Abstract
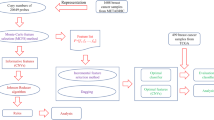
Genomic variation in DNA can cause many types of human cancer so the machine learning has important role in genomic medicine it can help to classify, predict and analysis of DNA sequence. Which is the most important biological characteristic? DNA copy number variations (CNVs) used to understand the difference between different human cancers and predict cancer causing from genetic sequence. But it’s not easy due to the high dimensionality of the CNV features. This paper presents approach to computationally classify a set of human cancer types. We use machine learning to train and test various models on set of human cancer using the CNV level values of 23,082 genes (features) for 2916 instances to construct the classifier. Then the genes are selected according to their importance by the filter feature selection method. We compare the performance of seven classifiers Support vector Machine, Random Forest, j48, Neural Network, Logistic Regression, Bagging and Dagging with other benchmark using 10-fold cross validation. The best performance achieved accuracy value 0.859 and ROC value 0.965 which are promising results. The classification models developed in this research could provide a reasonable prediction of the cancer patients’ stage based on their CNV level values. The proposed model confirmed that genes from chromosome 3 have in developing human cancers. It also predicted new genes not studied so far as important ones for the prediction of human cancers.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Barutcuoglu, Z., Schapire, R.E., Troyanskaya, O.G.: Hierarchical multi-label prediction of gene function. Bioinformatics 22(7), 830–836 (2006)
Tsuda, K., Shin, H.J., Scholkopf, B.: Fast protein classi_cation with multiple networks. Bioinformatics 21(2), 59–65 (2005). Joint Meeting of the 4th European Conference on Computational Biology/6th Meeting of the Spanish-Bioinformatics-Network, Madrid, Spain, 28 Sept–01 Oct (2005)
Li, J., Li, X., Su, H., Chen, H., Galbraith, D.W.: Framework of integrating gene relations from heterogeneous data sources: an experiment on Arabidopsis thaliana. Bioinformatics 22(16), 2037–2043 (2006)
Friedberg, E.C., Walker, G.C., Siede, W., Wood, R.D.: DNA Repair and Mutagenesis. American Society for Microbiology Press, Washington (2005)
Ciriello, G., Miller, M.L., Aksoy, B.A., Senbabaoglu, Y., Schultz, N., Sander, C.: Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 45, 1127–1133 (2013)
Cerami, E., Gao, J., Dogrusoz, U., Gross, B.E., Sumer, S.O., Aksoy, B.A., Jacobsen, A., Byrne, C.J., Heuer, M.L., Larsson, E.: The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012)
Gao, J., Aksoy, B.A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S.O., Sun, Y., Jacobsen, A., Sinha, R., Larsson, E.: Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, l1 (2013)
Cortes, C., Vapnik, V.: Support-vector networks. Mach. Learn. 20(3), 273–297 (1995)
Schölkopf, B., Smola, A.J., Williamson, R.C., Bartlett, P.L.: New support vector algorithms. Neural Comput. 12(5), 1207–1245 (2000)
Zhang, N., et al.: Classification of cancers based on copy number variation landscapes. Biochimica et Biophysica Acta (BBA) Gen. Subj. 1860(11), 2750–2755 (2016)
Freedman, D.A.: Statistical Models: Theory and Practice, p. 128. Cambridge University Press, Cambridge (2009)
Walker, S.H., Duncan, D.B.: Estimation of the probability of an event as a function of several independent variables. Biometrika 54(1–2), 167–179 (1967)
Frank, B., Bermejo, J.L., Hemminki, K., Sutter, C., Wappenschmidt, B., Meindl, A., Kiechle-Bahat, M., Bugert, P., Schmutzler, R.K., Bartram, C.R.: Copy number variant in the candidate tumor suppressor gene MTUS1 and familial breast cancer risk. Carcinogenesis 28, 1442–1445 (2007)
Elia, J., Gai, X., Xie, H., Perin, J., Geiger, E., Glessner, J.: M. D’arcy, E. Frackelton, C. Kim, F. Lantieri, Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 15, 637–646 (2010)
Li, X.C., Liu, C., Huang, T., Zhong, Y.: The occurrence of genetic alterations during the progression of breast carcinoma. Biomed. Res. Int. 2016, 5237827 (2016)
Curtis, C., Shah, S.P., Chin, S.-F., Turashvili, G., Rueda, O.M., Dunning, M.J., Speed, D., Lynch, A.G., Samarajiwa, S., Yuan, Y., Gräf, S., Ha, G., Haffari, G., Bashashati, A., Russell, R., McKinney, S., Langerød, A., Green, A., Provenzano, E., Wishart, G., Pinder, S., Watson, P., Markowetz, F., Murphy, L., Ellis, I., Purushotham, A., Børresen-Dale, A.-L., Brenton, J.D., Tavaré, S., Caldas, C., et al.: The genomic and transcriptomic architecture of 2,000 breast tumors reveals novel subgroups. Nature 486, 346–352 (2012)
Ali, H.R., Rueda, O.M., Chin, S.-F., Curtis, C., Dunning, M.J., Aparicio, S.A., Caldas, C.: Genome-driven integrated classification of breast cancer validated in over 7,500 samples. Genome Biol. 15, 431 (2014)
List, M., Hauschild, A.-C., Tan, Q., Kruse, T.A., Mollenhauer, J., Baumbach, J., Batra, R.: Classification of breast cancer subtypes by combining gene expression and DNA methylation data. J. Integr Bioinform. 11, 236 (2014)
Hall, M.A.: Correlation-based feature selection for machine learning. Technical report, Department of Computer Science, University of Waikato (1998)
Chizi, B., Maimon, O.: Dimension reduction and feature selection. In: Data Mining and Knowledge Discovery Handbook, pp. 83–100. Springer, New York (2010)
Chinnadurai, G.: The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 69, 731–734 (2009)
Huang, M.-Y., Wang, J.-Y., Chang, H.-J., Kuo, C.-W., Tok, T.-S., Lin, S.-R.: CDC25A, VAV1, TP73, BRCA1 and ZAP70 gene overexpression correlates with radiation response in colorectal cancer. Oncol. Rep. 25, 1297–1309 (2011)
Cristiana, L.N.: New insights into P53 signalling and cancer: implications for cancer therapy. J. Tumor 2 (2014)
Wen, H., Li, Y., Xi, Y., Jiang, S., Stratton, S., Peng, D., Tanaka, K., Ren, Y., Xia, Z., Wu, J.: ZMYND11 links histone H3. 3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 (2014)
Lorincz, A.T.: Cancer diagnostic classifiers based on quantitative DNA methylation. Expert. Rev. Mol. Diagn. 14, 293–305 (2014)
Sengupta, N., Yau, C., Sakthianandeswaren, A., Mouradov, D., Gibbs, P., Suraweera, N., Cazier, J.-B., Polanco-Echeverry, G., Ghosh, A., Thaha, M.: Analysis of colorectal cancers in British Bangladeshi identifies early onset, frequent mucinous histotype and a high prevalence of RBFOX1 deletion. Mol. Cancer 12, 1 (2013)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Elsadek, S.F.A., Makhlouf, M.A.A., Aldeen, M.A. (2019). Supervised Classification of Cancers Based on Copy Number Variation. In: Hassanien, A., Tolba, M., Shaalan, K., Azar, A. (eds) Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2018. AISI 2018. Advances in Intelligent Systems and Computing, vol 845. Springer, Cham. https://doi.org/10.1007/978-3-319-99010-1_18
Download citation
DOI: https://doi.org/10.1007/978-3-319-99010-1_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-99009-5
Online ISBN: 978-3-319-99010-1
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)