Abstract
Information processes in the brain, such as gene and protein expression, learning, memory, perception, cognition, consciousness are all spatio- and/or spectro temporal. Modelling such processes would require sophisticated information science methods and the best ones could be the brain-inspired ones, that use the same brain information processing principles. Spatio and spectro-temporal data (SSTD) are also the most common types of data collected in many domain areas, including engineering, bioinformatics, neuroinformatics, ecology, environment, medicine, economics, etc. However, there is lack of methods for the efficient analysis of such data and for spatio-temporal pattern recognition (STPR). The brain functions as a spatio-temporal information processing machine and deals extremely well with spatio-temporal data. Its organization and functions have been the inspiration for the development of new methods for SSTD analysis and STPR. Brain-inspired spiking neural networks (SNN) are considered the third generation of neural networks and are a promising paradigm for the creation of new intelligent ICT for SSTD. This new generation of computational models and systems is potentially capable of modeling complex information processes due to the ability to represent and integrate different information dimensions, such as time, space, frequency, and phase, and to deal with large volumes of data in an adaptive and self-organizing manner. This chapter reviews methods and systems of SNN for SSTD analysis and STPR, including single neuronal models, evolving spiking neural networks (eSNN), and computational neurogenetic models (CNGM). Software and hardware implementations and some pilot applications for audio-visual pattern recognition, EEG data-analysis, cognitive robotic systems, BCI, neurodegenerative diseases, and others are discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Spatio and Spectro-Temporal Data Modeling and Pattern Recognition
Most problems in nature require spatio or/and spectro-temporal data (SSTD) that include measuring spatial or/and spectral variables over time. SSTD is described by a triplet (X, Y, F), where X is a set of independent variables measured over consecutive discrete time moments t, Y is the set of dependent output variables, and F is the association function between whole segments (chunks) of the input data, each sampled in a time window d t, and the output variables belonging to Y
where
It is important for a computational model to capture and learn whole spatio and spectro-temporal patterns from data streams in order to predict most accurately future events for new input data. Examples of problems involving SSTD are: brain cognitive state evaluation based on spatially distributed EEG electrodes [47.3,4,5,6,7,8] (Fig. 47.1a); fMRI data [47.9] (Fig. 47.1b); moving object recognition from video data [47.10,11,12] (Fig. 47.15); spoken word recognition based on spectro-temporal audio data [47.13,14]; evaluating risk of disease, e.g., heart attack [47.15]; evaluating response of a disease to treatment based on clinical and environmental variables, e.g., stroke [47.16], prognosis of outcome of cancer [47.17], modeling the progression of a neurodegenerative disease, such as Alzheimerʼs disease [47.18,19], and modeling and prognosis of the establishment of invasive species in ecology [47.20,21]. The prediction of events in geology, astronomy, economics and many other areas also depends on accurate SSTD modeling.
Moving object recognition with the use of AER. (a) Disparity map of a video sample; (b) address event representation (AER) of the above video sample (after [47.10])
The commonly used models for dealing with temporal information based on hidden Markov models (HMM) [47.22] and traditional artificial neural networks (ANN) [47.23] have limited capacity to achieve the integration of complex and long temporal spatial/spectral components because they usually either ignore the temporal dimension or over-simplify its representation. A new trend in machine learning is currently emerging and is known as deep machine learning [47.24,24,25,26,27]. Most of the proposed models still learn SSTD by entering single time point frames rather than learning whole SSTD patterns. They are also limited in addressing adequately the interaction between temporal and spatial components in SSTD.
The human brain has the amazing capacity to learn and recall patterns from SSTD at different time scales, ranging from milliseconds to years and possibly to millions of years (e.g., genetic information, accumulated through evolution). Thus the brain is the ultimate inspiration for the development of new machine learning techniques for SSTD modeling. Indeed, brain-inspired spiking neural networks (SNN) [47.28,29,30] have the potential to learn SSTD by using trains of spikes (binary temporal events) transmitted among spatially located synapses and neurons. Both spatial and temporal information can be encoded in an SNN as locations of synapses and neurons and the time of their spiking activity, respectively. Spiking neurons send spikes via connections that have a complex dynamic behavior, collectively forming an SSTD memory. Some SNN employ specific learning rules such as spike-time-dependent-plasticity (STDP) [47.31] or spike driven synaptic plasticity (SDSP) [47.32]. According to the STDP a connection weight between two neurons increases when the pre-synaptic neuron spikes before the post-synaptic one. Otherwise, the weight decreases.
Models of single neurons as well as computational SNN models, along with their respective applications, have been already developed [47.29,30,33,34,35,36], including evolving connectionist systems and evolving spiking neural networks (eSNN), in particular where an SNN learns data incrementally by one-pass propagation of the data via creating and merging spiking neurons [47.37,38]. In [47.38] an eSNN is designed to capture features and to aggregate them into audio and visual perceptions for the purpose of person authentication. It is based on four levels of feed-forward connected layers of spiking neuronal maps, similarly to the way the cortex works when learning and recognizing images or complex input stimuli [47.39]. It is an SNN realization of some computational models of vision, such as the five-level HMAX model inspired by the information processes in the cortex [47.39].
However, these models are designed for (static) object recognition (e.g., a picture of a cat), but not for moving object recognition (e.g., a cat jumping to catch a mouse). If these models are to be used for SSTD, they will still process SSTD as a sequence of static feature vectors extracted in single time frames. Although an eSNN accumulates incoming information carried in each consecutive frame from a pronounced word or a video, through the increase of the membrane potential of output spike neurons, they do not learn complex spatio/spectro-temporal associations from the data. Most of these models are deterministic and do not allow to model complex stochastic SSTD.
In [47.40,41] a computational neurogenetic model (CNGM) of a single neuron and SNN are presented that utilize information about how some proteins and genes affect the spiking activities of a neuron, such as fast excitation, fast inhibition, slow excitation, and slow inhibition. An important part of a CNGM is a dynamic gene regulatory network (GRN) model of genes/proteins and their interaction over time that affect the spiking activity of the neurons in the SNN. Depending on the task, the genes in a GRN can represent either biological genes and proteins (for biological applications) or some system parameters including probability parameters (for engineering applications). Recently some new techniques have been developed that allow the creation of new types of computational models, e.g., probabilistic spiking neuron models [47.42,43], probabilistic optimization of features and parameters of eSNN [47.21,44], reservoir computing [47.36,45], and personalized modeling frameworks [47.46,47]. This chapter reviews methods and systems for SSTD that utilize the above and some other contemporary SNN techniques along with their applications.
Single Spiking Neuron Models
A Biological Neuron
A single biological neuron and the associated synapses is a complex information processing machine, that involves short-term information processing, long-term information storage, and evolutionary information stored as genes in the nucleus of the neuron (Fig. 47.2).
Single Neuron Models
Some of the-state-of-the-art models of a spiking neuron include: early models by Hodgkin and Huxley [47.48]; more recent models by Maas, Gerstner, Kistler, Izhikevich and others, e.g., spike response models (SRM) [47.29,30]; the integrate-and-fire model (IFM) [47.29,30]; Izhikevich models [47.49,50,51,52], adaptive IFM, and others.
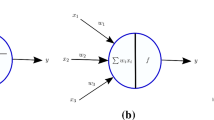
The most popular model for both biological modeling and engineering applications is the IFM. The IFM has been realized on software-hardware platforms for the exploration of patterns of activities in large scale SNN under different conditions and for different applications. Several large scale architectures of SNN using IFM have been developed for modeling brain cognitive functions and engineering applications. Figure 47.3a,b illustrates the structure and the functionality of the leaky IFM (LIFM), respectively. The neuronal post-synaptic potential (PSP), also called membrane potential u(t), increases with every input spike at a time t multiplied to the synaptic efficacy (strength) until it reaches a threshold. After that, an output spike is emitted and the membrane potential is reset to an initial state (e.g., 0). Between spikes, the membrane potential leaks, which is defined by a parameter.
An important part of a model of a neuron is the model of the synapses. Most neuronal models assume scalar synaptic efficacy parameters that are subject to learning, either on-line or off-line (batch mode). There are models of dynamics synapses (e.g., [47.43,53,54]), where the synaptic efficacy depends on synaptic parameters that change over time, representing both long-term memory (the final efficacy after learning) and short-term memory – the changes of the synaptic efficacy over a shorter time period not only during learning, but during recall as well.
One generalization of LIFM and the dynamic synaptic models is the probabilistic model of a neuron [47.42] as shown in Fig. 47.4a, which is also a biologically plausible model [47.30,43,55]. The state of a spiking neuron n i is described by the sum PSP i (t) of the inputs received from all m synapses. When PSP i (t) reaches a firing threshold ϑ i (t), neuron n i fires, i.e., it emits a spike. Connection weights (w j,i , j = 1, 2,… , m) associated with the synapses are determined during the learning phase using a learning rule. In addition to the connection weights w j,i (t), the probabilistic spiking neuron model has the following three probabilistic parameters:
-
1.
A probability p cj,i (t) that a spike emitted by neuron n j will reach neuron n i at a time moment t through the connection between n j and n i . If p cj,i (t) = 0, no connection and no spike propagation exist between neurons n j and n i . If p cj,i (t) = 1 the probability for propagation of spikes is 100%.
-
2.
A probability p sj,i (t) for the synapse s j,i to contribute to the PSP i (t) after it has received a spike from neuron n j .
-
3.
A probability p i (t) for the neuron n i to emit an output spike at time t once the total PSP i (t) has reached a value above the PSP threshold (a noisy threshold).
The total PSP i (t) of the probabilistic spiking neuron n i is now calculated using the following formula [47.42]
where e j is 1, if a spike has been emitted from neuron n j , and 0 otherwise; f 1(p cj,i (t)) is 1 with a probability p cji (t), and 0 otherwise; f 2(p sj,i (t)) is 1 with a probability p sj,i (t), and 0 otherwise; t 0 is the time of the last spike emitted by n i ; and η(t − t 0) is an additional term representing decay in PSP i . As a special case, when all or some of the probability parameters are fixed to ``1ʼʼ, the above probabilistic model will be simplified and will resemble the well-known IFM. A similar formula will be used when an LIFM is used as a fundamental model, where a time decay parameter is introduced.
It has been demonstrated that SNN that utilize the probabilistic neuronal model can learn better SSTD than traditional SNN with simple IFM, especially in a nosy environment [47.56,57]. The effect of each of the above three probabilistic parameters on the ability of a SNN to process noisy and stochastic information was studied in [47.56]. Figure 47.4b presents the effect of different types of nosy thresholds on the neuronal spiking activity.
A Neurogenetic Model of a Neuron
A neurogenetic model of a neuron was proposed in [47.41] and studied in [47.40]. It utilizes information about how some proteins and genes affect the spiking activities of a neuron such as fast excitation, fast inhibition, slow excitation, and slow inhibition. Table 47.1 shows some of the proteins in a neuron and their relation to different spiking activities. For a real case application, apart from the GABAB receptor some other metabotropic and other receptors could be also included. This information is used to calculate the contribution of each of the different synapses, connected to a neuron n i , to its post-synaptic potential PSP i (t)
where τ decay/rise synapse are time constants representing the rise and fall of an individual synaptic PSP, A is PSPʼs amplitude, ε ij synapse represents the type of activity of the synapse between neuron j and neuron i that can be measured and modeled separately for a fast excitation, fast inhibition, slow excitation, and slow inhibition (it is affected by different genes/proteins). External inputs can also be added to model background noise, background oscillations or environmental information.
An important part of the model is a dynamic gene/protein regulatory network (GRN) model of the dynamic interactions between genes/proteins over time that affect the spiking activity of the neuron. Although biologically plausible, a GRN model is only a highly simplified general model that does not necessarily take into account the exact chemical and molecular interactions. A GRN model is defined by:
-
1.
A set of genes/proteins, G = (g 1, g 2,… , g k )
-
2.
An initial state of the level of expression of the genes/proteins G(t = 0)
-
3.
An initial state of a connection matrix L = (L 11,… , L kk ), where each element L ij defines the known level of interaction (if any) between genes/proteins g j and g i
-
4.
Activation functions f i for each gene/protein g i from G. This function defines the gene/protein expression value at time (t + 1) depending on the current values G(t), L(t) and some external information E(t)
(47.4)
Learning and Memory in a Spiking Neuron
General Classification
A learning process has an effect on the synaptic efficacy of the synapses connected to a spiking neuron and on the information that is memorized. Memory can be:
-
1.
Short-term, represented as a changing PSP and temporarily changing synaptic efficacy
-
2.
Long-term, represented as a stable establishment of the synaptic efficacy
-
3.
Genetic (evolutionary), represented as a change in the genetic code and the gene/protein expression level as a result of the above short-term and long-term memory changes and evolutionary processes.
Learning in SNN can be:
-
1.
Unsupervised – there is no desired output signal provided
-
2.
Supervised – a desired output signal is provided
-
3.
Semi-supervised.
Different tasks can be learned by a neuron, e.g.,
-
1.
Classification
-
2.
Input–output spike pattern association.
Several biologically plausible learning rules have been introduced so far, depending on the type of the information presentation:
-
1.
Rate-order learning, which is based on the average spiking activity of a neuron over time [47.58,59,60]
-
2.
Temporal learning, that is based on precise spike times [47.61,62,63,64,65]
-
3.
Rank-order learning, that takes into account the order of spikes across all synapses connected to a neuron [47.65,66].
Rate-order information representation is typical for cognitive information processing [47.58]. Temporal spike learning is observed in the auditory [47.13], the visual [47.67], and motor control information processing of the brain [47.61,68]. Its use in neuroprosthetics is essential, along with applications for a fast, real-time recognition and control of sequence of related processes [47.69]. Temporal coding accounts for the precise time of spikes and has been utilized in several learning rules, the most popular being spike-time dependent plasticity (STDP) [47.31,70] and SDSP [47.32,69]. Temporal coding of information in SNN makes use of the exact time of spikes (e.g., in milliseconds). Every spike matters and its time matters too.
The STDP Learning Rule
The STDP learning rule uses Hebbian plasticity [47.71] in the form of long-term potentiation (LTP) and depression (LTD) [47.31,70]. Efficacy of synapses is strengthened or weakened based on the timing of post-synaptic action potential in relation to the pre-synaptic spike (an example is given in Fig. 47.5a). If the difference in the spike time between the pre-synaptic and post-synaptic neurons is negative (pre-synaptic neuron spikes first) then the connection weight between the two neurons increases, otherwise it decreases. Through STDP, connected neurons learn consecutive temporal associations from data. Pre-synaptic activity that precedes post-synaptic firing can induce long-term potentiation (LTP); reversing this temporal order causes long-term depression (LTD).
(a) An example of synaptic change in an STDP learning neuron (after[47.31]). (b) Rank-order LIF neuron
Spike Driven Synaptic Plasticity (SDSP)
SDSP is an unsupervised learning method [47.32,69], a modification of the STDP that directs the change of the synaptic plasticity of a synapse w 0 depending on the time of spiking of the pre-synaptic neuron and the post-synaptic neuron. increases or decreases, depending on the relative timing of the pre-synaptic and post-synaptic spikes.
If a pre-synaptic spike arrives at the synaptic terminal before a postsynaptic spike within a critical time window, the synaptic efficacy is increased (potentiation). If the post-synaptic spike is emitted just before the pre-synaptic spike, synaptic efficacy is decreased (depression). This change in synaptic efficacy can be expressed as
where Δt spk is the pre-synaptic and post-synaptic spike time window.
The SDSP rule can be used to implement a supervised learning algorithm, when a teacher signal, which copies the desired output spiking sequence, is entered along with the training spike pattern, but without any change of the weights of the teacher input.
The SDSP model is implemented as an VLSI analog chip [47.72]. The silicon synapses comprise bistability circuits for driving a synaptic weight to one of two possible analog values (either potentiated or depressed). These circuits drive the synaptic-weight voltage with a current that is superimposed on that generated by the STDP and which can be either positive or negative. If, on short time scales, the synaptic weight is increased above a set threshold by the network activity via the STDP learning mechanism, the bistability circuits generate a constant weak positive current. In the absence of activity (and hence learning) this current will drive the weight toward its potentiated state. If the STDP decreases the synaptic weight below the threshold, the bistability circuits will generate a negative current that, in the absence of spiking activity, will actively drive the weight toward the analog value, encoding its depressed state. The STDP and bistability circuits facilitate the implementation of both long-term and short-term memory.
Rank-Order Learning
The rank-order learning rule uses important information from the input spike trains – the rank of the first incoming spike on each synapse (Fig. 47.5b). It establishes a priority of inputs (synapses) based on the order of the spike arrival on these synapses for a particular pattern, which is a phenomenon observed in biological systems as well as an important information processing concept for some STPR problems, such as computer vision and control [47.65,66].
This learning makes use of the extra information of spike (event) order. It has several advantages when used in SNN, mainly: fast learning (as it uses the extra information of the order of the incoming spikes) and asynchronous data entry (synaptic inputs are accumulated into the neuronal membrane potential in an asynchronous way). The learning is most appropriate for AER input data streams [47.10] as the events and their addresses are entered into the SNN one by one, in the order of their happening.
The post-synaptic potential of a neuron i at a time t is calculated as
where mod is a modulation factor, j is the index for the incoming spike at synapse j, i and w j,i is the corresponding synaptic weight, and order(j) represents the order (the rank) of the spike at the synapse j, i among all spikes arriving from all m synapses to the neuron i. The order(j) has a value 0 for the first spike and increases according to the input spike order. An output spike is generated by neuron i if the PSP(i, t) becomes higher than a threshold PSPTh(i).
During the training process, for each training input pattern (sample, example) the connection weights are calculated based on the order of the incoming spikes [47.66]
Combined Rank-Order and Temporal Learning
In [47.11] a method for a combined rank-order and temporal (e.g., SDSP) learning is proposed and tested on benchmark data. The initial value of a synaptic weight is set according to the rank-order learning based on the first incoming spike on this synapse. The weight is further modified to accommodate following spikes on this synapse with the use of a temporal learning rule – SDSP.
STPR in a Single Neuron
In contrast to the distributed representation theory and to the widely popular view that a single neuron cannot do much, some recent results showed that a single neuronal model can be used for complex STPR.
A single LIF neuron, for example, with simple synapses can be trained with the STDP unsupervised learning rule to discriminate a repeating pattern of synchronized spikes on certain synapses from noise (from [47.1]) – see Fig. 47.6.
A single LIF neuron with simple synapses can be trained with the STDP unsupervised learning rule to discriminate a repeating pattern of synchronized spikes on certain synapses from noise (after [47.73]) ◂
Single neuron models have been introduced for STPR, for example, Temportron [47.74], Chronotron [47.75], ReSuMe [47.76], SPAN [47.77,78]. Each of them can learn to emit a spike or a spike pattern (spike sequence) when a certain STP is recognized. Some of them can be used to recognize multiple STP per class and multiple classes [47.76,77,78]. Figure 47.7c,d shows the use of a single SPAN neuron for the classification of five STP belonging to five different classes [47.78]. The accuracy of classification is rightly lower for the class 1 (the neuron emits a spike at the very beginning of the input pattern) as there is no sufficient input data – Fig. 47.7d [47.78].
(a) The SPAN model (after [47.78]). (b) The Widrow–Hoff delta learning rule applied to learn to associate an output spike sequence to an input STP (after [47.32,78]). (c) , (d) The use of a single SPAN neuron for the classification of 5 STP belonging to five different classes (after [47.78]). The accuracy of classification is rightly lower for the class 1 – spike at the very beginning of the input pattern as there is no sufficient input data (d)
Evolving Spiking Neural Networks
Despite the ability of a single neuron to conduct STPR, a single neuron has a limited power and complex STPR tasks will require multiple spiking neurons.
One approach is proposed in the evolving spiking neural networks (eSNN) framework [47.37,79]. eSNN evolve their structure and functionality in an on-line manner, from incoming information. For every new input pattern, a new neuron is dynamically allocated and connected to the input neurons (feature neurons). The neurons connections are established for the neuron to recognize this pattern (or a similar one) as a positive example. The neurons represent centers of clusters in the space of the synaptic weights. In some implementations similar neurons are merged [47.37,38]. This makes it possible to achieve very fast learning in an eSNN (only one pass may be necessary), both in a supervised and in an unsupervised mode.
In [47.77] multiple SPAN neurons are evolved to achieve a better accuracy of spike pattern generation than with a single SPAN – Fig. 47.8a.
In [47.69] the SDSP model from [47.32] was successfully used to train and test a SNN for 293 character recognition (classes). Each character (a static image) is represented as a 2000 bit feature vector, and each bit is transferred into spike rates, with 50 Hz spike burst to represent 1 and 0 Hz to represent 0. For each class, 20 different training patterns are used and 20 neurons are allocated, one for each pattern (altogether 5860) (Fig. 47.8b) and trained for several hundreds of iterations.
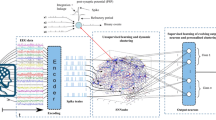
A general framework of eSNN for STPR is shown in Fig. 47.9. It consists of the following blocks:
-
1.
Input data encoding block
-
2.
Machine learning block (consisting of several sub-blocks)
-
3.
Output block.
The eSNN framework for STPR (after [47.85])
In the input block continuous value input variables are transformed into spikes. Different approaches can be used:
-
1.
Population rank coding [47.61] – Fig. 47.10a
Fig. 47.10 (a) Population rank order coding of input information. (b) AER of the input information (after [47.10])
-
2.
Thresholding the input value, so that a spike is generated if the input value (e.g., pixel intensity) is above a threshold
-
3.
Address event representation (AER) – thresholding the difference between two consecutive values of the same variable over time as it is in the artificial cochlea [47.14] and artificial retina devices [47.10] – Fig. 47.10b.
The input information is entered either on-line (for on-line, real-time applications) or as batch data. The time of the input data is in principle different from the internal SNN time of information processing.
Long and complex SSTD cannot be learned in simple one-layer neuronal structures as the examples in Fig. 47.8a,b. They require neuronal buffers as shown in Fig. 47.11a. In [47.80] a 3D buffer was used to store spatio-temporal chunks of input data before the data is classified. In this case, the size of the chunk (both in space and time) is fixed by the size of the reservoir. There are no connections between the layers in the buffer. Still, the system outperforms traditional classification techniques, as it is demonstrated on sign language recognition, where the eSNN classifier was applied [47.37,38]. Reservoir computing [47.36,45] has already become a popular approach for SSTD modeling and pattern recognition. In the classical view a reservoir is a homogeneous, passive 3D structure of probabilistically connected and fixed neurons that in principle has no learning and memory, nor has it an interpretable structure – Fig. 47.11b. A reservoir, such as a liquid state machine (LSM) [47.36,81], usually uses small world recurrent connections that do not facilitate capturing explicit spatial and temporal components from the SSTD in their relationship, which is the main goal of learning SSTD. Despite difficulties with the LSM reservoirs, it was shown on several SSTD problems that they produce better results than using a simple classifier [47.7,12,36,82]. Some publications demonstrated that probabilistic neurons are suitable for reservoir computing especially in a noisy environment [47.56,57]. In [47.83] an improved accuracy of the LSM reservoir structure on pattern classification of hypothetical tasks is achieved when STDP learning is introduced into the reservoir. The learning is based on comparing the liquid states for different classes and adjusting the connection weights so that same class inputs have closer connection weights. The method is illustrated on the phone recognition task of the TIMIT data base phonemes – spectro-temporal problem. 13 MSCC are turned into trains of spikes. The metric of separation between liquid states representing different classes is similar to Fisherʼs t-test [47.84].
After the presentation of an input data example (or a chink of data) the state of the SNN reservoir S(t) is evaluated in an output module and used for classification purposes (both during training and recall phase). Different methods can be applied to capture this state:
-
1.
Spike rate activity of all neurons at a certain time window: The state of the reservoir is represented as a vector of n elements (n is the number of neurons vector of n elements (n is the number of neurons in the reservoir), each element representing the spiking probability of the neuron within a time window. Consecutive vectors are passed to train/recall an output classifier.
-
2.
Spike rate activity of spatio-temporal clusters C 1, C 2,… C k of close (both in space and time) neurons: The state of each cluster C i is represented by a single number, reflecting on the spiking activity of the neurons in the cluster in a defined time window (this is the internal SNN time, usually measured in ms). This is interpreted as the current spiking probability of the cluster. The states of all clusters define the current reservoir state S(t). In the output function, the cluster states are used differently for different tasks.
-
3.
Continuous function representation of spike trains: In contrast to the above two methods that use spike rates to evaluate the spiking activity of a neuron or a neuronal cluster, here the train of spikes from each neuron within a time window, or a neuronal cluster, is transferred into a continuous value temporal function using a kernel (e.g., α-kernel). These functions can be compared and a continuous value error measured.
In [47.82] a comparative analysis of the three methods above is presented on a case study of Brazilian sign language gesture recognition (Fig. 47.18) using a LSM as a reservoir.
A single sample for each of the 15 classes of the LIngua BRAsileira de Sinais (LIBRAS) – the official Brazilian sign language is shown. Color indicates the time frame of a given data point (black/white corresponds to earlier/later time points) (after [47.82])
Different adaptive classifiers can be explored for the classification of the reservoir state into one of the output classes, including: statistical techniques, e.g., regression techniques; MLP; eSNN; nearest-neighbor techniques; and incremental LDA [47.86]. State vector transformation, before classification can be done with the use of adaptive incremental transformation functions, such as incremental PCA [47.87].
Computational Neurogenetic Models (CNGM)
Here, the neurogenetic model of a neuron [47.40,41] is utilized. A CNGM framework is shown in Fig. 47.12 [47.19].
A schematic diagram of a CNGM framework, consisting of: input encoding module; output function for SNN state evaluation; output classifier; GRN (optional module). The framework can be used to create concrete models for STPR or data modeling (after [47.19])
The CNGM framework comprises a set of methods and algorithms that support the development of computational models, each of them characterized by:
-
1.
eSNN at the higher level and a gene regulatory network (GRN) at the lower level, each functioning at a different time-scale and continuously interacting between each other.
-
2.
Optional use of probabilistic spiking neurons, thus forming an epSNN.
-
3.
Parameters in the epSNN model are defined by genes/proteins from the GRN.
-
4.
Ability to capture in its internal representation both spatial and temporal characteristics from SSTD streams.
-
5.
The structure and the functionality of the model evolve in time from incoming data.
-
6.
Both unsupervised and supervised learning algorithms can be applied in an on-line or in a batch mode.
-
7.
A concrete model would have a specific structure and a set of algorithms depending on the problem and the application conditions, e.g., classification of SSTD; modeling of brain data.
The framework from Fig. 47.12 supports the creation of a multi-modular integrated system, where different modules, consisting of different neuronal types and genetic parameters, represent different functions (e.g., vision, sensory information processing, sound recognition, and motor-control) and the whole system works in an integrated mode.
The neurogenetic model from Fig. 47.12 uses as a main principle the analogy with biological facts about the relationship between spiking activity and gene/protein dynamics in order to control the learning and spiking parameters in an SNN when SSTD is learned. Biological support of this can be found in numerous publications (e.g., [47.40,88,89,90]).
The Allen Human Brain Atlas [47.91] of the Allen Institute for Brain Science [47.92] has shown that at least 82% of human genes are expressed in the brain. For 1000 anatomical sites of the brains of two individuals 100 mln data points are collected that indicate gene expressions of each of the genes and underlies the biochemistry of the sites.
In [47.58] it is suggested that both the firing rate (rate coding) and spike timing as spatio-temporal patterns (rank order and spatial pattern coding) play a role in fast and slow, dynamic and adaptive sensorimotor responses, controlled by the cerebellar nuclei. Spatio-temporal patterns of a population of Purkinji cells are shaped by activities in the molecular layer of interneurons. In [47.88] it is demonstrated that the temporal spiking dynamics depend on the spatial structure of the neural system (e.g., different for the hippocampus and the cerebellum). In the hippocampus the connections are scale free, e.g., there are hub neurons, while in the cerebellum the connections are regular. The spatial structure depends on genetic pre-determination and on the gene dynamics. Functional connectivity develops in parallel with structural connectivity during brain maturation. A growth-elimination process (synapses are created and eliminated) depend on gene expression [47.88], e.g., glutamatergic neurons issued from the same progenitors tend to wire together and form ensembles, also for the cortical GABAergic interneuron population. Connections between early developed neurons (mature networks) are more stable and reliable when transferring spikes than the connections between newly created neurons (thus the probability of spike transfer). Postsynaptic AMPA-type glutamate receptors (AMPARs) mediate most fast excitatory synaptic transmissions and are crucial for many aspects of brain function, including learning, memory, and cognition [47.40,93].
Kasabov et al. [47.94] show the dramatic effect of a change of single gene, that regulates the τ parameter of the neurons, on the spiking activity of the whole SNN of 1000 neurons – see Fig. 47.13.
A GRN interacting with a SNN reservoir of 1000 neurons. The GRN controls a single parameter, i.e., the τ parameter of all 1000 LIF neurons, over a period of 5 s. The top diagram shows the evolution of τ. The response of the SNN is shown as a raster plot of spike activity. A black point in this diagram indicates a spike of a specific neuron at a specific time in the simulation. The bottom diagram presents the evolution of the membrane potential of a single neuron from the network (green curve) along with its firing threshold ϑ (red curve). Output spikes of the neuron are indicated as black vertical lines in the same diagram (after [47.94])
The spiking activity of a neuron may affect the expressions of genes as feedback [47.95]. As pointed out in [47.90] on longer time scales of minutes and hours the function of neurons may cause changes of the expression of hundreds of genes transcribed into mRNAs and also in microRNAs, which makes the short-term, long-term, and the genetic memories of a neuron linked together in a global memory of the neuron and further – of the whole neural system.
A major problem with the CNGM from Fig. 47.12 is how to optimize the numerous parameters of the model. One solution could be to use evolutionary computation, such as PSO [47.57,96] and the recently proposed quantum inspired evolutionary computation techniques [47.21,44,97]. The latter can deal with a very large dimensional space as each quantum-bit chromosome represents the whole space, each point to certain probability. Such algorithms are faster and lead to a close solution to the global optimum in a very short time. In one approach it may be reasonable to use same parameter values (same GRN) for all neurons in the SNN or for each of different types of neurons (cells) that will results in a significant reduction of the parameters to be optimized. This can be interpreted as the average parameter value for neurons of the same type. This approach corresponds to the biological notion to use one value (average) of a gene/protein expression for millions of cells in bioinformatics.
Another approach to define the parameters of the probabilistic spiking neurons, especially when used in biological studies, is to use prior knowledge about the association of spiking parameters with relevant genes/proteins (neurotransmitter, neuroreceptor, ion channel, neuromodulator) as described in [47.19]. Combination of the two approaches above is also possible.
SNN Software and Hardware Implementations to Support STPR
Software and hardware realizations of SNN are already available to support various applications of SNN for STPR. Among the most popular software/hardware systems are [47.99,100,101]:
- 1.
-
2.
Software simulators, such as Brian [47.100], Nestor, NeMo [47.103], etc.
-
3.
Silicon retina camera [47.10]
-
4.
Silicon cochlea [47.14]
-
5.
SNN hardware realization of LIFM and SDSP [47.72,104,105,106]
- 6.
-
7.
FPGA implementations of SNN [47.109]
-
8.
The recently announced IBM LIF SNN chip.
Figure 47.14 shows a hypothetical engineering system using some of the above tools (from [47.11,104]).
A hypothetical neuromorphic SNN application system (after [47.98]) ▸
Current and Future Applications of eSNN and CNGM for STPR
The applications of eSNN for STPR are numerous. Here only few are listed:
- 1.
-
2.
EEG data modeling and pattern recognition [47.3,4,5,6,7,111,112,113] directed to practical applications, such as: BCI [47.5], classification of epilepsy [47.112,113,114] – (Fig. 47.16).
-
3.
Robot control through EEG signals [47.115] (Fig. 47.17) and robot navigation [47.116].
-
4.
Sign language gesture recognition (e.g., the Brazilian sign language – Fig. 47.18) [47.82].
-
5.
Risk of event evaluation, e.g., prognosis of establishment of invasive species [47.21] – Fig. 47.19; stroke occurrence [47.16], etc.
- 6.
-
7.
Neurorehabilitation robots [47.117].
- 8.
-
9.
Knowledge discovery from SSTD [47.120].
-
10.
Neurogenetic robotics [47.121].
-
11.
Modeling the progression or the response to treatment of neurodegenerative diseases, such as Alzheimerʼs disease [47.18,19] – Fig. 47.20. The analysis of the GRN model obtained in this case could enable the discovery of unknown interactions between genes/proteins related to brain disease progression and how these interactions can be modified to achieve a desirable effect.
-
12.
Modeling financial and economic problems (neuroeconomics) where at a lower level the GRN represents the dynamic interaction between time series variables (e.g., stock index values, exchange rates, unemployment, GDP, the price of oil), while the higher level epSNN states represents the state of the economy or the system under study. The states can be further classified into pre-defined classes (e.g., buy, hold, sell, invest, likely bankruptcy) [47.122].
-
13.
Personalized modeling, which is concerned with the creation of a single model for an individual input data [47.17,46,47]. Here a whole SSTD pattern is taken as individual data rather than a single vector.
Robot control and navigation through EEG signals (from [47.110])
A prognostic system for ecological modeling (after [47.21])
Hierarchical CNGM (after [47.19])
Abbreviations
- AER:
-
address event representation
- AMPA:
-
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR:
-
(amino-methylisoxazole-propionic acid) receptor
- ANN:
-
artificial neural network
- BCI:
-
brain-computer interface
- CLC:
-
chloride channel
- CNGM:
-
computational neurogenetic model
- EEG:
-
electroencephalography
- FPGA:
-
field-programmable gate array
- GABAAR:
-
GABAA receptor
- GABABR:
-
GABAB receptor
- GABRA:
-
GABAA receptor
- GABRB:
-
GABAB receptor
- GDP:
-
guanosine diphosphate
- GRN:
-
gene regulatory network
- HMM:
-
hidden Markov model
- IBM:
-
individual-based model
- IFM:
-
integrate-and-fire model
- KCN:
-
kalium (potassium) voltage-gated channel
- LDA:
-
linear discriminant analysis
- LIF:
-
leaky integrate-and-fire neuron
- LIFM:
-
leaky IFM
- LSM:
-
liquid state machine
- LTD:
-
long-term depression
- LTP:
-
long-term potentiation
- MLP:
-
multilayer perceptron
- NMDA:
-
N-methyl-d-aspartate
- NMDR:
-
(N-methyl-d-aspartate acid) NMDA receptor
- PCA:
-
principle component analysis
- PSO:
-
particle swarm optimization
- PSP:
-
post-synaptic potential
- ReSuMe:
-
remote supervised method
- SCN:
-
sodium voltage-gated channel
- SDSP:
-
spike driven synaptic plasticity
- SNN:
-
spiking neural network
- SRM:
-
spike response model
- SSTD:
-
spatio and spectro-temporal data
- STDP:
-
spike-timing dependent plasticity
- STPR:
-
spatio-temporal pattern recognition
- eSNN:
-
evolving spiking neural network
- fMRI:
-
functional magnetic resonance imaging
- mRNA:
-
messenger RNA
References
Emotiv: http://www.emotiv.com
The FMRIB Centre, University of Oxford, http://www.fmrib.ox.ac.uk
D.A. Craig, H.T. Nguyen: Adaptive EEG thought pattern classifier for advanced wheelchair control, Proc. Eng. Med. Biol. Soc. – EMBSʼ07 (2007) pp. 2544–2547
A. Ferreira, C. Almeida, P. Georgieva, A. Tomé, F. Silva: Advances in EEG-based biometry, LNCS 6112, 287–295 (2010)
T. Isa, E.E. Fetz, K. Müller: Recent advances in brain-machine interfaces, Neural Netw. 22(9), 1201–1202 (2009)
F. Lotte, M. Congedo, A. Lécuyer, F. Lamarche, B. Arnaldi: A review of classification algorithms for EEG-based brain–computer interfaces, J. Neural Eng. 4(2), R1–R15 (2007)
S. Schliebs, N. Nuntalid, N. Kasabov: Towards spatio-temporal pattern recognition using evolving spiking neural networks, LNCS 6443, 163–170 (2010)
B. Schrauwen, J. Van Campenhout: BSA, a fast and accurate spike train encoding scheme, Neural Netw. 2003, Proc. Int. Jt. Conf., Vol. 4 (IEEE 2003) pp. 2825–2830
D. Sona, H. Veeramachaneni, E. Olivetti, P. Avesani: Inferring cognition from fMRI brain images, LNCS 4669, 869–878 (2007)
T. Delbruck: JAER open source project (2007) http://jaer.wiki.sourceforge.net
K. Dhoble, N. Nuntalid, G. Indivery, N. Kasabov: Online spatio-temporal pattern recognition with evolving spiking neural networks utilising address event representation, rank order, and temporal spike learning, Int. Joint Conf. Neural Netw. (IJCNN) (IEEE 2012)
N. Kasabov, K. Dhoble, N. Nuntalid, A. Mohemmed: Evolving probabilistic spiking neural networks for spatio-temporal pattern recognition: A preliminary study on moving object recognition, 7064, 230–239 (2011)
A. Rokem, S. Watzl, T. Gollisch, M. Stemmler, A.V.M. Herz, I. Samengo: Spike-timing precision underlies the coding efficiency of auditory receptor neurons, J. Neurophys. 95(4), 2541–2552 (2005)
A. van Schaik, L. Shih-Chii: AER EAR: A matched address event representation interface, Proc. ISCAS – IEEE Int. Symp. Circuits Syst., Vol. 5 (2005) pp. 4213–4216
P.J. Cowburn, J.G.F. Cleland, A.J.S. Coats, M. Komajda: Risk stratification in chronic heart failure, Eur. Heart J. 19, 696–710 (1996)
S. Barker-Collo, V.L. Feigin, V. Parag, C.M.M. Lawes, H. Senior: Auckland stroke outcomes study, Neurology 75(18), 1608–1616 (2010)
N. Kasabov: Global, local and personalised modelling and profile discovery in Bioinformatics: An integrated approach, Pattern Recogn. Lett. 28(6), 673–685 (2007)
R. Schliebs: Basal forebrain cholinergic dysfunction in Alzheimerʼs disease – interrelationship with β-amyloid, inflammation and neurotrophin signaling, Neurochem. Res. 30, 895–908 (2005)
N. Kasabov, R. Schliebs, H. Kojima: Probabilistic computational neurogenetic framework: From modelling cognitive systems to Alzheimerʼs disease, IEEE Trans. Auton. Ment. Dev. 3(4), 1–12 (2011)
C.R. Shortall, A. Moore, E. Smith, M.J. Hall, I.P. Woiwod, R. Harrington: Long-term changes in the abundance of flying insects, Insect Conserv. Divers. 2(4), 251–260 (2009)
S. Schliebs, M. Defoin-Platel, S. Worner, N. Kasabov: Integrated feature and parameter optimization for evolving spiking neural network: Exploring heterogeneous probabilistic models, Neural Netw. 22, 623–632 (2009)
L.R. Rabiner: A tutorial on hidden Markov models and selected applications in speech recognition, Proceedings of IEEE 77(2), 257–285 (1989)
N. Kasabov: Foundations of Neural Networks, Fuzzy Systems, and Knowledge Engineering (MIT Press, Cambridge 1996) p. 550
I. Arel, D.C. Rose, T.P. Karnowski: Deep machine learning: A new frontier artificial intelligence research, Comput. Intell. Mag. 5(4), 13–18 (2010)
I. Arel, D. Rose, B. Coop: DeSTIN: A deep learning architecture with application to high-dimensional robust pattern, Proc. 2008 AAAI Workshop Biologically Inspired Inspired Cognitive Architectures (BICA) (2008)
Y. Bengio: Learning deep architectures for AI, Found. Trends. Mach. Learn. 2(1), 1–127 (2009)
I. Weston, F. Ratle, R. Collobert: Deep learning via semi-supervised embedding, Proc. 25th Int. Conf. Mach. Learn. (2008) pp. 1168–1175
W. Gerstner: Time structure of the activity of neural network models, Phys. Rev. 51, 738–758 (1995)
W. Gerstner: Whatʼs different with spiking neurons?. In: Plausible Neural Networks for Biological Modelling, ed. by H. Mastebroek, H. Vos (Kluwer, Dordrecht 2001) pp. 23–48
G. Kistler, W. Gerstner: Spiking neuron models – single neurons. In: Populations, Plasticity (Cambridge Univ. Press, Cambridge 2002)
S. Song, K. Miller, L. Abbott: Competitive Hebbian learning through spike-timing-dependent synaptic plasticity, Nat. Neurosci. 3, 919–926 (2000)
S. Fusi, M. Annunziato, D. Badoni, A. Salamon, D. Amit: Spike-driven synaptic plasticity: Theory, simulation, VLSI implementation, Neural Comput. 12(10), 2227–2258 (2000)
A. Belatreche, L.P. Maguire, M. McGinnity: Advances in design and application of spiking neural networks, Soft Comput. 11(3), 239–248 (2006)
F. Bellas, R.J. Duro, A. Faiña, D. Souto: Multilevel Darwinisb Brain (MDB): Artificial evolution in a cognitive architecture for real robots, IEEE Trans. Auton. Ment. Dev. 2, 340–354 (2010)
S. Bohte, J. Kok, J. LaPoutre: Applications of spiking neural networks, Inf. Proc. Lett. 95(6), 519–520 (2005)
W. Maass, T. Natschlaeger, H. Markram: Real-time computing without stable states: A new framework for neural computation based on perturbations, Neural Comput. 14(11), 2531–2560 (2002)
N. Kasabov: Evolving Connectionist Systems: The Knowledge Engineering Approach (Springer, London 2007)
S. Wysoski, L. Benuskova, N. Kasabov: Evolving spiking neural networks for audiovisual information processing, Neural Netw. 23(7), 819–835 (2010)
M. Riesenhuber, T. Poggio: Hierarchical model of object recognition in cortex, Nat. Neurosci. 2, 1019–1025 (1999)
L. Benuskova, N. Kasabov: Computational Neuro-Genetic Modelling (Springer, New York 2007) p. 290
N. Kasabov, L. Benuskova, S. Wysoski: A computational neurogenetic model of a spiking neuron, IJCNN 2005 Conf. Proc., Vol. 1 (IEEE 2005) pp. 446–451
N. Kasabov: To spike or not to spike: A probabilistic spiking neuron model, Neural Netw. 23(1), 16–19 (2010)
W. Maass, H. Markram: Synapses as dynamic memory buffers, Neural Netw. 15(2), 155–161 (2002)
S. Schliebs, N. Kasabov, M. Defoin-Platel: On the probabilistic optimization of spiking neural networks, Int. J. Neural Syst. 20(6), 481–500 (2010)
D. Verstraeten, B. Schrauwen, M. DʼHaene, D. Stroobandt: An experimental unification of reservoir computing methods, Neural Netw. 20(3), 391–403 (2007)
N. Kasabov, Y. Hu: Integrated optimisation method for personalised modelling and case study applications, Int. J. Funct. Inf. Personal. Med. 3(3), 236–256 (2010)
N. Kasabov: Data analysis and predictive systems and related methodologies – personalised trait modelling system, NZ Patent PCT/NZ2009/000222 (2009)
A.L. Hodgkin, A.F. Huxley: A quantitative description of membrane current and its application to conduction and excitation in nerve, J. Physiol. 117, 500–544 (1952)
E. Izhikevich: Simple model of spiking neurons, IEEE Trans. Neural Netw. 14(6), 1569–1572 (2003)
E.M. Izhikevich: Which model to use for cortical spiking neurons?, Neural Netw. 15(5), 1063–1070 (2004)
E.M. Izhikevich, G.M. Edelman: large-scale model of mammalian thalamocortical systems, Proc. Natl. Acad. Sci. USA 105, 3593–3598 (2008)
E. Izhikevich: Polychronization: Computation with spikes, Neural Comput. 18, 245–282 (2006)
Z.P. Kilpatrick, P.C. Bresloff: Effect of synaptic depression and adaptation on spatio-temporal dynamics of an excitatory neural networks, Physica D 239, 547–560 (2010)
W. Maass, A.M. Zador: Computing and learning with dynamic synapses. In: Pulsed Neural Networks (MIT Press, Cambridge 1999) pp. 321–336
J.R. Huguenard: Reliability of axonal propagation: The spike doesnʼt stop here, Proc. Natl. Acad. Sci USA 97(17), 9349–9350 (2000)
S. Schliebs, A. Mohemmed, N. Kasabov: Are probabilistic spiking neural networks suitable for reservoir computing?, Int. Jt. Conf. Neural Netw. (IJCNN) (IEEE 2011) pp. 3156–3163
H. Nuzly, A. Hamed, N. Kasabov, S. Shamsuddin: Probabilistic evolving spiking neural network optimization using dynamic quantum inspired particle swarm optimization, Aust. J. Intell. Inf. Process. Syst. 11(1), 1074 (2010), available online at http://cs.anu.edu.au/ojs/index.php/ajiips/article/viewArticle/1074
S.J. Thorpe: Spike-based image processing: Can we reproduce biological vision in hardware, LNCS 7583, 516–521 (2012)
W. Gerstner, A.K. Kreiter, H. Markram, A.V.M. Herz: Neural codes: Firing rates and beyond, Proc. Natl. Acad. Sci. USA 94(24), 12740–12741 (1997)
J.J. Hopfield: Neural networks and physical systems with emergent collective computational abilities, Proc. Natl. Acad. Sci. USA 79, 2554–2558 (1982)
S.M. Bohte: The evidence for neural information processing with precise spike-times: A survey, Nat. Comput. 3(2), 195–206 (2004)
J. Hopfield: Pattern recognition computation using action potential timing for stimulus representation, Nature 376, 33–36 (1995)
H.G. Eyherabide, I. Samengo: Time and category information in pattern-based codes, Front. Comput. Neurosci. 4, 145 (2010)
F. Theunissen, J.P. Miller: Temporal encoding in nervous rigorous definition, J. Comput. Neurosci. 2(2), 149–162 (1995)
S. Thorpe, A. Delorme, R. VanRullen: Spike-based strategies for rapid processing, Neural Netw. 14(6–7), 715–725 (2001)
S. Thorpe, J. Gautrais: Rank order coding, Comput. Neurosci. 13, 113–119 (1998)
M.J. Berry, D.K. Warland, M. Meister: The structure and precision of retinal spiketrains, Proc. Natl. Acad. Sci. USA 94(10), 5411–5416 (1997)
P. Reinagel, R.C. Reid: Precise firing events are conserved across neurons, J. Neurosci. 22(16), 6837–6841 (2002)
J. Brader, W. Senn, S. Fusi: Learning real-world stimuli in a neural network with spike-driven synaptic dynamics, Neural Comput. 19(11), 2881–2912 (2007)
R. Legenstein, C. Naeger, W. Maass: What can a neuron learn with spike-timing-dependent plasticity?, Neural Comput. 17(11), 2337–2382 (2005)
D. Hebb: The Organization of Behavior (Wiley, New York 1949)
G. Indiveri, F. Stefanini, E. Chicca: Spike-based learning with a generalized integrate and fire silicon neuron, IEEE Int. Symp. Circuits Syst. (ISCAS 2010) (2010) pp. 1951–1954
T. Masquelier, R. Guyonneau, S. Thorpe: Spike timing dependent plasticity finds the start of repeating patterns in continuous spike trains, PlosONE 3(1), e1377 (2008)
R. Gutig, H. Sompolinsky: The tempotron: A neuron timing-based decisions, Nat. Neurosci. 9(3), 420–428 (2006)
R.V. Florian: The chronotron: A neuron that learns to fire temporally-precise spike patterns, Nature Precedings (2010), available online at http://precedings.nature.com/documents/5190/version/1
F. Ponulak, A. Kasinski: Supervised learning in spiking neural networks with ReSuMe: Sequence learning, Neural Comput. 22(2), 467–510 (2010)
A. Mohemmed, S. Schliebs, S. Matsuda, N. Kasabov: Evolving spike pattern association neurons and neural networks, Neurocomputing 107, 3–10 (2013)
A. Mohemmed, S. Schliebs, S. Matsuda, N. Kasabov: SPAN: Spike pattern association neuron for learning spatio-temporal sequences, Int. J. Neural Syst. 22(4), 1–16 (2012)
M. Watts: A decade of Kasabovʼs evolving connectionist systems: A Review, IEEE Trans. Syst. Man Cybern. C 39(3), 253–269 (2009)
H. Nuzlu, N. Kasabov, S. Shamsuddin, H. Widiputra, K. Dhoble: An extended evolving spiking neural network model for spatio-temporal pattern classification, Proc. IJCNN (IEEE 2011) pp. 2653–2656
E. Goodman, D. Ventura: Spatiotemporal pattern recognition via liquid state machines, Int. Jt. Conf. Neural Networks (IJCNN) ʼ06 (2006) pp. 3848–3853
S. Schliebs, H.N.A. Hamed, N. Kasabov: A reservoir-based evolving spiking neural network for on-line spatio-temporal pattern learning and recognition, 18th Int. Conf. Neural Inf. Proc. ICONIP 2011 (Springer, Shanghai 2011)
D. Norton, D. Ventura: Improving liquid state machines through iterative refinement of the reservoir, Neurocomputing 73, 2893–2904 (2010)
R.A. Fisher: The use of multiple measurements in taxonomic problems, Ann. Eugen. 7, 179–188 (1936)
EU FP7 Marie Curie project EvoSpike (2011–2012), http://ncs.ethz.ch/projects/evospike
S. Pang, S. Ozawa, N. Kasabov: Incremental linear discriminant analysis for classification of data streams, IEEE Trans. SMC-B 35(5), 905–914 (2005)
S. Ozawa, S. Pang, N. Kasabov: Incremental learning of chunk data for on-line pattern classification systems, IEEE Trans. Neural Netw. 19(6), 1061–1074 (2008)
J.M. Henley, E.A. Barker, O.O. Glebov: Routes, destinations and advances in AMPA receptor trafficking, Trends Neurosci. 34(5), 258–268 (2011)
Y.C. Yu, R.S. Bultje, X. Wang, S.H. Shi: Specific synapses develop preferentially among sister excitatory neurons in the neocortex, Nature 458, 501–504 (2009)
V.P. Zhdanov: Kinetic models of gene expression including non-coding RNAs, Phys. Rep. 500, 1–42 (2011)
BrainMap Project: www.brain-map.org
Allen Institute for Brain Science: www.alleninstitute.org
Gene and Disease (2005) NCBI, http://www.ncbi.nlm.nih.gov
N. Kasabov, S. Schliebs, A. Mohemmed: Modelling the effect of genes on the dynamics of probabilistic spiking neural networks for computational neurogenetic modelling, Proc. 6th Meet. Comp. Intell. Bioinfor. Biostat. (CIBB) 2011 (Springer 2011)
M. Barbado, K. Fablet, M. Ronjat, M. De Waard: Gene regulation by voltage-dependent calcium channels, Biochim. Biophys. Acta 1793, 1096–1104 (2009)
A. Mohemmed, S. Matsuda, S. Schliebs, K. Dhoble, N. Kasabov: Optimization of spiking neural networks with dynamic synapses for spike sequence generation using PSO, Proc. Int. Joint Conf. Neural Netw. (IEEE, San Jose 2011) pp. 2969–2974
M. Defoin-Platel, S. Schliebs, N. Kasabov: Quantum-inspired evolutionary algorithm: A multi-model EDA, IEEE Trans. Evol. Comput. 13(6), 1218–1232 (2009)
Neuromorphic Cognitive Systems Group, Institute for Neuroinformatics, ETH and University of Zurich, http://ncs.ethz.ch
R. Douglas, M. Mahowald: Silicon neurons. In: The Handbook of Brain Theory and Neural Networks, ed. by M. Arbib (MIT, Cambridge 1995) pp. 282–289
R. Brette, M. Rudolph, T. Carnevale, M. Hines, D. Beeman, J.M. Bower, M. Diesmann, A. Morrison, P.H. Goodman, F.C. Harris, M. Zirpe, T. Natschläger, D. Pecevski, B. Ermentrout, M. Djurfeldt, A. Lansner, O. Rochel, T. Vieville, E. Muller, A.P. Davison, S.E. Boustani, A. Destexhe: Simulation of networks of spiking neurons: A review of tools and strategies, J. Comput. Neurosci. 23, 349–398 (2007)
S. Furber, S. Temple: Neural systems engineering, Interface J. R. Soc. 4, 193–206 (2007)
jAER Open Source Project: http://jaer.wiki.sourceforge.net
NeMo spiking neural network simulator, http://www.doc.ic.ac.uk/∼akf/nemo/index.html
G. Indiveri, B. Linares-Barranco, T. Hamilton, A. Van Schaik, R. Etienne-Cummings, T. Delbruck, S. Liu, P. Dudek, P. Häfliger, S. Renaud: Neuromorphic silicon neuron circuits, Front. Neurosci. 5, 1–23 (2011)
G. Indiveri, E. Chicca, R.J. Douglas: Artificial cognitive systems: From VLSI networks of spiking neurons to neuromorphic cognition, Cogn. Comput. 1(2), 119–127 (2009)
G. Indiviery, T. Horiuchi: Frontiers in neuromorphic engineering, Front. Neurosci. 5, 118 (2011)
A.D. Rast, X. Jin, F. Galluppi, L.A. Plana, C. Patterson, S. Furber: Scalable event-driven native parallel processing: The SpiNNaker neuromimetic system, Proc. ACM Int. Conf. Comput. Front. (ACM 2010) pp. 21–29
X. Jin, M. Lujan, L.A. Plana, S. Davies, S. Temple, S. Furber: Modelling spiking neural networks on SpiNNaker, Comput. Sci. Eng. 12(5), 91–97 (2010)
S.P. Johnston, G. Prasad, L. Maguire, T.M. McGinnity: FPGA Hardware/software co-design methodology – towards evolvable spiking networks for robotics application, Int. J. Neural Syst. 20(6), 447–461 (2010)
KEDRI: http://www.kedri.aut.ac.nz
R. Acharya, E.C.P. Chua, K.C. Chua, L.C. Min, T. Tamura: Analysis and automatic identification of sleep stages using higher order spectra, Int. J. Neural Syst. 20(6), 509–521 (2010)
S. Ghosh-Dastidar, H. Adeli: A new supervised learning algorithm for multiple spiking neural networks with application in epilepsy and seizure detection, Neural Netw. 22(10), 1419–1431 (2009)
S. Ghosh-Dastidar, H. Adeli: Improved spiking neural networks for EEG classification and epilepsy and seizure detection, Integr. Comput.-Aided Eng. 14(3), 187–212 (2007)
A.E.P. Villa, Y. Asai, I. Tetko, B. Pardo, M.R. Celio, B. Schwaller: Cross-channel coupling of neuronal activity in parvalbumin-deficient mice susceptible to epileptic seizures, Epilepsia 46(6), 359 (2005)
G. Pfurtscheller, R. Leeb, C. Keinrath, D. Friedman, C. Neuper, C. Guger, M. Slater: Walking from thought, Brain Res. 1071(1), 145–152 (2006)
E. Nichols, L.J. McDaid, N.H. Siddique: Case study on self-organizing spiking neural networks for robot navigation, Int. J. Neural Syst. 20(6), 501–508 (2010)
X. Wang, Z.G. Hou, A. Zou, M. Tan, L. Cheng: A behavior controller for mobile robot based on spiking neural networks, Neurocomputing 71(4–6), 655–666 (2008)
D. Buonomano, W. Maass: State-dependent computations: Spatio-temporal processing in cortical networks, Nat. Rev. Neurosci. 10, 113–125 (2009)
T. Natschläger, W. Maass: Spiking neurons and the induction of finite state machines, Theor. Comput. Sci. Nat. Comput. 287(1), 251–265 (2002)
S. Soltic, N. Kasabov: Knowledge extraction from evolving spiking neural networks with rank order population coding, Int. J. Neural Syst. 20(6), 437–445 (2010)
Y. Meng, Y. Jin, J. Yin, M. Conforth: Human activity detection using spiking neural networks regulated by a gene regulatory network, Proc. Int. Jt. Conf. Neural Netw. (IJCNN) (IEEE, Barcelona 2010) pp. 2232–2237
R. Pears, H. Widiputra, N. Kasabov: Evolving integrated multi-model framework for on-line multiple time series prediction, Evol. Syst. 4(2), 99–117 (2013)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag
About this chapter
Cite this chapter
Kasabov, N. (2014). Brain-like Information Processing for Spatio-Temporal Pattern Recognition. In: Kasabov, N. (eds) Springer Handbook of Bio-/Neuroinformatics. Springer Handbooks. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-30574-0_47
Download citation
DOI: https://doi.org/10.1007/978-3-642-30574-0_47
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-30573-3
Online ISBN: 978-3-642-30574-0
eBook Packages: EngineeringEngineering (R0)