Abstract
Liver disease affects millions of people worldwide and auto-immune disease in particular has unmet needs for improvement of non-invasive methods for risk-stratification. Especially in cases where clinical markers are inconclusive. In this study we develop novel imaging features for quantitative MRI and show that these features improve the differentiation of AIH from biliary disease in challenging cases, where including imaging features with clinical markers improved the AUROC from 0.76 to 0.85.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
1 Introduction
Chronic liver disease already affects millions of people worldwide and the incidence is increasing rapidly [8]. Auto-immune liver diseases in particular, have unmet needs for improvement in non-invasive methods of diagnosis and in risk-stratification. This umbrella term compromises three distinct chronic conditions; auto-immune hepatitis (AIH), primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC). AIH is characterised by inflammation and damage to liver cells (hepatocytes) and thus clinical markers of liver cell damages, such as the enzyme alanine transferase (ALT) are used clinically in diagnosis and monitoring of disease activity. Conversely, PBC and PSC affect the bile ducts rather than the liver parenchyma itself and thus biochemical markers of bile duct inflammation such as alkaline phosphatase (ALP) are more commonly seen in affected patients. However, differentiating between these conditions using biochemical tests is imperfect; ALP may not be particularly raised in early disease, ALT can be raised in pure biliary disorders and neither ALT or ALP are specific to these diseases only. Additionally, overlap between AIH and biliary disorders can occur. It is in such situations of diagnostic uncertainty that a liver biopsy is traditionally indicated. Percutaneous liver biopsy is unpopular with patients and is invasive, with the risk of significant complications as well as sampling error [7]. Thus, the development of non-invasive methods of differentiating between these diseases has the potential to impact positively on patient care.
An additional problem is that, on imaging, liver disease has highly variable appearance, is heterogeneous, and there is often substantial variation over time [3]. This paper explores the use of corrected T1 (cT1) MRI imaging of the liver, an indicator of inflammation and fibrosis [2, 6] that has the potential to improve diagnosis.
The combination of cT1 imaging of the liver with ALT/ALP levels is applied in this paper in an attempt to distinguish AIH from the biliary disorders: PBC and PSC. The AIH, PSC and PBC cases treated in this work range from asymptomatic to cirrhotic. We will show how the combination of image-derived measurements with ALT/ALP values results in enhanced discrimination.
2 Methods
Patients with AIH, PSC and PBC were recruited from a single ambulatory practice. They were evaluated in a dedicated study visit that included both clinical and laboratory disease phenotyping, as well as quantitative MRI imaging. More precisely, as described in fuller detail elsewhere [2], a quantitative T1 image (pixels have the dimensions of time) was Myomaps, which is based on the shMOLLI (short-time modified look-locker inversion recovery) pulse sequence. As well, a T2* image, which is related to the iron content of the liver, was developed using the Dixon multi-echo pulse sequence. The T1 image was then “corrected” to take account of the T2* to give the final corrected T1 image (cT1). It has been shown that cT1 estimates inflammation and fibrosis, particularly in early liver disease [2].
The region of the cT1 image that corresponds to the liver (with larger vessels excluded) was automatically segmented using a fully convolutional method [4]. This was performed on each slice independently. Examples of the liver segmentation are shown in Fig. 1.
In practice, the variation of PSC/PBC/AIH is such that the mean cT1, which relates to the overall burden of inflammation and fibrosis, is not sufficient to characterise disease differences. For this reason, we developed a number of additional features aiming to capture the heterogeneity of the disease. These included measures of the distribution of cT1 values in the liver region such including skewness and kurtosis, and local regional variance.
Skewness and kurtosis have been used previously to characterise disease such as progression of Gliobastomas [1]. Here we apply the metrics to quantitative imaging of the liver cross section. Skewness and kurtosis are defined as follows:
where \(p_i\) are the pixel values in the liver mask, \(\bar{p}\) is the mean, s is the standard deviation, and N is the number of superpixels. Kurtosis describes the deviation from a normal distribution. \(k=3\) is a normal distribution, and a stronger peak and a heavier tail leads to a greater kurtosis (see Fig. 1). Skewness describes the asymmetry of the distribution.
Local Variation. As well as skewness and kurtosis, the liver segmentation was parcellated into superpixels using the m-SLIC method [5]. The m-SLIC variant to SLIC creates evenly distributed superpixels inside an irregular mask. The variance in each superpixel provides a measure of texture in the liver parenchyma that is not affected by vessel and liver border transitions, and aims to capture the patterns of inflammatory liver disease in the liver parenchyma.
3 Experimental Setup
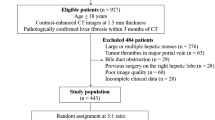
186 patients were recruited, 62 with AIH, 124 with either PBC or PSC. Mean age was 50 years (range 18–84). Scans were acquired at the Queen Elizabeth Hospital, Birmingham on a Siemens Magnetom Verio 3T MRI. Cases were scanned with the short Modified Look-Locker inversion recovery (MOLLI) sequence and cT1 was calculated [2]. 180 cases were available at the time of processing. The dataset was split randomly 60/40 into a training and test set. Parameters were optimised using Leave-One-Out cross validation (LOOCV) on the training set. The test set was only used to present the final results. The disease classification was made by hepatologists based on a combination of image characteristics and blood tests.
Linear discriminant analysis (LDA), random forests, and support vector machines were evaluated using LOOCV on the training set. LDA, the simplest classifier, outperformed the other classifiers. We assume that this was due to overfitting by the more complex classifiers, and so it was used in this analysis. The compactness and size of the superpixels were chosen based on the training set using a grid search of the parameters. We used forward feature selection with LOOCV on the training set to choose features that were effective for discriminating disease. We found that ALP had the biggest impact on the AUC, followed by kurtosis and local variation as shown in Fig. 2.
4 Results
ALT was moderately associated with clinical diagnosis (AUROC 0.75) with improvement in this when MRI derived features with machine learning was applied (AUROC 0.84). ALP alone was a good discriminator between biliary disease and auto-immune hepatitis (AUROC 0.89) with a modest improvement in this with the addition of quantitative MRI analysis (AUROC 0.91); Fig. 4. ALP is a key sign of biliary disease as shown in Fig. 3. However, in more challenging cases i.e. patients with an ALP < 200, ALP alone (25 AIH and 29 biliary cases in the validation set) was moderately correlated with diagnosis (AUROC 0.76). Addition of MRI analysis improved this considerably (AUROC 0.85, see Fig. 4).
5 Discussion
Quantitative non-biased MRI imaging features have the ability to aid in the discrimination between AIH and biliary disease, particularly in cases with lower ALP values which are more difficult to diagnose without the requirement for invasive liver biopsy. This could potentially change how liver disease is evaluated and classified in the future. In future, to further improve the results, we will also consider artefact exclusion as this could have an effect on the variation measured in the liver.
In this initial investigation, we chose features that captured both variation across the liver and local texture. We chose a limited number of features due to the size of the dataset, and, therefore, to avoid possible overfitting leading to poorer performance. However, with a larger dataset, there is potential to explore more complex radiomics-style features or learn features.
References
Baek, H.J., Kim, H.S., Kim, N., Choi, Y.J., Kim, Y.J.: Percent change of perfusion skewness and kurtosis: a potential imaging biomarker for early treatment response in patients with newly diagnosed glioblastomas. Radiology 264(3), 834–843 (2012). https://doi.org/10.1148/radiol.12112120. pMID: 22771885
Banerjee, R., et al.: Multiparametric magnetic resonance for the non-invasive diagnosis of liver disease. J. Hepatol. 60(1), 69–77 (2014)
Chundru, S., Kalb, B., Arif-Tiwari, H., Sharma, P., Costello, J., Martin, D.: MRI of diffuse liver disease: characteristics of acute and chronic diseases. Diagn. Interv. Radiol. 20(3), 200–208 (2014)
Irving, B., et al.: Deep quantitative liver segmentation and vessel exclusion to assist in liver assessment. In: Valdés Hernández, M., González-Castro, V. (eds.) MIUA 2017. CCIS, vol. 723, pp. 663–673. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-60964-5_58
Irving, B., et al.: maskSLIC: regional superpixel generation with application to local pathology characterisation in medical images. CoRR abs/1606.09518 (2017). http://arxiv.org/abs/1606.09518
Pavlides, M., et al.: Multiparametric magnetic resonance imaging predicts clinical outcomes in patients with chronic liver disease. J. Hepatol. 64(2), 308–315 (2016)
Regev, A., et al.: Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am. J. Gastroenterol. 97(10), 2614 (2002)
Wang, F.S., Fan, J.G., Zhang, Z., Gao, B., Wang, H.Y.: The global burden of liver disease: the major impact of China. Hepatology 60(6), 2099–2108 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Arndtz, K. et al. (2018). Novel Quantitative Magnetic Resonance Imaging Features with Liver Function Tests to Distinguish Parenchymal and Biliary Disease. In: Nixon, M., Mahmoodi, S., Zwiggelaar, R. (eds) Medical Image Understanding and Analysis. MIUA 2018. Communications in Computer and Information Science, vol 894. Springer, Cham. https://doi.org/10.1007/978-3-319-95921-4_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-95921-4_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-95920-7
Online ISBN: 978-3-319-95921-4
eBook Packages: Computer ScienceComputer Science (R0)







