Abstract
Cardiac resynchronization therapy (CRT) has shown its efficiency to treat patients with left-sided heart failure, however with 30% of them not responding to the therapy. One way to optimize CRT is to pre-operatively plan the implantation of the CRT device and especially the positioning of the stimulation lead pacing the left ventricle (LV), which is implanted through the coronary veins. Indeed, it has been shown that this lead should target LV sites with a late mechanical activation and without fibrosis. Additional imaging modalities should therefore be part of CRT’s planning, in order to describe the anatomy, mechanical activation, and tissue characteristics of the LV. We developed a full workflow to process, register, and fuse CT images, ultrasound (US) images, and MRI, including cine-MRI and late gadolinium enhancement (LGE) MRI. It results in a 3D patient-specific model, describing the anatomy of the LV and of the coronary veins, the electro-mechanical delays, and the presence of fibrosis. The process includes a semi-automatic segmentation of CT images to extract the LV cavity and the veins. 2D US images are processed using speckle tracking echography (STE) to estimate the mechanical strains. LGE-MRI is segmented to extract macroscopic fibrosis. All these images are registered using CT as the anatomical reference. Registration methods have thus been developed to register STE to CT, LGE to cine-MRI, and cine-MRI to CT. This whole process furnishes to the physician, before the CRT implantation, a patient-specific 3D model representing all the information needed to select the most appropriate LV pacing sites. Results obtained on patients undergoing CRT are presented.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
3.1 Introduction
Multimodal imaging has the ability to provide enhanced descriptions of organs by combining descriptors related to anatomy, function, and metabolism. Considering cardiac imaging, many different modalities are clinically used, including computed tomography (CT) imaging, magnetic resonance imaging (MRI), ultrasound (US) imaging, and nuclear imaging. All these modalities provide complementary information, opening the way towards a combined analysis of heart multiphysics that may be useful to improve the diagnosis of cardiac pathologies, plan and assist their treatment, and perform its follow-up.
Chronic heart failure (HF), affecting an estimated 26 million people worldwide [1], corresponds to the heart’s inability to pump enough blood to fulfill the body’s needs. Cardiac dyssynchrony, i.e., spatial inhomogeneity in timing of contractions, is frequently observed in these patients and leads to a poor outcome if left untreated. Cardiac resynchronization therapy (CRT) is a non-pharmacological treatment based on the implantation of a pacemaker with leads stimulating the heart. Generally, a biventricular pacing is used, with the implantation of three leads: two endocardial leads in the right atrium and ventricle, and one transvenous lead to pace the left ventricle (LV) [19]. CRT has shown its efficiency to treat patients with left-sided heart failure associated with dyssynchrony despite pharmacological therapy [23]. However, the percentage of non-responders remains high: about 30% [37]. A suboptimal position of the LV lead, that is the most difficult one to implant, may be one factor explaining non-response. Indeed, in current practice, the LV stimulation site is selected during the implantation, based on the per-operative angiographic images providing a very poor description of the left ventricle and of the coronary venous access.
On the other hand, it has been shown that cardiac sites with well-defined specific characteristics are better responders to the therapy [4, 10]. Especially, stimulating the site associated with the latest mechanical contraction improved patient outcome [28]. On the contrary, stimulating a fibrotic tissue (i.e., scarred tissue with degraded contractility) is associated with non-response to CRT [35]. In this context, generating an enhanced description of the heart would allow the clinicians to select the most appropriate pacing sites in order to plan the implantation. This description should include the mechanical contraction, the tissular properties but also a description of the LV anatomy and of the coronary veins which are used to implant the LV lead. These descriptions result from different imaging modalities, which have thus to be registered and fused.
Few works of the literature addressed the combination of multiple pre-operative modalities to plan CRT. This combination has mostly been done by using, independently for each modality, a division of the cardiac wall in segments (i.e., a 17-segment bulls-eye representation [11]) [2, 5, 13], thus providing a limited spatial accuracy. Some teams proposed to use a landmark-based registration to combine 3D echography and CT [33], or 2D echography, CT, and SPECT [31]. It provides more accurate characterizations, but is dependent on the selection of well-defined landmarks in the images. Performing an automatic registration method is challenging since the considered modalities are highly different in terms of dimension (2D vs 3D, static vs dynamic), resolution (e.g., 0.4 × 0.4 × 0.6 mm in CT versus 1.8 × 1.8 × 4 mm in cine-MRI in our case), represented information, and since the considered organ, the heart, is submitted to fast deformations, related to the cardiac contraction but also to respiration. It therefore needs the development of specific methods adapted to the characteristics of both the organ and the studied modalities.
We thus have developed a workflow to fuse multimodal cardiac images (i.e., CT, MRI and US) to generate, pre-operatively, a patient-specific model of the LV including different characteristics useful to plan CRT interventions. This model includes the anatomy of the LV and of the coronary veins, the mechanical contraction of the wall, and its tissular characteristics in terms of macroscopic fibrosis.
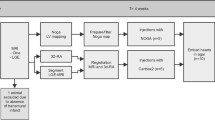
The global workflow is presented in Fig. 3.1. It is based on multimodal data processing, including the segmentation of CT images to extract the LV and veins, fibrosis characterization based on LGE-MRI, and processing of echographic data using speckle tracking echography to characterize the mechanical activation. Then, all these images are registered. These different steps have been previously presented in different publications [7,8,9, 32]. In this chapter, we summarize these processing and show how they can be integrated to generate a patient-specific 3D model which can help the clinician to select the optimal site(s) by considering the sites localized close to a vein, associated with a large mechanical delay and with the absence of fibrosis. The different processing steps are described below.
3.2 Multimodal Image Processing
The first step of the proposed workflow is to extract the descriptors of interest by processing the acquired images, namely the anatomy in CT images, mechanical activation in echography, and fibrosis in MRI. Table 3.1 summarizes the main characteristics of the acquired images.
3.2.1 Segmentation of LV and Veins in CT Images
CT imaging, because of its high spatial resolution, remains the reference modality for the anatomical description of the heart, especially considering the coronaries.
The considered CT images were acquired with a multislice computed tomography scanner (General Electric, LightSpeed VCT 64- slice system). Iodine contrast product was injected to enhance the left heart inner cavities (100 and 150 cm3 being at 300 mg/ml or 350 mg/ml). The acquisitions were retrospectively synchronized on the ECG, providing dynamic 3D+t images (10 or 20 phases describing the cardiac cycle). Since the contrast of the cavities is high, a simple region growing enables to segment the left cavities (atrium and ventricle). In case of segmentation leaks due to limited contrast, a manual correction was used to separate the cavities from neighboring structures.
The segmentation of the veins is much more challenging, but required since the LV lead implantation is done through the venous network. However, these structures are small, with a low contrast and with a variable anatomy among patients. Moreover, they are very close to arteries, making them difficult to identify. Many works have been proposed in the literature for the segmentation of vessels in MR or CT imaging [21]. However, most of them are focused on the arteries, which present a higher contrast than the veins. Moreover many methods fail to segment the vessels of medium and low diameter, which are of interest for the implantation. We thus used a semi-automatic process [9] including, for each slice containing a specific vein (coronary sinus, great cardiac vein, middle cardiac vein, posterior vein, or left marginal vein): the manual selection of a region of interest; a filtering to enhance tubular structures; a thresholding to identify the tubular objects; a connexity based object identification considering either the largest object or the one related an object previously segmented in the neighboring slices. In case of missing segments, the vein was finally interpolated.
Manual Selection of a Region of Interest
For each vein, the user defined a 2D region of interest on a slice selected such that the vein is visible. It is then extended to a volume of interest by including the three superior and inferior slices.
Tubular Structures Filter
The VOI was filtered using the Frangi filter [12], a filter based on the Hessian matrix of the image intensity, intended to enhance the tubular structures. In order to detect vessels of different sizes, a multi-scale approach was used by filtering the VOI with a set of Gaussian filters of different sizes (standard deviation in the range [1, 3] pixels). For each pixel, the highest value resulting from the different Gaussian smoothing was retained.
Thresholding and Connexity Based Object Identification
The resulting filtered ROI was thresholded using an interactively selected threshold T. The resulting binary region was analyzed to keep either the largest region (if the current slice is the first one to be processed), or the region connected to the vein segmented in the previous slice.
Interpolation of the Vein
In some cases, the whole vein could not be extracted using the previous process and some intermediate parts were missing between two segmented parts. Then, an interpolation was used to estimate the missing segment, based on the discretization of the contour using regularly spaced points and their interpolation with cubic splines.
3.2.2 Characterization of the Mechanical Activation Using Speckle Tracking Echography
Despite the increased attention gained by MRI for cardiac function assessment [14], the measurement of dyssynchrony remains mainly performed using echography, and especially with speckle tracking [10]. By tracking acoustic markers (speckles) in ultrasound images, speckle tracking echography (STE) provides a mechanical description of the LV contraction in terms of strain curves.
Transthoracic echocardiography (Vivid7, GE Healthcare, Horten, Norway) was acquired at rest. 2D acquisitions were realized using two- and four-chamber apical views over a cardiac cycle. On each view, the endocardium wall was manually segmented on diastole by the expert and tracked using speckle tracking (General Electric ECHOPAC workstation). The strain curves, computed on regularly spaced points on the endocardium, were then automatically computed by the speckle tracking software, and exported. Different descriptors can then be extracted from these strain curves, especially the maximal strain value, to identify areas with low contractility, and the delay between the peak of the QRS complex and the maximal strain (electro-mechanical delay) to identify the delayed segments.
3.2.3 Characterization of Fibrosis in LGE-MRI
LGE-MRI enables to identify macroscopic fibrosis (i.e., replacement of myocytes after cell damage) by using a gadolinium contrast agent injection and by imaging the increased amount of gadolinium in fibrous tissue [25].
LGE- and cine-MRI were acquired using cardiac SENSE coils (Philips Achieva®, Philips Medical Systems, Best, The Netherlands). Breath-hold cine (bTFE sequence) and LGE (TFE) were acquired for SAX and long axis views (2CH,4CH), using retrospective ECG synchronization. Both inversion recovery (IR) and phase-sensitive IR (PSIR) techniques were used to acquire LGE. 12 slices were acquired for SAX and 3–5 for 2CH/4CH. Multiview LGE was acquired 5–10 min after gadolinium injection (0.1 mmol/kg of Dotarem Guerbet, Roissy, France), at mid-diastole. 16 slices were acquired for SAX and 12 for 2CH/4CH.
If needed, motion artifacts were corrected using an in-house developed software presented in [7].
In order to segment fibrosis areas in LGE images, the inner and outer contours of the myocardium were firstly manually roughly segmented. Then, a fuzzy c-means algorithm was applied to the myocardium and blood pool to associate, to each voxel, a probability to belong to the gadolinium-enhanced class [18]. The membership probability was then automatically thresholded on the myocardium by using the threshold value providing the most stable output [3].
3.3 Multimodal Image Registration
Once all the images have been processed, they were registered and fused to generate the patient-specific 3D model incorporating all the descriptors. The CT image, providing the best spatial resolution, was considered as the anatomical reference. MRI and STE were therefore registered towards it. Since these three modalities embed dynamic data’s acquisition (cine sequence in MRI), the proposed registration process takes advantage of all the available information by combining a temporal synchronization with the spatial registration, thus enabling to register all the phases of the dynamic images in a single process.
3.3.1 STE to CT Registration
Few works have been proposed in the literature for cardiac multimodal registration considering 2D [24] or 3D US data [22, 27]. The goal of these works is mainly to compare pre- and per-operative data during surgical gestures guided by US [36], or to combine complementary descriptors to help in diagnosis [34]. The temporal synchronization of data is often the first problem to solve. Some works use the ECG, by considering only one instant related to the same cardiac phase [20], whereas others use a dynamic registration relying on an interpolation process [16, 34]. Then, to perform the geometrical matching, some authors considered either iconic methods based on mutual information [17], or geometrical methods based on geometrical landmarks [15, 30], or modelling methods based on the simulation of US images from CT data [34].
We developed a method based on Fourier descriptors to handle differences in spatial resolution and dynamic time warping (DTW) [29] to deal with differences of cardiac rates and ECG sampling frequencies.
The dynamic geometry of the ventricle was available in both STE and CT with a 2D dynamic contour for STE and a 3D dynamic surface for CT. The proposed process combined, in an iterative way, synchronization and registration [7, 32] (cf. Fig. 3.2a). At each iteration, the position of the echographic acquisition plane, expressed in the CT image coordinate system, was refined. This position enabled to generate a cut of the CT dynamic surface, resulting to a dynamic contour. The registration metric, used to express the quality of the registration, was thus defined between the two dynamic contours, from CT and from STE. In order to handle the differences between both contours (i.e., differences in smoothness), they were expressed in terms of Fourier descriptors (maximum order: 50). They were then synchronized by using DTW applied to the Fourier decompositions of both 2D dynamic contours. The final registration metric was, after synchronization, the sum of the differences of the Fourier descriptors.
3.3.2 LGE-MRI to CT Registration
LGE-MRI was registered to CT images by considering cine-MRI as an intermediate (cf. Fig. 3.2b). Indeed, cine-MRI is consistent with LGE-MRI and, by including the temporal dimension, provides more information than LGE-MRI for the registration towards CT images. LGE-MRI was rigidly registered towards the closest phase in cine-MRI by using normalized mutual information (NMI).
The registration of cine-MRI and CT is challenging mainly because of the temporal synchronization. If the ECG is used during the acquisition of both images, it may be deformed by the MRI magnetic field and is not always available after the acquisition.
Thus, cine-MRI and CT images were registered in two steps [8]: (1) temporal synchronization based on image information; (2) rigid registration of the synchronized sequences using a multi-image metric.
The temporal synchronization was performed by computing normalized cross-correlation (NCC) curves of input image sequences. One NCC curve was firstly computed for each image sequence, defined by the normalized cross-correlation between the first image of the sequence and each image of the same sequence. The two resulting curves have proven to globally describe the cardiac dynamics in dynamic CT and cine-MRI [6, 26]. The nonlinear synchronization of corresponding cardiac dynamics was then estimated by warping the time axes of these curves, using a DTW procedure. A multi-image rigid registration procedure, using normalized mutual information (NMI) as the metric, was finally used to align dynamic CT and cine-SAX (short axis) with synchronized cardiac dynamics.
3.4 Patient-Specific 3D Model Generation
The patient-specific model results from the registration of STE and LGE-MRI towards the planning CT image. Thus, the anatomical description (LV endocardium and veins) is enhanced by the mechanical activation (delay to the peak of strain and peak amplitude, defined on the 2D ultrasound acquisition planes) from STE and by segmented fibrosis provided by LGE-MRI.
The 2D mechanical descriptors have been represented by spheres, positioned on the endocardium along the US acquisition planes, and whose color represents the associated value. Considering the fibrosis, it is represented using a 3D volume corresponding to the localization and extent of fibrosis in the myocardium.
3.5 Results
The different steps presented above have been evaluated:
-
on simulated data: for STE to CT registration [32];
-
on real data:
Further evaluations could be performed, especially considering CT and fibrosis segmentations, by comparing the results with manual delineations from experts.
Hereafter, the results obtained on the real data of two patients undergoing a CRT procedure are presented. All the processing results, including CT and MRI segmentations, STE processing and images registrations have been validated by an clinical expert. All the images have been acquired 2 days before the CRT implantation (day D-2) and processed the day before (D-1), in order to be presented to the cardiologist before the implantation.
Figures 3.3, 3.4, and 3.5 illustrate the workflow’s different steps for the first patient. Figure 3.3 shows one slice of the CT image and the segmentation result with the left cavities (ventricle and atrium) and the veins. The veins of interest for the LV lead implantation are represented in green, the others in blue. Figure 3.4 presents the echographic image and the speckle tracking result. Figure 3.5 illustrates the MRI data (cine-MRI and LGE-MRI) and the result of the fibrosis segmentation process.
The developed registration workflow results in the illustrated models in Figs. 3.6 and 3.7 for the two patients. While the first patient has a large area of fibrosis, which has to be avoided for the left lead implantation, the second patient shows no tissular defects. The model thus enables the clinician to identify the cardiac sites accessible through a venous access and associated with no fibrosis and with a delayed mechanical contraction.
3.6 Conclusion
Multimodal cardiac imaging, by combining complementary descriptors (e.g., anatomical, mechanical, and tissular), has the ability to provide an improved characterization of the heart which may be useful in the management of cardiac diseases. However, some specific processes including segmentation, characterization, registration, and fusion have to be developed, evaluated, and combined to provide data management workflows adapted to the clinical needs and requirements.
In this work, we developed a workflow to assist the clinicians in planning CRT procedures. It involves the processing and registration of CT, MRI, and STE images, and the generation of a pre-operative patient-specific model. This model provides different information needed to define cardiac sites likely to respond positively to the stimulation: anatomical description of the left ventricle and the veins to assess the venous access, mechanical contraction to assess the delayed sites, and fibrosis detection to avoid the sites with poor tissular characteristics for CRT.
This process has been implemented in clinical conditions. It should allow the clinician to target the best pacing sites and to evaluate their accessibility through the venous network. This project needs further clinical evaluation, e.g., by evaluating if the proposed planning platform enables to reduce the intervention time and the rate of non-responding patients.
In the future, this workflow may be tuned and enriched to be extended to other pathologies and interventional gestures, such as the ablation of ventricular tachycardia or of atrial fibrillation.
References
A.P. Ambrosy, G.C. Fonarow, J. Butler, O. Chioncel, S.J. Greene, M. Vaduganathan, S. Nodari, C.S. Lam, N. Sato, A.N. Shah, M. Gheorghiade, The global health and economic burden of hospitalizations for heart failure: lessons learned from hospitalized heart failure registries. J. Am. Coll. Cardiol. 63(12), 1123–1133 (2014)
Z. Bakos, H. Markstad, E. Ostenfeld, M. Carlsson, A. Roijer, R. Borgquist, Combined preoperative information using a bullseye plot from speckle tracking echocardiography, cardiac CT scan, and MRI scan: targeted left ventricular lead implantation in patients receiving cardiac resynchronization therapy. Eur. Heart J. Cardiovasc. Imaging 15(5), 523–531 (2014)
N. Baron, N. Kachenoura, P. Cluzel, F. Frouin, A. Herment, P. Grenier, G. Montalescot, F. Beygui, Comparison of various methods for quantitative evaluation of myocardial infarct volume from magnetic resonance delayed enhancement data. Int. J. Cardiol. 167(3), 739–744 (2013)
J.M. Behar, S. Claridge, T. Jackson, B. Sieniewicz, B. Porter, J. Webb, R. Rajani, S. Kapetanakis, G. Carr-White, C.A. Rinaldi, The role of multi modality imaging in selecting patients and guiding lead placement for the delivery of cardiac resynchronization therapy. Expert. Rev. Cardiovasc. Ther. 15(2), 93–107 (2017)
M. Bertini, D. Mele, M. Malagù, A. Fiorencis, T. Toselli, F. Casadei, T. Cannizzaro, C. Fragale, A. Fucili, E. Campagnolo et al., Cardiac resynchronization therapy guided by multimodality cardiac imaging. Eur. J. Heart Fail. 18(11), 1375–1382 (2016)
J. Betancur, A. Simon, F. Tavard, B. Langella, C. Leclercq, M. Garreau, Segmentation-free MRI to CT 3d registration for cardiac resynchronization therapy optimization, in Computing in Cardiology Conference (IEEE, Piscataway, 2012), pp. 701–704
J. Betancur, A. Simon, E. Halbert, F. Tavard, F. Carré, A. Hernández, E. Donal, F. Schnell, M. Garreau, Registration of dynamic multiview 2D ultrasound and late gadolinium enhanced images of the heart: application to hypertrophic cardiomyopathy characterization. Med. Image Anal. 28, 13–21 (2016)
J. Betancur, A. Simon, B. Langella, C. Leclercq, A. Hernández, M. Garreau, Synchronization and registration of cine magnetic resonance and dynamic computed tomography images of the heart. IEEE J. Biomed. Health Informatics 20(5), 1369–1376 (2016)
S. Bruge, A. Simon, M. Lederlin, J. Betancur, A. Hernandez, E. Donal, C. Leclercq, M. Garreau, Multi-modal data fusion for cardiac resynchronization therapy planning and assistance, in 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (IEEE, Piscataway, 2015), pp. 2391–2394
P. Carità, E. Corrado, G. Pontone, A. Curnis, L. Bontempi, G. Novo, M. Guglielmo, G. Ciaramitaro, P. Assennato, S. Novo et al., Non-responders to cardiac resynchronization therapy: insights from multimodality imaging and electrocardiography. A brief review. Int. J. Cardiol. 225, 402–407 (2016)
M.D. Cerqueira, N.J. Weissman, V. Dilsizian, A.K. Jacobs, S. Kaul, W.K. Laskey, D.J. Pennell, J.A. Rumberger, T. Ryan, M.S. Verani et al., Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. Circulation 105(4), 539–542 (2002)
A.F. Frangi, W.J. Niessen, K.L. Vincken, M.A. Viergever, Multiscale vessel enhancement filtering, in Medical Image Computing and Computer-Assisted Intervention (MICCAI) (Springer, Berlin, 1998), pp. 130–137
O. Goitein, J.M. Lacomis, J. Gorcsan, D. Schwartzman, Left ventricular pacing lead implantation: potential utility of multimodal image integration. Heart Rhythm 3(1), 91–94 (2006)
B. Heydari, M. Jerosch-Herold, R.Y. Kwong, Imaging for planning of cardiac resynchronization therapy. JACC: Cardiovasc. Imaging 5(1), 93–110 (2012)
J. Hong, K. Konishi, H. Nakashima, S. Ieiri, K. Tanoue, M. Nakamuta, M. Hashizume, Integration of MRI and ultrasound in surgical navigation for robotic surgery, in World Congress on Medical Physics and Biomedical Engineering, pp. 3052–3055 (2007)
X. Huang, N. Hill, J. Ren, G. Guiraudon, T. Peters, Intra-cardiac 2D US to 3D CT image registration, in SPIE Medical Imaging, vol. 6509, pp. 65092E–1 (2007)
X. Huang, J. Moore, G. Guiraudon, D. Jones, D. Bainbridge, J. Ren, T. Peters, Dynamic 2D ultrasound and 3D CT image registration of the beating heart. IEEE Trans. Med. Imaging 28(8), 1179–1189 (2009)
N. Kachenoura, A. Redheuil, A. Herment, E. Mousseaux, F. Frouin, Robust assessment of the transmural extent of myocardial infarction in late gadolinium-enhanced MRI studies using appropriate angular and circumferential subdivision of the myocardium. Eur. Radiol. 18(10), 2140–2147 (2008)
C. Leclercq, S. Cazeau, P. Ritter, C. Alonso, D. Gras, P. Mabo, A. Lazarus, J. Daubert, A pilot experience with permanent biventricular pacing to treat advanced heart failure. Am. Heart J. 140(6), 862–870 (2000)
M. Ledesma-Carbayo, J. Kybic, M. Desco, A. Santos, M. Suhling, P. Hunziker, M. Unser, Spatio-temporal nonrigid registration for ultrasound cardiac motion estimation. IEEE Trans. Med. Imaging 24(9), 1113–1126 (2005)
D. Lesage, E.D. Angelini, I. Bloch, G. Funka-Lea, A review of 3D vessel lumen segmentation techniques: models, features and extraction schemes. Med. Image Anal. 13(6), 819–845 (2009)
F. Li, P. Lang, M. Rajchl, E. Chen, G. Guiraudon, T. Peters, Towards real-time 3D US-CT registration on the beating heart for guidance of minimally invasive cardiac interventions, in SPIE Medical Imaging, pp. 831615–831615 (2012)
C. Linde, W.T. Abraham, M.R. Gold, M.S.J. Sutton, S. Ghio, C. Daubert, Randomized trial of cardiac resynchronization in mildly symptomatic heart failure patients and in asymptomatic patients with left ventricular dysfunction and previous heart failure symptoms. J. Am. Coll. Cardiol. 52(23), 1834–1843 (2008)
T. Mäkelä, P. Clarysse, O. Sipila, N. Pauna, Q. Pham, T. Katila, I. Magnin, A review of cardiac image registration methods. IEEE Trans. Med. Imaging 21(9), 1011–1021 (2002)
N. Mewton, C.Y. Liu, P. Croisille, D. Bluemke, J.A. Lima, Assessment of myocardial fibrosis with cardiovascular magnetic resonance. J. Am. Coll. Cardiol. 57(8), 891–903 (2011)
D. Perperidis, R.H. Mohiaddin, D. Rueckert, Spatio-temporal free-form registration of cardiac MR image sequences. Med. Image Anal. 9(5), 441–456 (2005)
E. Puyol-Antón, M. Sinclair, B. Gerber, M.S. Amzulescu, H. Langet, M. De Craene, P. Aljabar, P. Piro, A.P. King, A multimodal spatiotemporal cardiac motion atlas from MR and ultrasound data. Med. Image Anal. 40, 96–110 (2017)
S. Saba, J. Marek, D. Schwartzman, S. Jain, E. Adelstein, P. White, O.A. Oyenuga, T. Onishi, P. Soman, J. Gorcsan, Echocardiography-guided left ventricular lead placement for cardiac resynchronization therapy results of the speckle tracking assisted resynchronization therapy for electrode region trial. Circ. Heart Fail. 6(3), 427–434 (2013)
H. Sakoe, S. Chiba, Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans. Acoust. Speech Signal Process. 26(1), 43–49 (1978)
A. Savi, M. Gilardi, G. Rizzo, M. Pepi, C. Landoni, C. Rossetti, G. Lucignani, A. Bartorelli, F. Fazio, Spatial registration of echocardiographic and positron emission tomographic heart studies. Eur. J. Nucl. Med. Mol. Imaging 22(3), 243–247 (1995)
A. Sommer, M.B. Kronborg, B.L. Nørgaard, S.H. Poulsen, K. Bouchelouche, M. Böttcher, H.K. Jensen, J.M. Jensen, J. Kristensen, C. Gerdes et al., Multimodality imaging-guided left ventricular lead placement in cardiac resynchronization therapy: a randomized controlled trial. Eur. J. Heart Fail. 18(11), 1365–1374 (2016)
F. Tavard, A. Simon, C. Leclercq, E. Donal, A.I. Hernández, M. Garreau, Multimodal registration and data fusion for cardiac resynchronization therapy optimization. IEEE Trans. Med. Imaging 33(6), 1363–1372 (2014)
F. Tournoux, R.C. Chan, R. Manzke, M.D. Hanschumacher, A.A. Chen-Tournoux, O. Gérard, J. Solis-Martin, E.K. Heist, P. Allain, V. Reddy et al., Integrating functional and anatomical information to guide cardiac resynchronization therapy. Eur. J. Heart Fail. 12(1), 52–57 (2010)
W. Wein, A. Khamene, D. Clevert, O. Kutter, N. Navab, Simulation and fully automatic multimodal registration of medical ultrasound, in Medical Image Computing and Computer-Assisted Intervention (MICCAI) (Springer, Berlin, 2007), pp. 136–143
J.A. Wong, R. Yee, J. Stirrat, D. Scholl, A.D. Krahn, L.J. Gula, A.C. Skanes, P. Leong-Sit, G.J. Klein, D. McCarty et al., Influence of pacing site characteristics on response to cardiac resynchronization therapy. Circ. Cardiovasc. Imaging 6(4), 542–550 (2013)
Q. Zhang, R. Eagleson, T. Peters, Real-time visualization of 4D cardiac MR images using graphics processing units, in IEEE International Symposium on Biomedical Imaging: Nano to Macro, pp. 343–346 (2006)
Q. Zhang, Y. Zhou, C.-M. Yu, Incidence, definition, diagnosis, and management of the cardiac resynchronization therapy nonresponder. Curr. Opin. Cardiol. 30(1), 40–49 (2015)
Acknowledgements
This work was supported by the French National Research Agency (ANR) in the framework of the Investissement d’Avenir Program through Labex CAMI (ANR-11-LABX-0004). It was conducted in part in the experimental platform TherA-Image (Rennes, France), supported by Europe FEDER.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Bruge, S. et al. (2018). Multimodal Image Fusion for Cardiac Resynchronization Therapy Planning. In: Abreu de Souza, M., Remigio Gamba, H., Pedrini, H. (eds) Multi-Modality Imaging. Springer, Cham. https://doi.org/10.1007/978-3-319-98974-7_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-98974-7_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-98973-0
Online ISBN: 978-3-319-98974-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)