Abstract
The absolute and relative supplies of nutrients such as nitrogen and phosphorus can affect ecosystem properties and microbial biodiversity. More recently, the theory of biological stoichiometry has advanced connections between ecosystem ecology and cellular/molecular biology by proposing a link between biochemical features of microbial cells and their nutrient ratios. Specifically, the growth rate hypothesis (GRH) postulates that cellular stoichiometry varies according to growth rate due to increased allocation to P-rich ribosomal RNA to support rapid growth. Expanding on the GRH, it is predicted that microbes have a suite of genomic features that determine the ability to achieve rapid growth and, hence, influence biomass N:P. These genomic features include codon usage bias, number of rRNA and tRNA genes, and genome size, all of which have been individually linked to growth rate and fitness. This chapter discusses two experiments conducted at the Churince system in Lagunita pond to test the GRH. Churince is an ideal location for this because surface waters in this region have highly imbalanced N:P stoichiometry (TN:TP atomic ratio >100), where P is likely to be strongly limiting. The first experiment was a replicated in situ mesocosm experiment comparing three different nutrient treatments with varying N:P to a control treatment. The second experiment was a whole-pond perturbation, fertilizing with nutrients of N:P = 16 and using metagenomics to compare responses to replicated, internal control mesocosms. We discuss changes in microbial biomass N:P, species composition, and genomic features of the microbes in response to these perturbations of nutrient supplies and N:P.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
The famous clarity of the pozas of Cuatro Ciénegas is a direct manifestation of its low-nutrient conditions (“oligotrophy ”). That is, very low concentrations of key nutrient elements (e.g., nitrogen (N) and especially phosphorus (P)) constrain the proliferation of microbes, algae, and aquatic vegetation. N and P limitation of ecosystem production is widespread globally (Elser et al. 2007), and, indeed, overcoming the low N and P status of soils via fertilizer application was essential in the “Green Revolution” that greatly enhanced agricultural production during the last century. Furthermore, not only are nutrient concentrations at CCB low, they are also imbalanced. The overall ratios of N and P are very high (often 75:1 or higher, by moles here and throughout), considerably higher than those seen, for example, in the world’s oceans (16) or greater than those thought to drive ecosystem P limitation (~28–30; Downing and McCauley 1992). So, in most of CCB’s ecosystems, we expect strong P limitation to be operating both in ecological as well as evolutionary time. While the effects of N and P limitation on both aquatic and terrestrial systems are very well-studied in various habitats, their impacts on the evolution of living things are less well-investigated. Some evidence of impacts of nutrient limitation on evolutionary processes can be seen in a variety of studies and in particular in patterns of biomass C:N:P ratios in various biota. For example, there are phylogenetic differences in the C:N:P ratios of terrestrial insects (Woods et al. 2004; Fagan et al. 2002), poleward trends in P content (C:P ratio) in congeneric taxa of Daphnia (Elser et al. 2000a), and even macroevolutionary patterns in the investment of elements (C, N, and S) in the amino acids making up microbial proteomes (Elser et al. 2011). However, evolutionary studies within a stoichiometric framework are just in their infancy. The extreme stoichiometric imbalance and pronounced P limitation at CCB provide an excellent opportunity to test impacts of P limitation on genomic evolution. But what expectations can be developed for the evolutionary underpinnings of stoichiometric variation in biota?
To explain observed variation in the N:P ratios of various biota (such as low N:P Daphnia vs high N:P copepods) and thus, potentially, their susceptibility and responses to N or P limitation, the growth rate hypothesis (GRH) was proposed (Elser et al. 2000b). In GRH, low N:P ratios in living things are attributed to increased overall allocation (in terms of percent of dry mass) to P-rich ribosomal RNA needed to meet the protein synthesis demands of fast growth rate. The hypothesis was developed to apply primarily to small-bodied heterotrophic organisms, excluding large-bodied animals due to the importance of P-rich bone in vertebrates (Elser et al. 1996) and excluding plants and other phototrophs due to their considerable capacity for luxury uptake and storage of elements such as P. A large number of experimental and observational tests of the GRH has emerged subsequently, generally lending support for the GRH in both intraspecific (physiological, ontogenetic) and interspecific comparisons. More specifically, Hessen et al. (2013) completed a meta-analysis of more than 70 published experiments testing the GRH in various invertebrate taxa and in bacteria. In these studies, authors reported statistically significant (p < 0.05) positive correlations between growth rate and RNA content, growth rate and P content, and RNA content and P content in 67–71% of the individual assessments. Thus, we can conclude that the GRH has been established as a sound foundation for understanding variation in biomass C:N:P ratios in microbes and small consumers, critical heterotrophic components of most ecosystems. But what is the genomic basis of the GRH and how has it evolved?
With recent increases in genomic information, the GRH has expanded to include a set of genomic characteristics that support growth rate. Elser et al. (2000b) hypothesized that one key genetic component contributing to production and support of elevated ribosome content is the number of copies of the ribosomal RNA (rRNA) operon in the genome. This is because rates of protein synthesis can be limited by the number of RNA polymerase molecules transcribing the rRNA gene. This limitation is overcome by increasing the rRNA gene dosage in the genome and the strategic placement of the gene(s) near the origin of replication (Wagner 1994). Support for this idea comes from various lab studies. For example, in E. coli, which typically has seven copies of rRNA genes in its genome, a prolonged lag time in response to nutrient availability was observed when three or four copies of the rRNA gene are deleted (Condon et al. 1995). High rRNA gene copy number is required for Bacillus subtilis to successfully grow after germination of spores (Yano et al. 2013). Exploring the growth rate of 184 strains of environmental isolates, Roller et al. (2016) identified a positive relationship between maximum growth rate and rRNA gene copy number. Furthermore, copiotrophic bacteria typically found in feast and famine condition or early successional species tend to have higher rRNA gene copy number than their phylogenetic counterparts in oligotrophic environments (Klappenbach et al. 2000; Lauro et al. 2009; Nemergut et al. 2016). However, increasing the amount of ribosomes (the enzyme) can only influence protein synthesis rates if it is accompanied by increased availability of the substrate, tRNA. To support the tRNA requirement, the abundance of tRNA genes in the genome is positively correlated with the rRNA gene copy number (Lee et al. 2009). Maintaining a high density of ribosomes and associated tRNA molecules may not on its own be sufficient to maintain maximally efficient protein synthesis if the genome codes for amino acids that are not well-matched to the organism’s own genomic tRNA pool. Thus, there can be selection for use of amino acids that achieve faster and more accurate translation rate, especially in highly expressed genes such as those that code for ribosomal proteins (Rocha 2004).
The genomic capabilities to support high growth rate just described come with a cost. For example, it is hypothesized that bacteria with increased rRNA gene copy number are less efficient at resource utilization and have a higher basal nutrient requirement. In E. coli, the rRNA gene P2 promoter is a constitutive promoter that is weakly regulated and maintains a low basal level of rRNA synthesis (Maeda et al. 2016; Paul et al. 2004). If a similar promoter structure is present in other bacteria, we can expect bacteria with higher rRNA gene copy number to have a higher basal level of rRNA synthesis and potentially higher cellular ribosome content and P requirement. The higher basal level is important for rapid response but will be more costly to maintain in a low-nutrient environment. The higher gene dosage also means that the cell needs to maintain a larger genome size and thus will have a higher P demand. A positive correlation between rRNA gene copy number and genome size exists for eukaryotes, although the underlying mechanisms are still unclear (Prokopowich et al. 2003). A more recent analysis of eukaryotic genomes also found larger genomes to have a higher proportion of duplicated genes than smaller genomes (Elliott and Gregory 2015). Similarly, bacteria with fewer rRNA genes tend to have smaller genomes while bacteria with higher rRNA gene copy number have larger genomes that also carry more biosynthetic pathways (Roller et al. 2016). Therefore, high growth rate capacity may become a burden to the cell in environments with chronically low-nutrient conditions.
Vieira-Silva and Rocha (2010) have synthesized these genomic traits as components of the “systemic imprint” of growth, allowing us to expand the GRH to include a broader genomic component. In this synthetic view, microbes with high rRNA and tRNA gene copy number have a higher capacity for ribosome production to support increased protein synthesis demands of rapid growth. Furthermore, there will be selection for efficient translation in such fast-growing taxa, leading to increased codon usage bias. Finally, carrying increased numbers of genes for ribosome production itself can contribute to increase in genome size. Thus, increased pools of ribosomes and tRNA molecules along with larger genomes all require investment of P-rich nucleic acids, which will be manifested as low biomass C:P and N:P ratios of such taxa and will imply that these taxa will perform poorly in low-nutrient, low N:P environments. These combined genomic and physiological adaptations have been investigated primarily at the organismal level (Gyorfy et al. 2015; Fu and Gong 2017). Furthermore, a recent study by Hartman et al. (2017) demonstrated a support for this hypothesis, showing that soil microbes associated with increased carbon turnover (faster growth) had higher rRNA gene copy number and that this relationship is constrained by inorganic P availability.
Over the course of two summers (2011 and 2012), we performed experiments to test the GRH and the genomic signature concept at the community level. The results we synthesize here are reported in Lee et al. (2015, 2017) and Okie et al. (in review). Specifically, we tested how these genomic signatures change as community structure shifts in response to changes in nutrient supply and N:P stoichiometry. We expect the microbial community in unenriched conditions to have a genomic signature indicating dominance of species with a low growth rate lifestyle. However, nutrient enrichment will induce a shift to microbes that have genomic signatures indicative of high growth rate and low biomass C:P and N:P ratios.
Experiments/Tests
Study site:
We tested these ideas in a pair of field experiments in Lagunita Pond (latitude: 26° 50′ 53.19″ N, longitude: 102° 8′ 29.98″ W). Lagunita is a shallow pond (~25 cm maximum depth) located adjacent to a larger lagoon (Laguna Intermedia) in the western region of the Cuatro Ciénegas Basin (CCB; Fig. 3.1a). Lee et al. (2015) report detailed physical and chemical characteristics of the pond. Briefly, Lagunita’s waters are high in conductivity and dominated by Ca2+, SO42−, and CO32−, while phosphorus (P) concentrations are moderate (total P: 1.79 μmol/L) but highly stoichiometrically imbalanced with nitrogen (N) with average total N:P ratios of ~122 (molar). The pond experiences strong evaporation during summer, and thus, in both experiments, water depths declined significantly during the study.
(a) Lagunita is a small pond located to the west of Laguna Intermedia in the Churince drainage . (b) Construction of the steel scaffold that allowed Lagunita mesocosms to be sampled without disturbing the sediments. (c) Administering the nutrient enrichment treatments in Experiment 1. (d) An experimental block from Experiment 1 on day 6 showing immediate response of plankton biomass to fertilization (+P, P enrichment only; NP, N&P enrichment at 16:1 molar ratio; NNP, N&P enrichment at 75:1 molar ratio). (Photo credits: J. Elser)
Methods
Experiment 1:
Experiment 1 sought to test the GRH by manipulating P availability at different N:P ratios in small, replicate enclosures that were accessed by constructing a welded steel superstructure above the pond (Fig. 3.1b). Enclosures consisted of clear cylinders (40 cm diameter) inserted several centimeters into the sediments (Fig. 3.1c and d). There were four treatments: unenriched (no added nutrients), +P (P only added), +NP16 (N and P added at a 16:1 molar ratio), and +NP75 (N and P added at a 75:1 molar ratio). Each treatment was replicated four times, and the enclosures distributed in Lagunita according to a randomized block design. In each of the enriched treatments, P was added (as KH2PO4) every 3–4 days to achieve a target concentration of 1 μM after PO4 concentrations in each enclosure were checked by colorimetric analysis of soluble reactive P (SRP). In the NP16 and NP75 treatments, N was added (as NH4NO3) with P to achieve the target ratio. Nutrient enrichment was sustained until day 21, and the experiment ran for 42 days in total. Basic limnological variables were monitored, and on days 6, 21, and 42, measurements of chlorophyll biomass in the water column were made. On days 21 and 42, samples were collected for determination of microbial cell densities, major nutrient concentrations, and seston biomass and C:N:P ratios. For analysis of microbial communities, samples of both surficial sediments and of the water column were obtained and flash frozen for later analysis of 16S and 18S profiling via 454 sequencing after PCR amplification. Finally, water column DNA samples were subjected to qPCR analysis for assessment of 16S rRNA gene copy numbers; these data were later normalized to cell abundance from cell counts.
Experiment 2:
Experiment 2 sought to test the GRH at a larger scale and involved a whole-ecosystem manipulation with replicated internal controls coupled to metagenomic analyses. Briefly, five 40 cm transparent cylinders were inserted into the pond at the beginning of the experiment. Initial samples of the water column and surficial sediments were then taken in each of the five cylinders as well as at five locations in the pond itself. The pond was then fertilized with N and P at 16:1 ratio to achieve P concentration of 1 μM. SRP concentrations were then checked every 3–4 days, and the pond was re-enriched to return SRP to 1 μM. The experiment ran for 32 days. At 16 and 32 days, samples for analysis of seston C, N, and P concentrations were taken. At 32 days, samples of the water column were taken within each of the five enclosures and at five locations in the pond itself. (Sediment samples were also taken, but these were not analyzed.) DNA was extracted and then subjected to metagenomic sequencing of each replicate using an Illumina MiSeq. Due to lack of read depth, two of the fertilized samples and one of the unfertilized samples were eliminated from further analyses. Thus, we had a total of seven metagenomes for analysis (four in the enriched pond, three in the unenriched enclosures). Sequences were analyzed for community composition, rRNA and tRNA gene copy numbers, genome size (using normalized core marker gene counts), and codon usage bias in ribosomal protein genes. These genomic traits constitute the “genomic signature” of growth (Vieira-Silva and Rocha 2010). Details of the analyses are presented in Okie et al. (in review).
Results
Experiment 1:
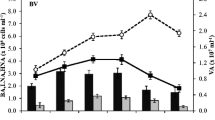
Fertilization led to large and rapid changes in water column biomass. Indeed, changes were visible to the naked eye (Fig. 3.1d) within 6 days of the start of the experiment, but the magnitude of response decreased somewhat by day 42 following 21 days without enrichment (Fig. 3.2a and b). Adding N along with P produced a larger response in chlorophyll (days 6 and 21) and seston (day 21), but this synergistic effect had disappeared by day 42 (Fig. 3.2a and b). Nutrient enrichment also lowered seston C:P ratios in the +P and +NP16 treatments but not in the +NP75 treatment (data not shown; see Lee et al. 2015), suggesting that enrichment at an N:P ratio of 75 did not alleviate P limitation. Nutrient enrichment strongly reorganized the water column microbial structure (Fig. 3.2c); among the enrichment treatments, enrichment N:P ratio also had a secondary effect on community structure (Fig. 3.2c), promoting the growth of previously rare microbial taxa at the expense of the initially abundant, and potentially endemic, taxa (data not shown; see Lee et al. 2017). Importantly, qPCR data indicate that taxa with increased copy numbers of ribosomal RNA genes were stimulated in the +P treatment (Fig. 3.2d), consistent with the GRH. No changes in ribosomal RNA gene copy number were observed in the +NP16 and +NP75 treatments, suggesting the ecosystem P limitation was not sufficiently relieved for P-demanding, high copy number taxa to proliferate in those two treatments.
Responses in Experiment 1. (a) Responses of autotrophic biomass (chlorophyll) to enrichment of P at different N:P ratios. Data are shown for sampling on days 6, 21, and 42 of the study. Horizontal bars join treatments that are not significantly different from each other. (b) Responses of total suspended organic C (seston) on days 21 and 42. (c) Multidimensional scaling results for the 16S community profiling data on day 21. Arrows indicate the overall effect of nutrient enrichment and the effects of increasing N:P ratio. (d) Cell number-normalized qPCR data for ribosomal RNA copy number on day 21. P enrichment along led to increased copy number, consistent with the GRH
Experiment 2:
Whole-pond fertilization with N and P led to large and rapid increases in chlorophyll and seston biomass in the water column (Fig. 3.3a) and increased seston P:C ratio (decreased C:P; Fig. 3.3c). Analyses of replicated metagenomic samples gave us an unusual opportunity to identify potential changes in microbial traits predicted by the GRH and its expansion, the “genomic signature” of growth. Fertilization did not change the relative proportions of major taxa in the community (i.e., Bacteria, Archaea, Eukarya, and viruses) although finer-scale changes in the abundance of various taxa led to major differences in the community composition of the fertilized pond relative to the unfertilized controls (Fig. 3.3b). Analysis of the replicated metagenomes resulted in consistent support for each component of the hypothesized genomic signature of growth (Fig. 3.3c). Members of the fertilized communities had consistently increased numbers of ribosomal RNA and tRNA genes, exhibited increased codon usage bias in the highly expressed ribosomal protein genes, and increased genome size. Larger genomes and increased numbers of rRNA and tRNA genes are consistent with increased production of P-rich ribosomes. Consequently, there is an observed increase in biomass P:C ratio in the fertilized treatment. It is not clear why N&P enrichment at 16:1 ratio was sufficient to shift ribosomal RNA copy numbers in this experiment but not in Experiment 1, when only the P alone treatment produced such a response.
Results of Experiment 2. (a) Response of autotroph biomass (chlorophyll) in Lagunita pond during fertilization (mean of five replicate samples) compared to control enclosures (means of five replicates). (b) Fertilization leads to major changes in microbial community structure (principal coordinates analysis) of 16S and 18S data from metagenomes. (c) Fertilization increased biomass P:C ratio and, consistent with the genomic signature of growth hypothesis, increased genome size, tRNA and rRNA gene copy number, and codon usage bias in the microbial community based on replicated metagenomic analysis. Circles indicate the treatment medians and bars indicate 75th and 25th percentile bounds. All responses were statistically significant (p < 0.05, one-tailed test)
Interpretations and Conclusions
The metagenomic analyses of our experiment provide some of the first field evidence supporting the predictions of an extended growth rate hypothesis in microbial assemblages. Fertilization consistently led to increased proliferation of taxa with a P-intensive lifestyle driven by genomic features that support increased capacity for production and high-speed operation of protein synthesis machinery (increased dosages of ribosomal RNA and transfer RNA genes, increased codon usage bias). This appears to come at a cost in P competitiveness, as taxa favored by fertilization also had larger genomes (made of P-rich DNA) and higher biomass P:C ratios. Overall, we propose that these data are supportive of a view of microbial life histories as lying on a stoichiometric “r/K continuum ” (Pianka 1972). On this continuum, “K-selected” species dominate under the normal, unenriched conditions in CCB, persisting with low-P biomass but modest capacities for growth. Upon nutrient enrichment, “r-selected” members of the rare biosphere with large capacity for growth and carrying P-rich protein synthesis machinery respond to sudden nutrient availability and become dominant, pushing CCB’s characteristic microbial taxa to the background.
Our experiments confirm the strong nutrient sensitivity of the aquatic ecosystems of Cuatro Ciénegas , adding to data from similar nutrient enrichment studies involving stromatolites in the Rio Mesquites (Elser et al. 2005a, b and Chap. 6). Enrichment itself has a major impact in restructuring the communities (Fig. 3.2c) with a secondary impact of N:P stoichiometry. Importantly, this nutrient sensitivity places the unique and potentially endemic microbial taxa at particular risk, as these are the taxa that have an extended evolutionary history under the low-nutrient, stoichiometrically imbalanced nutrient conditions in the basin and that seem to be ill-equipped for life under high nutrient conditions. This argues for strict management approaches that prevent inputs of N and P from human and livestock waste and from transport (aerial or waterborne) of agricultural fertilizers. Of particular concern is long-distance nutrient transport from industrial/urban sources (e.g., NOx compounds) and from agricultural fertilizer and fertilized soils in wind-borne dust, as these cannot be managed at the local scale but must be addressed regionally.
References
Condon C, Liveris D, Squires C et al (1995) Ribosomal RNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J Bacteriol 177:4152–4156
Downing JA, McCauley E (1992) The nitrogen: phosphorus relationship in lakes. Limnol Oceanogr 37:936–945. https://doi.org/10.4319/lo.1992.37.5.0936
Elliott TA, Gregory TR (2015) What’s in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philos Trans R Soc Lond B Biol Sci 370(1678):20140331. https://doi.org/10.1098/rstb.2014.0331
Elser JJ, Dobberfuhl DR, MacKay NA et al (1996) Organism size, life history, and N:P stoichiometry. Bioscience 46:674–684. https://doi.org/10.2307/1312897
Elser JJ, O’Brien J, Dobberfuhl DR (2000a) The evolution of ecosystem processes: growth rate and elemental stoichiometry of a key herbivore in temperate and arctic habitats. J Evol Biol 13:845–853. https://doi.org/10.1046/j.1420-9101.2000.00215.x
Elser JJ, Sterner RW, Gorokhova E et al (2000b) Biological stoichiometry from genes to ecosystems. Ecol Lett 3:540–550. https://doi.org/10.1111/j.1461-0248.2000.00185.x
Elser JJ, Schampel JH, Garcia-Pichel F et al (2005a) Effects of phosphorus enrichment and grazing snails on modern stromatolitic microbial communities. Freshw Biol 50:1808–1825. https://doi.org/10.1111/J.1365-2427.2005.01451.X
Elser JJ, Schampel JH, Kyle M et al (2005b) Response of grazing snails to phosphorus enrichment of modern stromatolitic microbial communities. Freshw Biol 50:1826–1835. https://doi.org/10.1111/j.1365-2427.2005.01453.x
Elser JJ, Bracken MES, Cleland EE et al (2007) Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10:1135–1142. https://doi.org/10.1111/J.1461-0248.2007.01113.X
Elser JJ, Acquisti C, Kumar S (2011) Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition. Trends Ecol Evol 26:38–44 https://doi.org/10.1016/j.tree.2010.10.006
Fagan WF, Siemann E, Mitter C et al (2002) Nitrogen in insects: implications for trophic complexity and species diversification. Am Nat 160:784–802. https://doi.org/10.1086/343879
Fu R, Gong J (2017) Single cell analysis linking ribosomal (r)DNA and rRNA copy numbers to cell size and growth rate provides insights into molecular protistan ecology. J Eukaryot Microbiol 64:885–896. https://doi.org/10.1111/jeu.12425
Gyorfy Z, Draskovits G, Vernyik V et al (2015) Engineered ribosomal RNA operon copy-number variants of E. coli reveal the evolutionary trade-offs shaping rRNA operon number. Nucleic Acids Res 43:1783–1794. https://doi.org/10.1093/nar/gkv040
Hartman WH, Ye R, Horwath WR et al (2017) A genomic perspective on stoichiometric regulation of soil carbon cycling. ISME J 11:2652–2665. https://doi.org/10.1038/ismej.2017.115
Hessen DO, Elser JJ, Sterner RW et al (2013) Ecological stoichiometry: an elementary approach using basic principles. Limnol Oceanogr 58:2219–2236. https://doi.org/10.4319/lo.2013.58.6.2219
Klappenbach JA, Dunbar JM, Schmidt TM (2000) rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol 66:1328–1333. https://doi.org/10.1128/aem.66.4.1328-1333.2000
Lauro FM, McDougald D, Thomas T et al (2009) The genomic basis of trophic strategy in marine bacteria. PNAS 106:15527–15533. https://doi.org/10.1073/pnas.0903507106
Lee ZM-P, Bussema C, Schmidt TM (2009) rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res 37(Suppl 1):D489–D493. https://doi.org/10.1093/nar/gkn689
Lee ZM, Steger L, Corman JR et al (2015) Response of a stoichiometrically imbalanced ecosystem to manipulation of nutrient supplies and ratios. PLoS One 10(4):e0123949
Lee ZM-P, Poret-Peterson AT, Siefert JL et al (2017) Nutrient stoichiometry shapes microbial community structure in an evaporitic shallow pond. Front Microbiol 8:949. https://doi.org/10.3389/fmicb.2017.00949
Maeda M, Shimada T, Ishihama A (2016) Strength and regulation of seven rRNA promoters in Escherichia coli. PLoS One 10(12):e0144697. https://doi.org/10.1371/journal.pone.0144697
Nemergut DR, Knelman JE, Ferrenberg S et al (2016) Decreases in average bacterial community rRNA operon copy number during succession. ISME J 10:1147–1156. https://doi.org/10.1038/ismej.2015.191
Paul BJ, Ross W, Gaal T, Gourse RL (2004) rRNA transcription in Escherichia coli. Annu Rev Genet 38:749–770. https://doi.org/10.1146/annurev.genet.38.072902.091347
Pianka ER (1972) r and K selection or b and d selection? Am Nat 106:581–588. https://doi.org/10.1086/282798
Prokopowich CD, Gregory TR, Crease TJ (2003) The correlation between rDNA copy number and genome size in eukaryotes. Genome 46:48–50. https://doi.org/10.1139/g02-103
Rocha EPC (2004) Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res 14:2279–2286. https://doi.org/10.1101/gr.2896904
Roller BRK, Stoddard SF, Schmidt TM (2016) Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat Microbiol 1:16160. https://doi.org/10.1038/nmicrobiol.2016.160
Vieira-Silva S, Rocha EPC (2010) The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet 6(1):e1000808. https://doi.org/10.1371/journal.pgen.1000808
Wagner R (1994) The regulation of ribosomal RNA synthesis and bacterial cell growth. Arch Microbiol 161:100–109. https://doi.org/10.1007/bf00276469
Woods HA, Fagan WF, Elser JJ et al (2004) Allometric and phylogenetic variation in insect phosphorus content. Funct Ecol 18:103–109. https://doi.org/10.1111/j.1365-2435.2004.00823.x
Yano K, Wada T, Suzuki S et al (2013) Multiple rRNA operons are essential for efficient cell growth and sporulation as well as outgrowth in Bacillus subtilis. Microbiology 159:2225–2236. https://doi.org/10.1099/mic.0.067025-0
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Elser, J., Okie, J., Lee, Z., Souza, V. (2018). The Effect of Nutrients and N:P Ratio on Microbial Communities: Testing the Growth Rate Hypothesis and Its Extensions in Lagunita Pond (Churince). In: García-Oliva, F., Elser, J., Souza, V. (eds) Ecosystem Ecology and Geochemistry of Cuatro Cienegas. Cuatro Ciénegas Basin: An Endangered Hyperdiverse Oasis. Springer, Cham. https://doi.org/10.1007/978-3-319-95855-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-95855-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-95854-5
Online ISBN: 978-3-319-95855-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)