Abstract
Ciliated protozoa are the principal component of the rumen microbiota. They contribute significantly the digestion of ruminants. As anaerobic fermentative microorganisms, rumen ciliated protozoa produce a significant amount of hydrogen and formate. Methanogenic archaea therefore associate closely with rumen ciliated protozoa. The presence of episymbiotic methanogens in rumen ciliated protozoa has been demonstrated as early as 1980s by microscopy. The number of ciliate-associated methanogens increases from the 100 level to 104/cell of ciliates after feeding. Enhancement of hydrogen and/or formate production from the ciliates by feeding attracts free-living methanogens. There are a couple of studies about the phylogeny of the ciliate-associated methanogens based on a molecular ecological approach. A range of methanogenic archaeal 16S rDNA, representing Methanobacteriales, Methanomicrobiales and Methanosarcinales, have been detected as ciliate-associated methanogens. However, it is still difficult to draw a conclusion about a potentially specific interaction between a particular ciliate species and a species of methanogenic archaea from these limited studies.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
1 The Rumen and Ciliated Protozoa
The rumen has a great capacity to digest plant polymers with the aid of anaerobic microbiota (Hungate 1966). This microbial ecosystem allowed ruminant animals to evolve into the predominant mammals in particular environments such as the semi-arid savannas (Hoffman 1973).
It is well established that the rumen ciliate protozoa have a significant impact on feed digestion in the ruminant animals, although the elimination of the ciliated protozoa does not impair the survival of the ruminant (Ushida et al. 1991).
As an anaerobic environment, the rumen microbial ecosystem requires an electron sink other than oxygen (Wolin 1975). Methane is the prevalent electron sink in this particular ecosystem (Hungate 1966). Fermentative microorganisms, therefore, create a specific relationship with hydrogenotrophic organisms to perform the different fermentation steps.
In the rumen, ciliated protozoa are known as potent hydrogen and formate producers. One cell of an axenic culture of rumen protozoa can produce 5 nmol of hydrogen per day (Ushida and Jouany 1996; Tokura et al. 1997). Potentially, this corresponds to a daily hydrogen production of approximately 50 L in the rumen of a sheep (Ushida et al. 1996). Besides hydrogen, one axenic ciliate cell produces 100 nmol of formate, which corresponds to a daily production of about 50 mol of formate in the rumen of a sheep. Such a concentration of hydrogen and formate attracts methanogens and makes the ciliate/methanogen consortium a predominant contributor for the ruminal methanogenesis. In fact, methanogenic bacteria associated with rumen ciliates were apparently responsible for 9–25% of methanogenesis in rumen fluid (Newbold et al. 1995). Consequently, elimination of the ciliated protozoa from the rumen, the defaunation, is associated with a 30–45% reduction of ruminal methanogenesis (Ushida et al. 1996).
In the case of rumen ciliates, the elimination of methanogens causes a decrease in the degradative capacities to some extent (Table 1) (Ushida and Jouany 1996). In particular, the elimination of methanogenesis increases the hydrogen and formate production from the ciliates at least by a factor of two but sometimes to a level several times higher. This slows the fermentation process down (Wolin 1975).
2 Methanogens Associated with Rumen Ciliates
Methanogenic archaea associate closely with the rumen ciliates to facilitate the interspecies hydrogen transfer in the form of an episymbiosis or an endosymbiosis.
No free-living methanogens were detected in the protozoal fraction prepared by sedimentation (Sharp et al. 1998). Therefore, it was believed that all methanogens that are metabolically associated with the ciliates are present inside the cell or intimately attached to the cell surface of the ciliates.
Episymbiotic methanogens of rumen ciliates were microscopically observed as early as 1980 by their characteristic F420 autofluorescence (Vogels et al. 1980). Endosymbiotic methanogens were observed by an archaea-specific oligonucleotide probe approach (Finlay et al. 1994). It has been shown that these endosymbiotic methanogens are localized in the cytoplasm, not in digestive vacuoles, and adjacent to the hydrogenosomes. Interestingly, the number of the endosymbiotic methanogens exceeds the number of those attached on the cell surfaces of ciliates.
The number of ciliate-associated methanogens increased from the level of 100 to 104 most probable number (MPN)/cell of ciliate after feeding (Tokura et al. 1997). When the ciliated protozoa engulfed and fermented feed particles, the number of ciliate-associated methanogens increased. Since the maximal level was recorded shortly (1–2 h) after feeding, it is unlikely that endosymbionts grow to this level in this short period of time. Accordingly, such a rapid increase in the numbers of ciliate-associated methanogens may reflect the active attachment or vigorous engulfment of free-living methanogens. Indeed, the hydrogen supply from the ciliates strongly attracted free-living methanogens (Stumm et al. 1982).
Ciliated protozoa predate and digest engulfed bacteria as a major prey. If engulfed methanogens would be the source of the endosymbiotic methanogens, these methanogens need to be resistant against protozoal lytic activity, or they may escape from the digestion within food vacuoles. This point may be supported by the fact that anaerobic ciliates, Metopus spp. and Nyctotherus spp., which harbour methanogenic symbionts closely related to the free-living organisms (Embley and Finley 1993; van Hoek et al. 2000). One study evaluated the resistance of methanogenic archaea against the lytic activity of rumen protozoa. It was found that some of the methanogens are relatively resistant against the protozoal lytic activities; i.e. Methanosarcina barkeri DSM 800 was more resistant than Methanobrevibacter sp. MF1 (Newbold et al. 1996). DSM 800 could establish the interspecies hydrogen transfer with Polyplastron multivesiculatum (Ushida et al. 1997).
3 Detection of Methanogens Associated with Ciliates
There are a couple of studies about the phylogeny of the ciliate-associated methanogens based on molecular phylogenetic approaches (Sharp et al. 1998; Tokura et al. 1999b; Chagan et al. 1999; Ohene-Adjei et al. 2007; Regensbogenova et al. 2004; Irbis and Ushida 2004; Tymensen et al. 2012; Belanche et al. 2014). However, little information is available about the methanogens isolated from ciliates. An isolation of methanogens from washed ciliated protozoa was tried, and the strain Methanobrevibacter sp. MB9 was isolated. This isolate was phylogenetically close to Methanobrevibacter ruminantium, on the basis of morphology and 16S rRNA phylogeny (Tokura et al. 1999a). Other attempts for the isolation of symbiotic methanogens have not been found in the literature probably due to the difficulty and tediousness of the isolation procedure. Even for the free-living methanogens, relatively few had been isolated from the rumen (Janssen and Kirs 2008). The isolate Methanobrevibacter sp. MB9 uses hydrogen, formate and small amounts of 2-propanol. This substrate use, 2-propanol, is not common for ruminal Methanobrevibacter species.
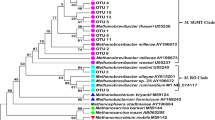
Partial sequences of 16S rDNA of ciliate-associated methanogens are available from studies in Japan and the UK (Chagan et al. 1999; Tokura et al. 1999b; Irbis and Ushida 2004; Regensbogenova et al. 2004). DNA was extracted from washed cells of ciliate protozoa. In some studies, DNA was extracted from a single cell of the ciliated protozoa. Table 2 shows the distribution of archaeal 16S sequences retrieved from the cells of ciliated rumen protozoa. The phylogenetic analyses are also shown in Fig. 1a, b. In this table, there are some unidentified strains of which strain 1Y is phylogenetically close to Methanobrevibacter gottschalkii, strain SM9 is close to Methanobrevibacter millerae, strain OCP is close to Methanobrevibacter olleyae and strain Z8 is close to Methanobrevibacter ruminantium (Rea et al. 2007; Evans et al. 2009). More recent studies (Tymensen et al. 2012) demonstrated that greater abundance of OTUs corresponding to the genus Methanobrevibacter in rumen protozoa associated methanogens (PAM) than free-living methanogens. In the latter, OTUs corresponding to the genus Methanomicrobium are present in abundance.
Neighbour-joining tree computed from partial 16S DNA of methanogens associated with rumen ciliated protozoa by MEGA 4.0 (Tamura et al. 2007) with 500 replicates for bootstrap. (a) Partial sequences (E. coli [DQ118017]16S rDNA position 781–1233) are used to analyse ciliate-associated archaea (AB189856-AB189868); (b) (E. coli [DQ118017] position 218–798) are used to analyse ciliate-associated archaea (AB022181-AB022186, AB026168-026175, AJ606400-AJ606419)
Methanobrevibacter-like sequences and those similar to Methanomicrobium were the predominant sequences detected in different studies. Methanobrevibacter sp. 1Y-like sequences were found in a range of protozoal species. Lastly, Methanomicrobium mobile (AY196679)-like sequences were detected in a range of protozoa both in Japanese and British studies. Methanogens belonging to the Methanobacteriales were detected predominantly in Japanese studies (Accession numbers start with AB; see Tokura et al. 1997, 1999a, b; Chagan et al. 1999; Irbis and Ushida 2004), while those belong to Methanomicrobiales were predominantly detected in a British study (Accession numbers start with AJ; see Regensbogenova et al. 2004). Interestingly enough, Ophryoscolex caudatum was studied in both the Japanese and British studies. This particular rumen protozoon harboured a variety of methanogens such as Methanobacteriales, Methanomicrobiales and Methanosarcinales. In addition to these methanogens, Ophryoscolex caudatum harboured also Thermoplasmatales. Other Entodiniomorphs like Polyplastron multivesiculatum, Eudiplodinium maggii, Diplodinium dentatum, Metadinium medium and Entodinium furca harboured relatively limited numbers of species of methanogens. In the case of holotrichs, Isotricha intestinalis harboured a phylogenetically broad range of methanogens similar to that shown in Ophryoscolex caudatum. An aquatic ciliate, Metopus contortus, can host a broad range of methanogens. Accordingly, this aquatic ciliate is defined as the generalist host for methanogens (Embley and Finlay 1993). Rumen ciliated protozoa like Isotricha intestinalis and Ophryoscolex caudatum can also be a generalist host for methanogens. However, it is still difficult to draw a conclusion about a potentially specific interaction between a particular ciliate species and a species of methanogenic archaea from these limited studies.
4 The Effect of Ciliated Protozoa on the Composition of Methanogenic Archaea in the Rumen
As indicated above, a cell of ciliated protozoa can harbour up to 104 methanogens.
This number may differ according to protozoal species, since protozoal size determined the number of PAM as big protozoa had 1.7–3.3 times more methanogen DNA than smaller protozoa (Belanche et al. 2014). Since the number of ciliates ranges from 105 to 106 cells/mL rumen fluid (Williams and Coleman 1991), they may encompass a methanogenic population as large as 1010 methanogens/mL. If there is a specific relationship between the ciliate species and their methanogenic symbionts, an increase in the number of ciliated protozoa should affect the methanogenic archaeal population as a selective pressure upon the methanogenic population. In an in vivo study, Ohene-Adjei et al. (2007) indicated that an inoculation of Polyplastron multivesiculatum into the rumen predominantly associated with the detection of methanogens closely related to Methanobrevibacter bryantii, Methanobrevibacter ruminantium and Methanosphaera stadtmanae. These authors also showed that inoculation of holotrich protozoa (Isotrichidae) into the rumen was primarily associated with the detection of methanogens closely related to Methanobrevibacter smithii. Although this Canadian in vivo study appears not to agree with the results shown in Table 2, it is likely that the presence of particular ciliate protozoa may promote the predominance of particular species of methanogens.
Again, the specificity of the host-methanogenic symbiont relationship is still difficult to be proven, because a long-term pure culture system for rumen ciliates has not been established so far. Without a pure culture of rumen ciliated protozoa, consisting of aposymbiotic ciliates, an inoculation study as reported for Trimyema compressum cannot be realized (Wagener et al. 1990; Holler and Pfennig 1991).
References
Belanche A, De la Fuente G, Newbold CJ (2014) Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol Ecol 90:663–677
Chagan I, Tokura M, Jouany JP, Ushida K (1999) Detection of methanogenic archaea associated with rumen ciliate protozoa. J Gen Appl Microbiol 45:305–308
Eadie JM (1967) Studies on the ecology of certain rumen ciliate protozoa. J Gen Microbiol 49:173–194
Embley TM, Finlay BJ (1993) Systematic and morphological diversity of endosymbiotic methanogens in anaerobic ciliates. Antonie van Leeuwenhoek 64:261–271
Evans PN, Hinds LA, Sly LI, McSweeney CS, Morrison M, Wright ADG (2009) Community composition and density of methanogens in the foregut of the Tammar Wallaby (Macropus eugenii). Appl Environ Microbiol 75:2598–2602
Finlay BJ, Esteban G, Clarke KJ, Williams AG, Embley TM, Hirt RP (1994) Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol Lett 117:157–161
Hoffman RR (1973) Ruminant stomach. East African Literature Bureau, Nairobi, Kenya
Holler S, Pfennig N (1991) Fermentation products of the anaerobic ciliate Trimyema compressum in monoxenic cultures. Arch Microbiol 156:327–334
Hungate RE (1966) The rumen and its microbes. Academic Press, New York
Irbis C, Ushida K (2004) Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J Gen Appl Microbiol 50:203–212
Janssen PH, Kirs M (2008) Structure of the archaeal community of the rumen. Appl Environ Microbiol 74:3619–3625
Newbold CJ, Lassalas B, Jouany JP (1995) The importance of methanogens associated with ciliate protozoa in ruminal methane production in vitro. Lett Appl Microbiol 21:230–234
Newbold CJ, Ushida K, Morvan B, Fonty G, Jouany JP (1996) The role of ciliate protozoa in the lysis of methanogenic bacteria in rumen fluid. Lett Appl Microbiol 23:421–425
Ohene-Adjei S, Teather RM, Ivanj M, Forster RJ (2007) Postinoculation protozoan establishment and association patterns of Methanogenic archaea in the ovine rumen. Appl Environ Microbiol 73:4609–4618
Rea S, Bowman JP, Popovski S, Pimm C, Wright ADG (2007) Methanobrevibacter millerae sp. nov. and Methanobrevibacter olleyae sp. nov., methanogens from the ovine and bovine rumen that can utilize formate for growth. Int J Syst Evol Microbiol 57:450–456
Regensbogenova M, McEwan NR, Javorsky P, Kisidayova S, Michalowski T, Newbold CJ, Hackstein JHP, Pristas P (2004) A re-appraisal of the diversity of the methanogens associated with the rumen ciliates. FEMS Microbiol Lett 238:307–313
Sharp R, Ziemer CJ, Stern MD, Stahl DA (1998) Taxon-specific associations between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiol Ecol 36:71–78
Stumm CK, Gijzen HJ, Vogels GD (1982) Association of methanogenic bacteria with ovine rumen ciliates. Br J Nutr 47:95–99
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tokura M, Ushida K, Miyazaki K, Kojima Y (1997) Methanogens associated with rumen ciliates. FEMS Microbiol Ecol 22:137–143
Tokura M, Tajima K, Ushida K (1999a) Isolation of Methanobrevibacter sp. as a ciliate-associated ruminal methanogen. J Gen Appl Microbiol 45:43–47
Tokura M, Chagan I, Ushida K, Kojima Y (1999b) Phylogenetic study of methanogens associated with rumen ciliates. Curr Microbiol 39:123–128
Tymensen LD, Beauchemin KA, McAllister TA (2012) Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes. Microbiology 158:1808–1817
Ushida K, Jouany JP (1996) Methane production from ciliated rumen protozoa and its effect on protozoal activity. Lett Appl Microbiol 23:129–132
Ushida K, Jouany JP, Demeyer DI (1991) Effects of presence or absence of rumen protozoa on the efficiency of utilization of concentrate and fibrous feeds. In: Tsuda T, Sasaki Y, Kawashima R (eds) Physiological aspects of digestion and metabolism in ruminants. Academic Press, San Diego, pp 626–655
Ushida K, Tokura M, Itabashi H, Takenaka A (1996) Ciliate protozoa and ruminal methanogenesis. In: Onodera R, Itabashi H, Ushida K, Yano H, Sasaki Y (eds) Rumen microbes and digestive physiology in ruminants. Japan Scientific Society Press/Karger, Tokyo, pp 209–220
Ushida K, Newbold CJ, Jouany JP (1997) Interspecies hydrogen transfer between the rumen ciliate Polyplastron multivesiculatum and Methanosarcina barkeri. J Gen Appl Microbiol 43:129–131
van Hoek AHAM, van Alen TA, Sprakel VSI, Leunissen JAM, Brigge T, Vogels GD, Hackstein JHP (2000) Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol Biol Evol 17:251–258
Vogels GD, Hoppe WF, Stumm CK (1980) Association of methanogenic bacteria with rumen ciliates. Appl Environ Microbiol 40:608–612
Wagener S, Bardele CF, Pfennig N (1990) Functional integration of Methanobacterium formicicum into the anaerobic ciliate Trimyema compressum. Arch Microbiol 153:496–501
Williams AG, Coleman GS (1991) The rumen protozoa. Springer, London
Wolin MJ (1975) Interactions between the bacterial species of the rumen. In: McDonald IW, Warner ACI (eds) Digestion and metabolism in the ruminant. University of New England Publ. Unit, Armidale, Australia
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Ushida, K. (2018). Symbiotic Methanogens and Rumen Ciliates. In: Hackstein, J. (eds) (Endo)symbiotic Methanogenic Archaea. Microbiology Monographs, vol 19. Springer, Cham. https://doi.org/10.1007/978-3-319-98836-8_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-98836-8_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-98835-1
Online ISBN: 978-3-319-98836-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)