion avoiding high stress concentrations [53]. Almost all methods of assessing lung function treat the lung as a single continuum, such as image registration [26, 44, 75, 88] or finite element simulation [1, 2, 37, 96], and fail to fully account for the discontinuity in the displacement field at the lobar fissures. The impact of neglecting discontinuities at the fissures is not understood as the degree of sliding in the lobes is relatively unaddressed in literature. There is value in quantifying the amount of lobar sliding if not for the sole purpose of analyzing its influence on current methods of assessing lung function.
Ding et al. quantified lobar sliding from lobe-by-lobe CT image registration by interpolating the displacement field on either side of the fissure to the fissure surface [33]. The difference in the displacement field represents the degree of sliding. Up to 20 mm of sliding was observed in the ventral portions of the fissure, increasing from nearly no sliding near carina. However, due to complexities in the algorithm only the left lung was considered since it contains only a single fissure. Other groups have also noted discontinuities in the displacement field along the fissures. For example, Cai et al. used grid-tagged MRI to obtain a displacement field within the lung [12]. They clearly demonstrate a discontinuity in the displacement field along fissure surfaces and note that discontinuities can be up to 20 mm.
Another approach to quantify lobar sliding is based on a finite element analysis of the displacement field using the previous defined stretch tensor \(\mathbf{U }\) [4]. A discontinuity in the displacement field will manifest itself as a region of elevated shear. If the amount of shear created due to sliding is much greater than the shear created by parenchymal distortion then shear can be considered a good indicator of relative sliding. Since large strain theory is used, we consider the shear components of the stretch tensor\(\mathbf{U }\). The eigenvalues of \(\mathbf{U }\) define the stretch in the principal directions of \(\mathbf{U }\) (see Eq. (7.22) and Sect. 7.4.5.1). It can be shown that the maximum shear component (denoted \(\gamma _{max}\)) at a point is half the difference between the maximum principal stretch and minimum principal stretch. Consider that \(\lambda _1 > \lambda _2 > \lambda _3\), the maximum shear is defined as
Figure 7.14 gives an example of maximum shear distribution from breath-hold maximum exhale to maximum inhale for one normal human subject. Figure 7.14a shows the distribution of maximum shear on a coronal slice. To obtain the image, each lobe and the chest wall are segmented and independently registered. The resulting displacement fields are then added to form a single image. In doing this, the discontinuity at the lobar boundaries is properly accounted for as no information outside of the respective lobe is included in the registration. Note that large sliding magnitudes are observed on the outer lung surface and lobar fissures. The sliding magnitude dissipates when moving medially along the fissure between the right upper lobe and right middle lobe. Figure 7.14b shows the 3D distribution of maximum shear within the lung region. Higher shear stretch (higher sliding) is observed in the dorsal region, while less sliding is observed near the carina.
5 Summary
Image registration can be used to determine the spatial locations of corresponding voxels in a sequence of pulmonary scans. The computed correspondences yield the displacement fields corresponding with the motion of the lung between a pair of images. In this chapter, we discussed a method of intensity-based registration designed to match lung images. Since image registration is inherently an ill-posed problem due to the fact that determining the unknown the displacements merely from the images is an underdetermined problem, it is necessary to validate registration methods in the context of physiologically plausible results. We provided several methods for validating lung registration performance. Accurate registrations can represent the underlying anatomical and physiological changes of lung tissue, and thus make post-analysis on motion estimation and lung mechanics meaningful. We also provided a systematic study of image registration based measurements of the regional lung mechanics and motion patterns. Local volume change, major deformation magnitude and direction, anisotropy deformation pattern, and shear stretch on lobar fissures and lung surface against chest wall were discussed. These measurements enable estimation of tissue motion and assessment of lung function at a regional level.
Notes
- 1.
However, we need to mention that this is not fully true due to the increased blood volume through inhalation [43].
References
Al-Mayah, A., Moseley, J., Velec, M., Brock, K.K.: Sliding characteristic and material compressibility of human lung: parametric study and verification. Med. Phys. 36(10), 4625–4633 (2009)
Al-Mayah, A., Moseley, J., Velec, M., Hunter, S., Brock, K.: Deformable image registration of heterogeneous human lung incorporating the bronchial tree. Med. Phys. 37(9), 4560–4571 (2010)
Amelon, R., Cao, K., Ding, K., Christensen, G.E., Reinhardt, J.M., Raghavan, M.L.: Three-dimensional characterization of regional lung deformation. J. Biomech. 44(13), 2489–2495 (2011)
Amelon, R., Cao, K., Reinhardt, J.M., Christensen, G.E., Raghavan, M.: Estimation of lung lobar sliding using image registration. Proc. SPIE 8317, 83171H (2012).
Amit, Y.: A non-linear variational problem for image matching. SIAM J. Sci. Comput. 15(1), 207–224 (1994)
Ashburner, J., Andersson, J., Friston, K.: High-dimensional image registration using symmetric priors. NeuroImage 9, 619–628 (1999)
Avants, B.B., Tustison, N.J., Song, G., Cook, P.A., Klein, A., Gee, J.C.: A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54(3), 2033–2044 (2011)
Bookstein, F.L.: Principal warps: thin-plate splines and the decomposition of deformations. IEEE Trans. Pattern Anal. Mach. Intell. 11(6), 567–585 (1989)
Broit, C.: Optimal registration of deformed images. Ph.D. Thesis, University of Pennsylvania, Philadelphia, PA, USA (1981)
Buck, R.C.: Advanced Calculus, 3rd edn. McGraw-Hill, St. Louis (1978)
Byrd, R.H., Lu, P., Nocedal, J., Zhu, C.: A limited memory algorithm for bound constrained optimization. SIAM J. Sci. Comput. 16(5), 1190–1208 (1995)
Cai, J., Sheng, K., Benedict, S.H., Read, P.W., Larner, J.M., Mugler, J.P., III, de Lange, E.E., Cates, G.D., Jr., Miller, G.W.: Dynamic MRI of grid-tagged hyperpolarized Helium-3 for the assessment of lung motion during breathing. Int. J. Radiat. Oncol. Biol. Phys. 75(1), 276–284 (2009)
Cao, K., Ding, K., Christense, G.E., Reinhardt, J.M.: Tissue volume and vesselness measure preserving nonrigid registration of lung CT images. Proc. SPIE 7623, 762309 (2010)
Cao, K., Ding, K., Christensen, G.E., Raghavan, M.L., Amelon, R.E., Reinhardt, J.M.: Unifying vascular information in intensity-based nonrigid lung CT registration. In: 4th International Workshop on Biomedical Image Registration, LCNS 6204, pp. 1–12. Springer, Berlin (2010)
Cao, K., Du, K., Ding, K., Reinhardt, J.M., Christensen, G.E.: Regularized nonrigid registration of lung CT images by preserving tissue volume and vesselness measure. In: Grand Challenges in Medical Image Analysis (2010)
Castillo, E., Castillo, R., Martinez, J., Shenoy, M., Guerrero, T.: Four-dimensional deformable image registration using trajectory modeling. Phys. Med. Biol. 55(1), 305 (2010)
Castillo, E., Castillo, R., Zhang, Y., Guerrero, T.: Compressible image registration for thoracic computed tomography images. J. Med. Biol. Eng. 29(5), 222-233 (2009)
Choi, Y., Lee, S.: Injectivity conditions of 2D and 3D uniform cubic B-spline functions. Graph. Models 62(6), 411–427 (2000)
Christensen, G.E.: Deformable shape models for anatomy. Ph.D. Thesis, Department of Electrical Engineering, Sever Institute of Technology, Washington University, St. Louis, MO 63130 (1994)
Christensen, G.E.: Consistent linear-elastic transformations for image matching. In: Kuba, A., Samal, M. (eds.) Information Processing in Medical Imaging, LCNS 1613, pp. 224–237. Springer, Berlin (1999)
Christensen, G.E., Johnson, H.J.: Consistent image registration. IEEE Trans. Med. Imaging 20(7), 568–582 (2001)
Christensen, G.E., Joshi, S.C., Miller, M.I.: Volumetric transformation of brain anatomy. IEEE Trans. Med. Imaging 16(6), 864–877 (1997)
Christensen, G.E., Rabbitt, R.D., Miller, M.I.: 3D brain mapping using a deformable neuroanatomy. Phys. Med. Biol. 39, 609–618 (1994)
Christensen, G.E., Rabbitt, R.D., Miller, M.I.: Deformable templates using large deformation kinematics. IEEE Trans. Image Proc. 5(10), 1435–1447 (1996)
Christensen, G.E., Rabbitt, R.D., Miller, M.I., Joshi, S., Grenander, U., Coogan, T., Essen, D.V.: Topological properties of smooth anatomic maps. In: Bizais, Y., Braillot, C., Paola, R.D. (eds.) Information Processing in Medical Imaging, vol. 3, pp. 101–112. Kluwer Academic, Boston (1995)
Christensen, G.E., Song, J.H., Lu, W., Naqa, I.E., Low, D.A.: Tracking lung tissue motion and expansion/compression with inverse consistent image registration and spirometry. Med. Phys. 34(6), 2155–2165 (2007)
Collignon, A., Maes, F., Delaere, D., Vandermeulen, D., Suetens, P., Marchal, G.: Automated multi-modality image registration based on information theory. In: Bizais, Y., Braillot, C., Paola, R.D. (eds.) Information Processing in Medical Imaging, vol. 3, pp. 263–274. Kluwer Academic, Boston (1995)
Cook, T.S., Tustison, N., Biederer, J., Tetzlaff, R., Gee, J.: How do registration parameters affect quantitation of lung kinematics? In: Proceedings of the 10th International Conference on Medical Image Computing and Computer-Assisted Intervention—MICCAI’07, vol. Part I, pp. 817–824. Springer, Berlin (2007)
Davis, M.H., Khotanzad, A., Flamig, D.P., Harms, S.E.: Elastic body splines: a physics based approach to coordinate transformation in medical image matching. In: Proceedings of the Eighth Annual IEEE Symposium on Computer-Based Medical Systems, CBMS ’95, pp. 81–88. IEEE Computer Society, Washington, DC, USA (1995)
Delmon, V., Rit, S., Pinho, R., Sarrut, D.: Direction dependent B-splines decomposition for the registration of sliding objects. In: Proceedings of the Fourth International Workshop on Pulmonary Image Analysis, pp. 45–55, Toronto, Canada (2011)
Ding, K., Bayouth, J.E., Buatti, J.M., Christensen, G.E., Reinhardt, J.M.: 4DCT-based measurement of changes in pulmonary function following a course of radiation therapy. Med. Phys. 37(3), 1261–1272 (2010)
Ding, K., Cao, K., Christensen, G.E., Hoffman, E.A., Reinhardt, J.M.: Registration-based regional lung mechanical analysis: retrospectively reconstructed dynamic imaging versus static breath-hold image acquisition. In: Proceedings of SPIE Conference on Medical Imaging, vol. 7262, p. 72620D (2009)
Ding, K., Yin, Y., Cao, K., Christensen, G.E., Lin, C.L., Hoffman, E.A., Reinhardt, J.M.: Evaluation of lobar biomechanics during respiration using image registration. In: Proceedings of International Conference on Medical Image Computing and Computer-Assisted Intervention 2009, vol. 5761, pp. 739–746 (2009)
Dunlap, N., McIntosh, A., Sheng, K., Yang, W., Turner, B., Shoushtari, A., Sheehan, J., Jones, D.R., Lu, W., Ruchala, K., Olivera, G., Parnell, D., Larner, J.L., Benedict, S.H., Read, P.W.: Helical tomotherapy-based STAT stereotactic body radiation therapy: dosimetric evaluation for a real-time SBRT treatment planning and delivery program. Med. Dosim. 35(4), 312–319 (2010)
Ehrhardt, J., Werner, R., Schmidt-Richberg, A., Handels, H.: Statistical modeling of 4D respiratory lung motion using diffeomorphic image registration. IEEE Trans. Med. Imaging 30(2), 251–265 (2011) (in press)
Enquobahrie, A., Ibanez, L., Bullitt, E., Aylward, S.: Vessel enhancing diffusion filter. Insight J (2007)
Eom, J., Shi, C., Xu, X., De, S.: Modeling respiratory motion for cancer radiation therapy based on patient-specific 4DCT data. In: Medical Imaging Computing and Computer Assisted Intervention, Lecture Notes in Computer Science, vol. 12, pp. 348–355 (2009)
Ferrant, M., Warfield, S.K., Nabavi, A., Jolesz, F.A., Kikinis, R.: Registration of 3D intraoperative MR images of the brain using a finite element biomechanical model. In: MICCAI ’00: Proceedings of the Third International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 19–28. Springer, London (2000)
Fitzpatrick, J., Hill, D., Maurer, C.: Image registration. In: Sonka, M., Fitzpatrick, J. (eds.) Handbook of Medical Imaging, vol. 2, chap. 8, pp. 447–513. SPIE Press, San Diego (2000)
Frangi, A.F., Niessen, W.J., Vincken, K.L., Viergever, M.A.: Multiscale vessel enhancement filtering. In: MICCAI, vol. 1496, pp. 130–137 (1998)
Gee, J.C., Sundaram, T., Hasegawa, I., Uematsu, H., Hatabu, H.: Characterization of regional pulmonary mechanics from serial MRI data. In: Proceedings of the 5th International Conference on Medical Image Computing and Computer-Assisted Intervention-Part I, MICCAI ’02, pp. 762–769. Springer, London (2002)
Gorbunova, V., Lo, P., Ashraf, H., Dirksen, A., Nielsen, M., de Bruijne, M.: Weight preserving image registration for monitoring disease progression in lung CT. In: MICCAI, vol. 5242, pp. 863–870 (2008)
Guerrero, T., Sanders, K., Castillo, E., Zhang, Y., Bidaut, L., Pan, T., Komaki, R.: Dynamic ventilation imaging from four-dimensional computed tomography. Phys. Med. Biol. 51(4), 777–791 (2006)
Guerrero, T., Sanders, K., Noyola-Martinez, J., Castillo, E., Zhang, Y., Tapia, R., Guerra, R., Borghero, Y., Komaki, R.: Quantification of regional ventilation from treatment planning CT. Int. J. Radiat. Oncol. Biol. Phys. 62(3), 630–634 (2005)
Guo, J., Fuld, M.K., Alford, S.K., Reinhardt, J.M., Hoffman, E.A.: Pulmonary analysis software suite 9.0: Integrating quantitative measures of function with structural analyses. In: First International Workshop on Pulmonary Image Analysis, pp. 283–292, New York (2008)
Hajnal, J.V.: Medical Image Registration (Biomedical Engineering), 1st edn. CRC Press, Cambridge (2001)
Han, X.: Feature-constrained nonlinear registration of lung CT images. In: Grand Challenges in Medical Image Analysis (2010)
Hermosillo, G., Chefd’Hotel, C., Faugeras, O.: Variational methods for multimodal image matching. Int. J. Comput. Vision 50, 329–343 (2002)
Hoffman, E.A., Ritman, E.L.: Effect of body orientation on regional lung expansion in dog and sloth. J. Appl. Physiol. 59(2), 481–491 (1985)
Holden, M., Hill, D., Denton, E., Jarosz, J., Cox, T., Rohlfing, T., Goodey, J., Hawkes, D.: Voxel similarity measures for 3-D serial MR brain image registration. IEEE Trans. Med. Imaging 19, 94–102 (2000)
Horn, B.K., Schunck, B.G.: Determining Optical Flow. Artif. Intell. 17, 185–203 (1981)
Hu, S., Hoffman, E.A., Reinhardt, J.M.: Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans. Med. Imaging 20, 490–498 (2001)
Hubmayr, R.D., Rodarte, J.R., Walters, B.J., Tonelli, F.M.: Regional ventilation during spontaneous breathing and mechanical ventilation in dogs. J. Appl. Physiol. 63(6), 2467–2475 (1987)
ICRU Report: Prescribing, recording and reporting photon beam therapy. ICRU, Bethesda, MD (1999). Supplement to ICRU report 50
Kabus, S., Lorenz, C.: Fast elastic image registration. In: Grand Challenges in Medical Image Analysis (2010)
Klein, S., Staring, M., Pluim, J.P.: Evaluation of optimization methods for nonrigid medical image registration using mutual information and B-splines. IEEE Trans. Image Process. 16(12), 2879–2890 (2007)
Klinder, T., Lorenz, C., Ostermann, J.: Free-breathing intra- and intersubject respiratory motion capturing, modeling, and prediction. In: SPIE Medical Imaging 2009: Image Processing, vol. 7259 (2009)
Knowlton, R.: Clinical applications of image registration, Handbook of Medical Imaging, pp. 613–621. Academic Press, Orlando (2000)
Lester, H., Arridge, S.: A survey of hierarchical non-linear medical image registration. Med. Image Anal. 32(1), 129–149 (1999)
Maintz, J., Viergever, M.: A survey of medical image registration. Med. Image Anal. 2(1), 1–36 (1998)
Mattes, D., Haynor, D., Vesselle, H., Lewellen, T., Eubank, W.: PET-CT image registration in the chest using free-form deformations. IEEE Trans. Med. Imaging 22(1), 120–128 (2003)
Metz, C., Klein, S., Schaap, M., van Walsum, T., Niessen, W.: Nonrigid registration of dynamic medical imaging data using \(\text{ nD }+\text{ tB }\)-splines and a groupwise optimization approach. Med. Image Anal. 15(2), 238–249 (2011)
Miller, M.I., Banerjee, A., Christensen, G.E., Joshi, S.C., Khaneja, N., Grenander, U., Matejic, L.: Statistical methods in computational anatomy. Stat. Methods Med. Res. 6, 267–299 (1997)
Modat, M., McClelland, J., Ourselin, S.: Lung registration using the Niftyreg package. In: Grand Challenges in Medical Image Analysis (2010)
Muenzing, S.E.A., van Ginneken, B., Pluim, J.P.W.: Knowledge driven regularization of the deformation field for PDE based non-rigid registration algorithms. In: Grand Challenges in Medical Image Analysis (2010)
Murphy, K., van Ginneken, B., Reinhardt, J.M., et al.: Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE Trans. Med. Imaging 30(11), 1901–1920 (2011)
Murphy, K., van Ginneken, B., Pluim, J., Klein, S., Staring, M.: Semi-automatic reference standard construction for quantitative evaluation of lung CT registration. In: MICCAI, vol. 5242, pp. 1006–1013 (2008)
Nocedal, J.: Updating quasi-Newton matrices with limited storage. Math. Comput. 35(151), 773–782 (1980)
Nocedal, J., Wright, S.J. (eds.): Numerical Optimization. Springer, New York (1999)
Ourselin, S., Roche, A., Subsol, G., Pennec, X., Ayache, N.: Reconstructing a 3D Structure from Serial Histological Sections. Image Vision Comput. 19(1–2), 25–31 (2000)
Pace, D., Enquobahrie, A., Yang, H., Aylward, S., Niethammer, M.: Deformable image registration of sliding organs using anisotropic diffusive regularization. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2011, pp. 407–413 (2011)
Penney, G., Weese, J., Little, J., Desmedt, P., Hill, D., Hawkes, D.: A comparison of similarity measures for use in 2-d-3-d medical image registration. IEEE Trans. Med. Imaging 17, 586–595 (1998)
Press, W.H., Teukolsky, S.A., Vetterling, W.T., Flannery, B.P.: Numerical recipes in C: the art of scientific computing, 2nd edn. Cambridge University Press, New York (1992)
Reinhardt, J.M., Ding, K., Cao, K., Christensen, G.E., Hoffman, E.A., Bodas, S.V.: Registration-based estimates of local lung tissue expansion compared to xenon-CT measures of specific ventilation. Med. Image Anal. 12(6), 752–763 (2008)
Reinhardt, J.M., Ding, K., Cao, K., Christensen, G.E., Hoffman, E.A., Bodas, S.V.: Registration-based estimates of local lung tissue expansion compared to xenon CT measures of specific ventilation. Med. Image Anal. 12(6), 752–763 (2008)
Rikxoort, E.M., Prokop, M., Hoop, B., Viergever, M.A., Pluim, J.P., Ginneken, B.: Automatic segmentation of the pulmonary lobes from fissures, airways, and lung borders: evaluation of robustness against missing data. In: Proceedings of the 12th International Conference on Medical Image Computing and Computer-Assisted Intervention: Part I, MICCAI ’09, pp. 263–271. Springer, Berlin (2009)
Rodarte, J.R., Hubmayr, R.D., Stamenovic, D., Walters, B.J.: Regional lung strain in dogs during deflation from total lung capacity. J. Appl. Physiol. 58(1), 164–172 (1985)
Ruan, D., Esedoglu, S., Fessler, J.: Discriminative sliding preserving regularization in medical image registration. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2009. ISBI ’09, pp. 430–433 (2009)
Rueckert, D., Sonoda, L., Hayes, C., Hill, D., Leach, M., Hawkes, D.: Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18(8), 712–721 (1999)
Ruhaak, J., Heldmann, S.: Variational lung registration with explicit boundary alignment. In: Grand Challenges in Medical Image Analysis (2010)
Sarrut, D., Boldea, V., Miguet, S., Ginestet, C.: Simulation of four-dimensional CT images from deformable registration between inhale and exhale breath-hold CT scans. Med. Phys. 33(3), 605–617 (2006)
Schmidt-Richberg, A., Ehrhardt, J., Werner, R., Handels, H.: Diffeomorphic diffusion registration of lung CT images. In: Grand Challenges in Medical Image Analysis (2010)
Schmidt-Richberg, A., Werner, R., Handels, H., Ehrhardt, J.: Estimation of slipping organ motion by registration with direction-dependent regularization. Med. Image Anal. 16(1), 150–159 (2012)
Simon, B.: Non-invasive imaging of regional lung function using X-ray computed tomography. J. Clin. Monit. Comput. 16(5–6), 433–442 (2000)
Simon, B.A., Marcucci, C.: Parameter estimation and confidence intervals for Xe-CT ventilation studies: a Monte Carlo approach. JAP 84(2), 709–716 (1998)
Song, G., Tustison, N., Avants, B., Gee, J.C.: Lung ct image registration using diffeomorphic transformation models. In: Grand Challenges in Medical Image Analysis (2010)
Staring, M., Klein, S., Reiber, J.H.C., Niessen, W.J., Stoel, B.C.: Pulmonary image registration with elastix using a standard intensity-based algorithm. In: Grand Challenges in Medical Image Analysis (2010)
Sundaram, T.A., Gee, J.C.: Towards a model of lung biomechanics: pulmonary kinematics via registration of serial lung images. Med. Image Anal. 9(6), 524–537 (2005)
Sundaram, T.A., Gee, J.C.: Quantitative comparison of registration-based lung motion estimates from whole-lung mr images and corresponding two-dimensional slices. In: Proceedings of ISMRM 15th Meeting, p. 3039 (2007)
Szeliski, R., Coughlan, J.: Spline-based image registration. Int. J. Comput. Vision 22(3), 199–218 (1997)
Thevenaz, P., Unser, M.: Spline pyramids for inter-modal image registration using mutual information. In. Proc. SPIE 3169, 236–247 (1997)
Ukil, S., Reinhardt, J.M.: Anatomy-guided lung lobar surface detection in X-ray CT images. IEEE Trans. Med. Imaging 28(2), 202–214 (2009)
Vandemeulebroucke, J., Rit, S., Kybic, J., Clarysse, P., Sarrut, D.: Spatiotemporal motion estimation for respiratory-correlated imaging of the lungs. Med. Phys. 38(1), 166–178 (2011)
Viola, P., III, Wells, M.W.: Alignment by maximization of mutual information. Int. J. Comput. Vision 24(2), 137–154 (1997)
Wang, H., Garden, A.S., Zhang, L., Wei, X., Ahamad, A., Kuban, D.A., Komaki, R., O’Daniel, J., Zhang, Y., Mohan, R., Dong, L.: Performance evaluation of automatic anatomy segmentation algorithm on repeat or four-dimensional computed tomography images using deformable image registration method. Int. J. Radiat. Oncol. Biol. Phys. 72(1), 210–219 (2008)
Werner, R., Ehrhardt, J., Schmidt, R., Handels, H.: Patient-specific finite element modeling of respiratory lung motion using 4D CT image data. Med. Phys. 36(5), 1500–1511 (2009)
West, J.B., Matthews, F.L.: Stress, strain, and surface pressures in the lung caused by its weight. J. Appl. Physiol. 32(3), 332–345 (1972)
Wikipedia: Histogram matching. Hypertext Document. http://en.wikipedia.org/wiki/Histogram_matching
Wu, G., Wang, Q., Lian, J., Shen, D.: Estimating the 4d respiratory lung motion by spatiotemporal registration and building super-resolution image. In: Proceedings of the 14th International Conference on Medical Image Computing and Computer-Assisted Intervention—vol. Part I, MICCAI’11, pp. 532–539 (2011)
Yang, D., Brame, S., Naqa, I.E., Aditya, A., Wu, Y., Goddu, S.M., Mutic, S., Deasy, J.O., Low, D.A.: Technical note: DIRART—a software suite for deformable image registration and adaptive radiotherapy research. Med. Phys. 38(1), 67–77 (2011)
Yin, Y., Hoffman, E.A., Lin, C.L.: Local tissue-weight-based nonrigid registration of lung images with application to regional ventilation. In: Hu, X.P., Clough, A.V. (eds.) SPIE Medical Imaging 2009: Biomedical Applications in Molecular, Structural, and Functional Imaging, vol. 7262, p. 72620C, Lake Buena Vista, FL (2009)
Yin, Y., Hoffman, E.A., Lin, C.L.: Mass preserving non-rigid registration of CT lung images using cubic B-spline. Med. Phys. 36(9), 4213–4222 (2009)
Younes, L., Arrate, F., Miller, M.I.: Evolutions equations in computational anatomy. NeuroImage 45(1 Suppl), S40–S50 (2009)
Zhu, C., Byrd, R.H., Lu, P., Nocedal, J.: Algorithm 778: L-BFGS-B: Fortran subroutines for large-scale bound-constrained optimization. ACM Trans. Math. Softw. 23(4), 550–560 (1997)
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Cao, K. et al. (2013). Intensity-Based Registration for Lung Motion Estimation. In: Ehrhardt, J., Lorenz, C. (eds) 4D Modeling and Estimation of Respiratory Motion for Radiation Therapy. Biological and Medical Physics, Biomedical Engineering. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-36441-9_7
Download citation
DOI: https://doi.org/10.1007/978-3-642-36441-9_7
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-36440-2
Online ISBN: 978-3-642-36441-9
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)