Abstract
This chapter reviews the recently described family Cohaesibacteraceae, their two genera (Cohaesibacter and Breoghania) and three species (Cohaesibacter gelatinilyticus Hwang and Cho 2008, Cohaesibacter marisflavi Qu et al. 2011, and Breoghania corrubedonensis Gallego et al. 2010). The type species for each genus are Cohaesibacter gelatinilyticus and Breoghania corrubedonensis, respectively. Cohaesibacteraceae was created to accommodate strains that were not genetically similar enough to be classified into existing families of the order Rhizobiales. Besides the genetic dissimilarity, the sole major respiratory quinone (Q-10) and the polar lipid composition differentiate Cohaesibacteraceae from the other families of this order.
The members of Cohaesibacteraceae are Gram-negative, facultative anaerobic (Cohaesibacter) or aerobic (Breoghania) rods and are motile by polar flagella. Cohaesibacter comprises strains isolated from coastal waters of the east coast of Korea and China, whereas Breoghania has its type strain isolated from coastal waters of northwest Spain. Nothing is known about their applications or ecological importance, so this chapter only provides phenotypic and genetic characterization. Additionally, the phylogenetic relationship with other families of the order Rhizobiales is presented. Finally, the formal taxonomical descriptions of both genera and their respective species are given.
Access provided by Autonomous University of Puebla. Download reference work entry PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction and General Characteristics
The family Cohaesibacteraceae is in the order Rhizobiales, which is one of the seven orders of Alphaproteobacteria. Cohaesibacteraceae is distinguished from the other 12 families of Rhizobiales (Rhizobiaceae, Bartonellaceae, Brucellaceae, Phyllobacteriaceae, Methylocystaceae, Beijerinckiaceae, Bradyrhizobiaceae, Hyphomicrobiaceae, Methylobacteriaceae, Rhodobiaceae, Aurantimonadaceae, and Xanthobacteraceae) by phylogenetic analysis of ribosomal DNA (16S). This recently proposed family includes two genera: Cohaesibacter and Breoghania. They are distinct lineages formed by strains from coastal marine environments that did not cluster with any other families in 16S phylogenetic analysis. In addition, the members of Cohaesibacteraceae have other genetic and phenotypic characteristics, which are discussed in detail in this chapter. The family and the type species of the genus Cohaesibacter, Cohaesibacter gelatinilyticus, had their emended description approved by the International Committee on Systematics of Prokaryotes and its judicial commission (Euzéby 2011).
Phylogeny
Phylogenetic analyses, including secondary structure information of 16S rRNA, from 189 species of the order Rhizobiales determined that strains from coastal seawaters of the east coast of Korea (CL-GR15 and CL-GR35) belong to that order but do not represent any previously described family. Thus, the family Cohaesibacteraceae was characterized. Later, another seawater strain (DQHS21) from the east coast of China showed greater similarity with 16S of Cohaesibacter gelatinilyticus Hwang and Cho 2008 than with the other Alphaproteobacteria genera. Hwang and Cho (2008) detected three signature nucleotides in the 16S gene that characterize the genus Cohaesibacter in a study describing the type species Cohaesibacter gelatinilyticus. In addition, a strain (UBF-B1) isolated from a beach in the northwest of Spain after an oil spill was phylogenetic compared with Rhizobiales sequences (including Cohaesibacteraceae strains) using the 16S fragment and other genes (atpD, pyrG, rpo and fusA). Results showed UBF-B1 clustering together with Cohaesibacter gelatinilyticus; however, they had only 92 % similarity. These data resulted in a new genus description: Breoghania.
Taxonomy
Family Cohaesibacteraceae, Hwang and Cho (2008) emend. Gallego et al. (2010)
Cohaesibacteraceae (Co.hae.si.bact.er.a’ce.ae. N.L. masc. n. Cohaesibacter, type genus of the family; -aceae ending to denote a family; N.L. fem. pl. n. Cohaesibacteraceae, the Cohaesibacter family) (Hwang and Cho 2008).
Cohaesibacteraceae is a family phylogenetically positioned in the order Rhizobiales in the class Alphaproteobacteria. It is most closely related with the families Brucellaceae (genera Ochrobactrum and Brucella: 90.9–92.5 % similarity); Rhizobiaceae (genus Sinorhizobium: 90.9–91.5 %); Bartonellaceae (genus Bartonella: 90.6–91.9 %); Rhodobiaceae (genus Rhodobium: 89.3–91.2 %), and Phyllobacteriaceae (genus Phyllobacterium: 89.3–91.2 %) (Fig. 8.1 ).
Phylogenetic position of the family Cohaesibacteraceae based on 16S rRNA and created using the neighbor-joining algorithm with the Jukes-Cantor correction. The sequence dataset and alignment were used according to the All-Species Living Tree Project (LTP) database (Yarza et al. 2010; http://www.arb-silva.de/projects/living-tree). The tree topology was stabilized with a representative set of nearly 750 high-quality type-strain sequences proportionally distributed among the different bacterial and archaeal phyla. In addition, a 40 % maximum frequency filter was applied in order to remove hypervariable positions and potentially misplaced bases from the alignment. Scale bar indicates estimated sequence divergence
The members of the family have varying capabilities for nitrate reduction and the DNA composition ranges from 52 to 64 mol% G+C. The sole major respiratory quinone (Q-10) differentiates Cohaesibacteraceae from the families Aurantimonadaceae, Phyllobacteriaceae, Rhodobiaceae, Bradyrhizobiaceae, Methylocystaceae and Hyphomicrobiaceae. Cohaesibacteraceae is also differentiated from 10 families in the order Rhizobiales (except the families Brucellaceae and Phyllobacteriaceae) by polar lipid composition (Table 8.1 ).
Genus Cohaesibacter, Hwang and Cho (2008) emend. Gallego et al. (2010)
Cohaesibacter [Co.hae.s’i.bac.ter. L. part. adj. cohaesus (from L. v. cohaereo) pressed together, clung together; N.L. masc. n. bacter a rod; N.L. masc. n. Cohaesibacter rods that appear cohesive with each other].
The members of this genus are Gram-negative, facultative anaerobic rods. They have oxidase activity and variable catalase activity. The predominant fatty acids are C18 : 1ω7c and C15 : 0 iso 2-OH and/or C16 : 1ω7c and C18 : 0. The respiratory quinone is ubiquinone 10 (Q-10). The DNA G+C content ranges from 53.0 to 55.2 mol%. The major polar lipids are phosphatidylcholine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylmonomethylethanolamine, an unidentified aminolipid (AL1) and an unidentified glycolipid. The species of the genus have genetic signatures that are two compensatory transversion mutations (positions 678: A and 712: T) and a single transversion mutation (position 194: T). The type species is Cohaesibacter gelatinilyticus.
Description of Cohaesibacter gelatinilyticus, Hwang and Cho (2008)
Cohaesibacter gelatinilyticus (ge.la.ti.ni.ly’ti.cus. N.L. n. gelatinum gelatin; Gr. adj. lutikos able to dissolve; N.L. adj. lyticus dissolving; N.L. masc. adj. gelatinilyticus gelatin-dissolving).
The cells are approximately 0.2–0.4 μm wide and 1.0–3.0 μm long. They are weakly motile by a polar flagellum. Reproduction occurs by bussing, binary fission, or asymmetric division. Rosette formation occurs. No growth occurs on trypticase soy agar (TSA), fivefold-diluted TSA, Czapek–Dox agar, MacConkey agar, blood agar, or the above media supplemented with either 3 % (w/v) NaCl or 3 % (w/v) sea salts. Circular, entire, convex and creamy white colonies appear on marine agar 2216 or R2A agar supplemented with 3 % (w/v) NaCl. At optimal growth conditions, colonies are approximately 2 mm in diameter after incubation for 1 week. Intracellular granules of poly-β-hydroxybutyrate are formed. Growth occurs on acetate, α-ketobutyric acid, citrate, d-fructose, d-glucose, mannitol, d-mannose, ribose, sorbitol, glutamic acid, glycerol, glycogen, inositol, inulin, l-arginine, l-asparagine, l-lysine, l-ornithine, N-acetylglucosamine, polyethylene glycol, l-pyruvate, sodium succinate, sucrose, thiamine, casamino acids, l-proline, peptone, tryptone and yeast extract. No growth occurs on acetamide, benzoate, dl-cysteine, cellobiose, d-galactose, raffinose, salicin, trehalose, l-xylose, ethanol, formic acid, glycine, 2-propanol, d-lactose, l-arabinose, ascorbate, l-rhamnose, maleic acid, oxalic acid, salicylate, tartrate or urea.
Growth occurs in temperatures between 15 °C and 31 °C (optimum, 25–30 °C), at pH 6–9 (optimum pH 8) and NaCl concentrations of 2–5 % (w/v, optimum 3 %). Decomposition of casein, cellulose, xanthine, hypoxanthine, and hydrolyses of gelatin and aesculin occur. Negative results are observed for nitrate reduction, indole production, arginine dihydrolase and urease. Alkaline phosphatase and trypsin are present. Esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, naphthol-phosphohydrolase, and N-acetyl-β-glucosaminidase activities are weakly present. Lipase (C14), valine arylamidase, cystine arylamidase, α-chymotrypsin, α- and β-glucosidase, α- and β-galactosidase, α-mannosidase and α-fucosidase activities are absent.
Acid production from glycerol, dl-arabinose, d-ribose, dl-xylose, d-glucose, d-fructose, d-mannose, inositol, d-mannitol, d-sorbitol, N-acetylglucosamine, d-lyxose, l-fucose, potassium gluconate, and potassium 5-ketogluconate were observed. Acid production was not observed from erythritol, d-adonitol, methyl β-d-xylopyranoside, d-galactose, l-sorbose, l-rhamnose, dulcitol, methyl α-d-mannopyranoside, methyl α-d-glucopyranoside, amygdalin, arbutin, aesculin, salicin, d-cellobiose, maltose, d-lactose, melibiose, sucrose, trehalose, inulin, melezitose, raffinose, starch, glycogen, xylitol, gentiobiose, turanose, d-tagatose, d-fucose, DL-arabitol, or potassium 2-ketogluconate. The cells are sensitive to (μg per disc) gentamicin (6), cephalexin (20), vancomycin (20), mitomycin C (0.6), kanamycin (20), penicillin (6), erythromycin (10), chloramphenicol (20), ciprofloxacin (3), and ampicillin (6). Cells are resistant to tetracycline (20 μg per disc), nalidixic acid (20 μg per disc) and streptomycin (6 μg per disc).
The major fatty acids are C18 : 1ω7c (54.3–55.1 %) and C15 : 0 iso 2-OH and/or C16 : 1ω7c (summed feature 3; 19.2–20.4 %), C20 : 1ω7c (9.6–11.1 %), C18 : 0 (3.1–3.3 %), C14 : 0 3-OH and/or C16 : 1 iso I (3.0 %), C18 : 0 3-OH (1.9–2.0 %), C17 : 1ω8c (1.1–1.5 %) and C16 : 0 (1.1 %). Other minor fatty acids (<1 %) are C9 : 0, C14 : 0, C17 : 0, C19 : 0, C20 : 0, C14 : 1ω5c, C19 : 0 cyclo ω8c, C16 : 0 3-OH, C17 : 0 iso 3-OH and C19 : 0 10-methyl. Besides the polar lipids of the genus, C. gelatinilyticus has also minor amounts of an unidentified aminolipid (AL2) and unidentified lipids (L1–4). The type strain, CL-GR15T (= KCCM 42319T = DSM 18289T), was isolated from coastal waters of the east coast of Korea.
Description of Cohaesibacter marisflavi, Qu et al. (2011)
Cohaesibacter marisflavi (ma.ris.fla’vi. L. neut. n. mare –is the sea; L. adj. flavus -a -um yellow; N.L. gen. n. marisflavi of the Yellow Sea, referring to the isolation of the type strain).
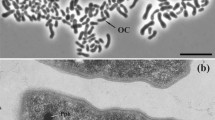
The cells are Gram-negative, catalase-negative, oxidase-positive, facultative anaerobic rods that are 0.6–0.7 mm wide and 1.8–2.0 mm long (Fig. 8.2 ). Cells are motile by a single polar flagella. They reproduce by binary fission or asymmetrical division. Colonies, which grow on 2216E agar medium, are white, smooth, circular, lightly transparent, and 1.0–2.0 μm in diameter after 3–5 days of cultivation at 30 °C. Intracellular poly-b-hydroxybutyrate granules are accumulated. Positive results were obtained in API 20NE tests for indole production, utilization of d-glucose, d-mannose, malic acid, N-acetylglucosamine, and trisodium citrate and weakly positive for utilization of β-galactosidase, maltose and l-arabinose; results were negative for gelatin hydrolysis, d-glucose fermentation, arginine dihydrolase, and utilization of d-mannitol, potassium gluconate, capric acid, adipic acid, and phenylacetic acid. In the Biolog GN2 system, the tests were positive for utilization of dextrin, d-fructose, raffinose, glycerol, l-arabinose, N-acetyl-d-glucosamine and sucrose. Growth occurs in temperatures between 10 °C and 38 °C (optimum 25–30 °C) at pH 4–9 (optimum pH 7–8) and in 0.5–15 % (w/v) NaCl (optimum 3 %).
Transmission electron micrograph of negatively stained cells of strain CL-GR15T. Bar, 1 μm (From Hwang and Cho (2008), with permission)
In the API ZYM test strip, the results were positive for esterase (C4), naphthol-AS-BIphosphohydrolase, and trypsin; weakly positive for acid phosphatase, alkaline phosphatase, esterase lipase (C8), and leucine aminopeptidase; and negative for cystine aminopeptidase, lipase (C14), N-acetyl-β-glucosaminidase, valine aminopeptidase, α-chymotrypsin, α-fucosidase, α-galactosidase, α-glucosidase, α-mannosidase, β-galactosidase, β-glucosidase, and β-glucuronidase. They also have urease and β-glucosidase (aesculin hydrolysis) activities and nitrate reduction. Cells are sensitive to (μg per disc, unless noted) ampicillin (10), carbenicillin (100), cefalexin (30), chloramphenicol (30), ciprofloxacin (5), doxycycline (30), erythromycin (15), kanamycin (30), nalidixic acid (30), neomycin (30), novobiocin (30), penicillin G (10 IU), rifampicin (5), streptomycin (10), and tetracycline (30). Cells are resistant to (μg per disc, unless noted) clindamycin (2), lincomycin (2), oxacillin (1), polymyxin B (300 IU), and vancomycin (30).
The major fatty acids are C18 : 1ω7c, C18 : 0, C16 : 0 and C16 : 1ω7c/iso-C15 : 0 2-OH. Ubiquinone 10 is the major quinone. The DNA G+C content is 55.2 mol%. The type strain, DQHS21T (=CGMCC 1.9157T = NCCB 100300T) was isolated from sediment sampled from a seawater pond used for sea cucumber culture at Jimo, Qingdao province, China, on the west coast of the Yellow Sea.
Genus Breoghania, Gallego et al. (2010)
Breoghania (Bre.o.gha’ni.a. N.L. fem. n. Breoghania, named after Breoghan, according to Celtic mythology (Leabhar Ghabhala, XII century), the first Celtic king of Gallaecia (actual Galicia), founder of the city of Brigantia (probably A. Coruña) who built a tower on the coast from where Eire (Ireland) could be seen.
The members of this genus are Gram-negative, aerobic rods that are motile by polar flagella. Cellular division occurs by binary fission or asymmetric division. They present oxidase and catalase activity. The DNA G+C content of the type strain is 63.9 mol%. The respiratory quinone is ubiquinone 10 (Q-10). The predominant fatty acid is C18:1ω7c (75.3 %); and other fatty acids found in smaller amounts are C19:0 cycloω8c and C16:0. The major polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylmonomethylethanolamine, and phosphatidylcholine. The type species is Breoghania corrubedonensis.
Description of Breoghania corrubedonensis, Gallego et al. (2010)
Breoghania corrubedonensis (Co.ru.be.do.nen’sis N.L. fem. adj. corrubedonensis, of or belonging to Corrubedo, northwest Spain, isolated from the beach of Corrubedo, the location of the sand sample that was used to inoculate the enrichment cultures from which strain UBF-P1T was isolated).
Cells are regular, irregular, or bulbous rods that are approximately 0.6–0.7 μm wide and 2–3.5 μm long (Fig. 8.3 ). Cells are motile by one or two subpolar flagella. Reproduction occurs by binary fission or asymmetric division. Circular, entire, convex, mucoid, and creamy white colonies appear on LB ASW, LB 3 % NaCl, or marine agar. After incubation for 5 days at optimal growth conditions, colonies are approximately 1 mm in diameter. Growth occurs on glucose, arabinose, mannitol, d-sorbitol, adipate, gluconate (w), phenyl acetate, acetate, succinate, malate, pyruvate, Casamino acids (Difco), acetone, and Tween 20. No growth occurs on d-xylose, mannose, maltose, starch, caprate, N-acetyl-glucosamine, citrate, lactate, oxalacetate, propionate, or methanol. Hydrolysis of gelatine or aesculin does not occur. Nitrate is reduced to nitrite, but nitrite reduction does not occur, nor does indole production or H2S formation. β-Glucosidase and β-galactosidase are not produced. Starch, DNA, and Tween 80 are not degraded. Growth occurs in temperatures between 15 °C and 40 °C (optimum 30 °C), at pH 5–8.5 (optimum 7.5) and with NaCl concentrations of 1–10 %. Aminopeptidase, arginine dihydrolase, ornithine decarboxylase, urease, catalase, oxidase, and nitrate reduction are observed. Acid production was weak produced from l-arabinose.
Transmission electron micrographs of negatively stained cells of strain UBFP1T (From Gallego et al. (2010), with permission)
The major fatty acids are C18:1ω7c (75.3 %), C19:0 cyclo ω8c (6.4 %), C16:0 (4.4 %), C16:1ω7c and/or C15:0 iso 2-OH (summed feature 3, 2.5 %), C18:0 3-OH (2.4 %), C18:0 (2.2 %), and C20:1ω9c (1.3 %). Other minor (<1 %) fatty acids are C14:0 3-OH or C16:1 iso I (summed feature 2), C17:1ω8c, C17:1ω6c, and C17:0. The major polar lipids are diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), phosphatidylmonomethylethanolamine (PME), and phosphatidylcholine (PC). The DNA G+C content is 63.9 mol%. The type strain was isolated from the beaches of Corrubedo in northwest Spain. It has been deposited in three culture collections under the numbers CECT 7622, LMG 25482, and DSM 23382.
Remarks
The whole genome sequencing of the type strain Cohaesibacter gelatinilyticus CL-GR15 is in progress. The South Korean project is sequencing the genome using Illumina Hiseq 2000, 454-GS-FLX (http://www.genomesonline.org/cgi-bin/GOLD/GOLDCards.cgi?goldstamp=Gi0034926).
References
Euzéby J (2011) Notification of changes in taxonomic opinion previously published outside the IJSEM. Int J Syst Evol Microbiol 61:6–7
Gallego S, Vila J, Nieto JM, Urdiain M, Rossello-Mora R, Grifolla M (2010) Breoghania corrubedonensis gen. nov sp nov., a novel alphaproteobacterium isolated from a Galician beach (NW Spain) after the prestige fuel oil spill, and emended description of the family Cohaesibacteraceae and the species Cohaesibacter gelatinilyticus. Syst Appl Microbiol 33:316–321
Hwang CY, Cho BC (2008) Cohaesibacter gelatinilyticus gen. nov., sp nov., a marine bacterium that forms a distinct branch in the order Rhizobiales, and proposal of Cohaesibacteraceae fam. nov. Int J Syst Evol Microbiol 58:267–277
Qu LY, Lai QL, Zhu FL, Hong XG, Sun XQ, Shao ZZ (2011) Cohaesibacter marisflavi sp. nov., isolated from sediment of a seawater pond used for sea cucumber culture, and emended description of the genus Cohaesibacter. Int J Syst Evol Microbiol 61:762–766
Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer K, Glöckner FO et al (2010) Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses. Syst Appl Microbiol 33:291–299
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this entry
Cite this entry
Rua, C.P.J., Thompson, F. (2014). The Family Cohaesibacteraceae: The Genera Cohaesibacter and Breoghania. In: Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-30197-1_374
Download citation
DOI: https://doi.org/10.1007/978-3-642-30197-1_374
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-30196-4
Online ISBN: 978-3-642-30197-1
eBook Packages: Biomedical and Life SciencesReference Module Biomedical and Life Sciences