Abstract
Carbapenem-resistant Enterobacteriaceae (CRE), especially carbapenemase-producing CRE (CP-CRE), have emerged as a major class of bacterial pathogens. They are frequently associated with high mortality and morbidity due to their unprecedented multi- or pan-drug resistance, in addition to the absence of standardized, clinically effective detection methods for early identification. Consequently, there is an urgent need for rapid and accurate detection of carbapenem resistance in clinical laboratories, as it is imperative for patient treatment, infection control, and epidemiological studies aimed at limiting further spread of CRE. A number of nucleic acid- and non-nucleic acid-based methods for rapid molecular detection of CRE are currently available or in development. Molecular detection of CP-CRE, in comparison with conventional culture-based phenotypic tests, offers several advantages, including the rapid turnaround time, the definitive identification of specific carbapenemase types, and, in some cases, the ability to test directly from clinical specimens without the need for culture. In this chapter, we will discuss the performance characteristics of these molecular technologies achieved to date on molecular detection for CRE and particularly CP-CRE.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Carbapenem-resistant Enterobacteriaceae (CRE)
- Carbapenemase-producing carbapenem-resistant Enterobacteriaceae (CP-CRE)
- Molecular detection
- Nucleic acid-based tests
- Next-generation sequencing
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) have emerged as a major class of bacterial pathogens which pose a significant threat to global public health. CRE are usually resistant to all β-lactam antibiotics and frequently carry additional resistance mechanisms against other antimicrobial agents, resulting in limited treatment options. One current clinical dilemma is that CRE infections are associated with high mortality (∼30–70%) in immunocompromised hosts, while identification of CRE by culture typically takes 2–3 days, leading to delays in appropriate therapy.
CRE are generally defined as Enterobacteriaceae that are non-susceptible (i.e., intermediate or resistant) to a carbapenem [1]. Resistance to carbapenems can arise from multiple mechanisms, including alterations in outer membrane permeability mediated by the loss of porins, the upregulation of efflux systems along with hyperproduction of AmpC β-lactamases or extended-spectrum β-lactamases (ESBLs), or, more commonly, the production of carbapenemases [2]. In its 2015 Update CRE Toolkit [1], CDC updated the definition of CRE to include Enterobacteriaceae that are (i) resistant to any carbapenem antimicrobials (i.e., minimum inhibitory concentrations (MICs) of ≥2 μg/ml against ertapenem or ≥4 μg/ml against doripenem, meropenem, or imipenem) or (ii) documented to produce carbapenemase through a phenotypic or molecular assay, regardless of in vitro carbapenem susceptibility. Of these two categories, carbapenemase-producing CRE (CP-CRE) have received more attention, as they are more widely disseminated in comparison with non-carbapenemase-producing CRE. This is primarily due to the fact that carbapenemase genes are frequently harbored by mobile elements found on large conjugative plasmids, thereby facilitating horizontal transfer of resistance into different bacterial strains and species. In addition, plasmids harbored by CP-CRE often carry additional resistance elements and thus have the potential to increase resistance to multiple drug classes.
Carbapenemase can be divided into different Ambler classes [3], primarily Ambler A, B, and D. Ambler classes A (e.g., KPC carbapenemases) and D (e.g., OXA-48-like carbapenemases) contain active serine sites, whereas Ambler class B (metallo-β-lactamases, or MBLs)—including IMP (active on imipenem), VIM (Verona integron-encoded MBL), and NDM (New Delhi metallo-β-lactamase)—requires zinc ions in their active sites. Among the aforementioned, KPC, NDM, and OXA-48 carbapenemases are the most common. KPCs are most frequently identified in Klebsiella pneumoniae in the USA, China, Colombia, Israel, Greece, and Italy, with NDMs primarily found in K. pneumoniae and Escherichia coli from the Indian subcontinent and OXA-48-like carbapenemases frequently seen in K. pneumoniae and E. coli from North Africa and Turkey [4]. In addition, KPC producers have been mostly identified among nosocomial isolates, whereas NDM and OXA-48 producers are associated with both nosocomial and community-acquired pathogens [4].
CP-CRE are currently disseminated throughout most global regions, wherein they are frequently associated with high mortality and morbidity due to their unprecedented multi- or pan-drug resistance, in addition to the absence of standardized, clinically effective detection methods for early identification [5]. Consequently, there is an urgent need for rapid and accurate detection of carbapenem resistance in clinical laboratories, as it is imperative for patient treatment, infection control, and epidemiological studies aimed at limiting further spread of CRE. In actual practice, however, clinical laboratories commonly struggle with how best to detect CRE and especially how to detect carbapenemase-producing isolates [6].
Molecular detection of CP-CRE, in comparison with conventional culture-based phenotypic tests, offers several advantages, including the rapid turnaround time, the definitive identification of specific carbapenemase types, and, in some cases, the ability to test directly from clinical specimens without the need for culture (Table 1) [9]. Molecular detection of carbapenemase genes is often regarded as the gold standard for studies evaluating detection methods for CP-CRE. In this chapter, we will discuss the progress achieved to date on molecular detection methods for CRE and particularly CP-CRE.
Based on the detection targets involved, molecular detection of CP-CRE can generally be divided into nucleic acid-based assays and non-nucleic acid-based assays, with the former being the most commonly used.
Rapid Nucleic Acid-Based Tests
As described above, carbapenemases are encoded by different β-lactamase genes, which allow for direct detection of the presence or absence of resistance genes using nucleic acid-based assays. These assays provide a rapid, sensitive, and specific tool for the recognition and identification of carbapenem resistance genes [10] and can provide molecular epidemiologic data which can be essential for infection control and outbreak investigations. Most of these techniques are based on PCR technology and may additionally be followed by Sanger DNA sequencing of the amplicon to identify sequence variations.
Conventional PCR Assays
The increasing frequency of carbapenemase-producing Gram-negative bacteria underlies the necessity of tools to monitor the emergence and spread of different classes of carbapenemase genes. As such, several conventional PCRs have been developed to detect and differentiate specific carbapenemase genes. In 2007, Ellington et al. [11] developed a multiplex PCR assay which successfully detects and distinguishes genes encoding five different acquired MBL families (VIM, IMP, SPM, GIM, and SIM) in a single reaction. The assay displayed excellent performance, correctly distinguishing and identifying 11 known reference MBL-producing strains producing IMP-1, IMP-2, IMP-4, IMP-7, IMP-12, VIM-1, VIM-2, VIM-7, SIM-1, GIM-1, and SPM-1, respectively.
In 2011, Poirel et al. [12] developed a multiplex PCR assay for detection of 11 carbapenemase genes belonging to different classes. The assay consisted of three multiplex PCR reactions and was able to detect several common carbapenemase genes belonging to Ambler classes A (KPC), B (NDM, IMP, and VIM), and D (OXA-48), as well as several newly identified carbapenemase genes encoding DIM-1, BIC-1, AIM-1, etc. The assay was rapid and reproducible and provided a convenient molecular tool for detection of both common and “minor” carbapenemase genes, thereby allowing for better evaluation of the prevalence of these clinically relevant carbapenemase genes.
Whereas the two assays described above focused solely on carbapenemase genes, other researchers have developed assays that also target AmpC β-lactamase and ESBL genes. In 2010, Dallenne et al. [13] developed a set of six multiplex PCRs and one simplex PCR for rapid detection of the most frequently encountered β-lactamase genes, including OXA-1-like broad-spectrum β-lactamases, ESBLs, plasmid-mediated AmpC β-lactamases, and class A, B, and D carbapenemases. An evaluation of the assay was performed using a collection of 31 clinical Enterobacteriaceae strains displaying resistance to broad-spectrum third-generation cephalosporins or carbapenems. Direct sequencing from PCR products was subsequently carried out to identify β-lactamase gene variations. Most PCR amplicons contained major substitutions, allowing the identification of different clusters of β-lactamase genes (e.g., differentiating broad-spectrum β-lactamase SHV genes from ESBL-type SHV genes).
In an updated version of the assay, Voets et al. [14] described an additional set of 7 multiplex PCR assays for detection of an additional 25 β-lactamase families, including plasmid-mediated AmpC β-lactamases (ACC, ACT, DHA, CMY, FOX, LAT, MIR, and MOX), metallo-carbapenemases (GIM, NDM, SIM, and SPM), serine carbapenemases (IMI, SME, and NMC-A), and OXA β-lactamases (OXA groups 1, 2, 4, 23, 24, 48, 51, and 58). The combination of the two PCR assays [13, 14] can therefore detect a wide range of β-lactamase genes using the same amplification conditions. This enables the identification of the majority of clinically important β-lactamases responsible for resistance to third-generation cephalosporins and carbapenems.
More recently, Lee et al. [15] developed a rapid and accurate PCR assay using 62 primer pairs, designed through an elaborate optimization process. To investigate the applicability of this large-scale bla detection method (named large-scaleblaFinder by the authors), the assays were performed on a number of previously reported bacterial control isolates/strains. With 100% specificity and 100% sensitivity in 189 control strains and 403 clinical isolates, the large-scaleblaFinder detected almost all clinically recognized bla genes, along with 24 previously unreported bla genes. This PCR-based system is therefore able to detect nearly all bla genes existing in a clinical isolate, providing an important aid for monitoring the emergence and dissemination of bla genes, and potentially minimizing the spread of resistant bacteria.
The aforementioned conventional PCR assays can be easily adapted by different laboratories around the world and require limited resources and have been widely used in CRE detection and epidemiological studies. However, conventional PCRs are performed on bacteria grown in pure culture and usually involve gel electrophoretic analysis of multiple bands, which is both time-consuming and less practical for direct detection of bla genes in primary clinical samples. By contrast, real-time PCR assays overcome most of these limitations and offer several advantages including greater sensitivity, minimal post-amplification analysis, and lower risk of PCR-based laboratory contamination.
Real-Time PCR Assays
In-house Real-Time PCR
At present, many clinical laboratories use “in-house” or laboratory-developed tests (LDT) involving real-time PCR-based methods, to overcome the limitations of phenotypic detection methods and conventional PCRs and to reduce detection time. Real-time PCR assays performed directly on bacterial colonies or primary specimens can generate results within hours, with excellent sensitivity and specificity. A number of multiplex real-time PCR methods have been described in the literature [16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41]. These real-time PCR assays typically use sequence-specific probes (e.g., molecular beacon or TaqMan probe) or nonspecific double-stranded DNA (dsDNA)-binding dyes (e.g., SYBR Green), followed by melting curve analysis, for the detection of amplified DNA products.
Mendes et al. [41], for example, described one of earliest multiplex real-time PCR assays for detection of metallo-β-lactamase-producing Gram-negative bacteria. The assay is a single-tube reaction, requiring a total of 2 h following colony selection. MBL identification is based on differentiation of characteristic amplicon melting curves. Shortly thereafter, Bisiklis et al. [42] reported another real-time PCR assay which is able to specifically detect blaVIM and blaIMP genes from Gram-negative bacteria within 1 h. The authors showed that melting curve analysis of the real-time PCR products clearly differentiates the target genes into four groups: (i) blaVIM-1-like, (ii) blaVIM-2-like, (iii) blaIMP-1-like, and (iv) blaIMP-2-like.
In 2011, Chen et al. [39] developed a multiplex real-time PCR assay using molecular beacons (MB-PCR) for rapid and accurate identification of blaKPC variants. The assay consists of six molecular beacons and two oligonucleotide primer pairs, allowing for detection and classification of a number of blaKPC variants (blaKPC-2 to blaKPC-11). The described real-time PCR can distinguish between different blaKPC variants and therefore provides information of both epidemiological and evolutionary significance. Subsequently, the same group [43] described a multiplex real-time PCR assay capable of identifying both the epidemic K. pneumoniae ST258 clone and blaKPC in a single reaction using molecular beacon probes. The assay displayed excellent sensitivity (100%) and specificity (100%), providing an effective tool for screening of KPC-producing K. pneumoniae isolates and surveillance of the epidemic ST258 clone. More recently, Chavda et al. [44] reported a multiplexed molecular beacon-based real-time PCR assay to identify prominent extended-spectrum-β-lactamases, plasmid-mediated AmpC β-lactamases (pAmpC), and carbapenemase genes directly from perianal swab specimens. The assay included two linear-after-the-exponential PCR (LATE-PCR) assays with melting curve analysis in order to improve the performance for single-mutation-based SHV- and TEM-ESBL detection. This assay is one of few real-time PCR methods able to detect SHV- or TEM-type ESBLs without further sequencing requirements. The assay was evaluated using 158 perianal swabs collected from hematopoietic stem cell transplant recipients and demonstrated that it was highly sensitive and specific for detection of CTX-M-, AmpC-, and KPC-producing Enterobacteriaceae compared to culture on chromogenic agar [44].
Commercial Real-Time PCRs
Over the past few years, commercial manufacturers have likewise recognized the need to have low-to-moderate complexity tests for carbapenemase detection available for rapid detection and institutional surveillance purposes. Several commercial real-time PCR-based platforms have been developed. A few have obtained FDA clearance for clinical testing, while the majority are available for research use only (RUO).
FDA-Approved Real-Time PCR Assays
-
(a)
BioFire FilmArray
BioFire Diagnostics, LLC (Salt Lake, UT, USA), now part of bioMérieux, has developed an integrated diagnostic platform known as the BioFire® FilmArray, which fully automates the detection and identification of multiple organisms from a single sample in about 1 h. An unprocessed clinical sample is subjected to nucleic acid purification, reverse transcription, a high-order nested multiplex PCR reaction, and amplicon melt curve analysis (Fig. 1a). Biochemical reactions are enclosed in a disposable pouch, minimizing PCR contamination risk [45]. Their FilmArray blood culture identification (BCID) panel can identify >25 pathogens and 4 antibiotic resistance genes from positive blood cultures in 1 h [46]. At the end of the run, a report is automatically generated which documents any detectable organism(s) as well as the antimicrobial resistance genes: mecA, vanA/B, or blaKPC. FilmArray BCID was the first FDA-cleared (June 2013) diagnostic test to directly query the blaKPC gene. A recent large multicenter study evaluated 2207 positive blood cultures (1568 clinical and 639 seeded) collected in 8 clinical microbiology laboratories in the USA [47]. The assay displayed both 100% sensitivity and specificity in detecting KPC gene from 6 clinical KPC-positive specimens and 33 seeded specimens. Recently, a research use-only antimicrobial resistance panel of FilmArray systems covering a wide range of resistance mechanisms in Gram-negative bacteria was also developed [48]. The panel consists of assays for 22 resistance determinants, including ESBLs (CTX-M, TEM, SHV), AmpCs (CMY, DHA, FOX), carbapenemases (KPC, NDM, VIM, IMP, OXA), and quinolone resistance determinants (gyrA, parC, QnrA/B/S/D, QepA). Currently, this panel is under optimization, and it is expected that implementation of the antimicrobial resistance Gram-negative panel will benefit clinical laboratories interested in rapid molecular detection of CRE.
-
(b)
GeneXpert Carba-R Test
In 2013, Cepheid described a GeneXpert® (Cepheid, Sunnyvale, CA, USA) as a real-time PCR platform with ready-to-use cartridges for rapid detection of clinically relevant carbapenemase genes (blaKPC, blaVIM, and blaNDM) directly from rectal swabs or perirectal swabs (Xpert MDRO assay) (Fig. 1a) [49]. An updated assay, the Xpert® Carba-R, was subsequently developed to allow for detection of blaKPC, blaNDM, blaVIM, blaIMP (subgroup 1), and blaOXA-48-like (e.g., blaOXA-48, blaOXA-162, blaOXA-163) carbapenemase genes, making it one of the earliest commercially available assays able to detect blaIMP-1. However, the Xpert Carba-R assay is unable to detect several important blaOXA-48 variants, e.g., blaOXA-181 and blaOXA-232 [50]; a later version (Xpert Carba-R version 2) was subsequently updated to allow for efficient detection of blaOXA-181 and blaOXA-232.
The Xpert Carba-R kit version 2 (v2) was tested on a collection of 150 well-characterized enterobacterial isolates, including several different blaOXA-48-like variants (20 blaOXA-48, 2 blaOXA-162, 9 blaOXA-181, 5 blaOXA-204, 3 blaOXA-232, and 2 blaOXA-244) [51]. The Xpert Carba-R v2 was able to detect all blaKPC, blaNDM, blaVIM, and blaOXA-48 variants, including blaOXA-181 and blaOXA-232. In addition, the assay’s performance was evaluated within the context of the daily workflow of a hygiene unit in a setting with low CP-CRE prevalence [52]. The Xpert® Carba-R v2 assay correctly detected 12 OXA-48-like, 1 KPC, and 1 OXA-48-like/NDM carriers with 100% sensitivity and 99.13% specificity and with 85.71% and 100% positive and negative predictive values, respectively [52]. This study demonstrated that the Xpert® Carba-R v2 kit is well adapted for rapid screening of high-risk patients in low-prevalence regions, with turnaround times of <1 h versus 24/48 h for culture [52].
In March 2016, the Xpert Carba-R assay obtained initial FDA clearance for detection and differentiation of carbapenemase genes in pure bacterial isolates, followed by expanded clearance in June 2016 for analysis of direct rectal swab specimens, thereby positioning the Xpert Carba-R kit as a valuable tool for identification of colonized patients and as an aid to infection control efforts.
Commercially Available Research Use-Only (RUO) Assays
The Check-Direct CPE kit (Check-Points, Wageningen, The Netherlands) is a new commercial multiplex real-time PCR assay designed to simultaneously detect the most prevalent and clinically important carbapenemase genes (blaVIM, blaOXA-48, blaNDM, and blaKPC) directly from rectal swabs. The Check-Direct CPE assay is able to differentiate between blaKPC, blaOXA-48, and blaVIM/NDM, obtaining results within 3 h; however, it is not able to differentiate between NDM and VIM, as the probes corresponding to these targets share the same fluorescent tags [24]. Also, IMP carbapenemases are not targeted by this assay [53]. In a multicenter evaluation of the Check-Direct CPE assay for direct screening of carbapenemase-producing Enterobacteriaceae from rectal swabs in Belgium, the assay showed 100% sensitivity and 94% specificity when compared with selective culture [54]. In another study, Lau et al. [55] evaluated the clinical performance of Check-Direct CPE for carbapenemase detection directly from 301 perirectal swabs (258 patients) in a non-outbreak setting. Check-Direct CPE demonstrated a sensitivity value, specificity value, positive predictive value (PPV), and negative predictive value (NPV) of 100% (all blaKPC), 88%, 21%, and 100%, respectively. False positives accounted for 79% (n = 34) of samples and were all due to targets with low incidence in the USA, such as blaNDM/VIM and blaOXA-48. The authors suggested that Check-Direct CPE will likely prove most useful in high-prevalence areas or outbreak settings where rapid carbapenemase detection is critical for infection control management [55].
Other carbapenemase surveillance assays currently in development or clinical trials include BD MAX™ CRE assay (Becton-Dickinson, USA), RenDx Carbaplex assay (Renshaw, UK), and Amplidiag CarbaR+VRE (Mobidiag, Espoo, Finland). These assays detect a variety of carbapenemases, typically including KPC, NDM, OXA-48, and VIM (VIM is not available for BD MAX). These commercial assays offer a reliable method to detect bacteria with clinically significant carbapenemases. Whether clinical laboratories choose to perform molecular testing, and the subsequent choice of test, will ultimately depend on the cost, intended throughput, target gene prevalence, and ability to fit into local workflows.
PCR/ESI-MS
Endimiani et al. developed a PCR-based PCR/electrospray ionization-mass spectrometry (PCR/ESI-MS) method for the detection and identification of blaKPC genes among Enterobacteriaceae in 2010 [56]. The PCR/ESI-MS technology measures the exact molecular mass of PCR products and interprets the data as DNA sequence information. As such, it is a promising genotyping system possessing high multiplexing capacity and can be used for detecting different genes present in a single strain. This system can also detect single-nucleotide polymorphisms, including mutations corresponding to changes in key amino acids. In their study, Endimiani and colleagues detected 100% of the KPC producers, and all blaKPC-2-possessing and blaKPC-3-possessing strains were correctly reported [56]. Given its rapid performance, the PCR/ESI-MS-based platform could be used in hospitals to improve the outcome of infected patients, as well as to perform epidemiological and infection control studies where isolates need to be rapidly detected.
Loop-Mediated Isothermal Amplification
Despite its unparalleled success as a molecular biology tool, there are inherent limitations associated with PCR, such as the cost involved in purchasing consumables and the inactivation of Taq polymerase by inhibitors (such as heparin) in crude biological samples. In order to overcome these deficiencies, the loop-mediated isothermal amplification (LAMP) assay, a relatively simple and field-adaptable platform which only requires a temperature-controlled water bath to ensure isothermal conditions, has been developed [57]. Autocycling strand displacement DNA synthesis is performed in the presence of the Bst DNA polymerase under isothermal conditions, using a set of four to eight primers that attach to unique sites on the target DNA sequence, ensuring highly specific amplification. Several in-house LAMP assays for carbapenemase genes, including blaOXA-48, blaVIM, blaIMP-14, and blaKPC, have been reported, all demonstrating high sensitivity and specificity and rapid turnaround time [38, 58,59,60].
The eazyplex® SuperBug CRE system (Amplex Biosystems GmbH, Giessen, Germany) is a commercially available LAMP assay, consisting of a freeze-dried, ready-to-use mixture which facilitates an isothermal amplification reaction that targets carbapenemase variants of the VIM, NDM, and KPC families, several members of the OXA family (OXA-48, OXA-23, OXA-40, and OXA-58 for eazyplex® SuperBug complete A and OXA-48, OXA-23, OXA-40, and OXA-181 for eazyplex® SuperBug complete B), as well as CTX-M-1 and CTX-M-9 ESBL families. The eazyplex® SuperBug CRE system can directly detect carbapenemase producers from bacterial colonies, rectal swabs, or positive blood cultures, allowing for detection within 15–30 min (depending on sample type) without DNA extraction. Amplification products are visualized by real-time fluorescence detection of a fluorescent dye bound to double-stranded DNA, using a portable Genie® II instrument. Garcia-Fernandez et al. tested a collection of 94 previously genotypically characterized and 45 prospectively collected carbapenemase-producing strains [61]. The eazyplex® SuperBug CRE system correctly detected bla carbapenemase genes with or without blaCTX-M genes in 100% of the molecularly characterized strains.
Microarray
DNA hybridization techniques in microarray formats allow for simultaneous detection of numerous sequences. Microarray technology utilizes a number of DNA probes that hybridize to DNA targets, including carbapenemase genes. Microarrays can be paired with PCR amplification of target genes or used to directly query DNA sequences within bacterial isolates. The advantage of an array platform compared to PCR assays is in the number of targets available for interrogation; while PCR can typically accommodate a maximum of four to five targets per assay, microarrays can include hundreds of targets, depending on the platform.
Check-Points has several commercially available microarray kits for epidemiological use (http://www.check-points.eu/). The Check-MDR CT101 array targets carbapenemase genes blaKPC and blaNDM, AmpC β-lactamase genes, and blaCTX-M, blaSHV, and blaTEM (both wild type and ESBLs). The Check-MDR CT102 array also includes carbapenemase genes blaVIM, blaIMP, and blaOXA-48 but omits the AmpC targets. Check-MDR CT103 XL contains all of the targets in Check-MDR CT101 and CT102 and includes additional carbapenemase genes typically identified in Acinetobacter baumannii or Pseudomonas aeruginosa, such as blaOXA-23, blaOXA-24/40, and blaOXA-58, as well as gene encoding some emerging carbapenemases (e.g., blaGIM, blaGES, blaSPM) and ESBLs (e.g., blaVEB, blaPER, blaBEL, blaGES), thereby making the Check-MDR CT103 XL array one of the most clinically relevant β-lactamase gene detection commercial panels. The assays usually work on bacterial cultures and involve DNA extraction, ligation-mediated PCR, amplification of ligated probes, and hybridization on the microarray. These assays provide highly accurate detection of known resistance genes within several hours, thereby facilitating rapid implementation of isolation measures and appropriate antibiotic treatment [62]. Bogaerts et al. [63] evaluated the performance of Check-MDR CT103 XL in 223 well-characterized Enterobacteriaceae, Pseudomonas spp., and Acinetobacter spp. strains. Specificity and sensitivity values of 100% were recorded for most bla genes, with a slightly lower signal observed for blaIMP.
The Verigene Gram-negative blood culture (BC-GN) assay (Nanosphere, Inc., Northfield, IL) is an FDA-approved (Jan 2014), automated, multiplexed nucleic acid microarray-based test for rapid Gram-negative bacterial speciation and antimicrobial resistance detection from blood cultures. A workflow of Verigene BC-GN assay is shown in Fig. 1b. The assay allows for detection of the eight most commonly isolated Gram-negative organisms, E. coli, K. pneumoniae, K. oxytoca, Enterobacter spp., Citrobacter spp., Proteus sp., Acinetobacter spp., and P. aeruginosa, as well as six classes of resistance genes: blaCTX-M, blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA (https://www.luminexcorp.com). The assay exhibits 92.2–100% positive agreement for speciation and 95.3–100% for resistance gene identification, when compared with the reference method (www.nanosphere.us). In a recent study, Walker et al. [64] evaluated the clinical impact of implementing the Verigene BC-GN assay for detection of Gram-negative bacteria in positive blood cultures obtained from hospitalized patients. The BC-GN panel yielded a positive identification in 87% of Gram-negative cultures and was accurate in 95/97 (98%) of the cases compared to results using conventional culture. Verigene BC-GN had significantly shorter turnaround times for organismal identification (mean, 10.9 h versus 37.9 h; P < 0.001). Moreover, length of ICU stay, 30-day mortality, and mortality associated with multidrug-resistant organisms were significantly lower in the Verigene BC-GN intervention group (P < 0.05). The results showed that the Verigene BC-GN assay is a valuable addition for early identification of Gram-negative organisms that cause bloodstream infections and can significantly impact patient care, particularly when resistance markers are detected. However, it is important to note that the BC-GP assay is not a target amplification assay; instead, amplification occurs within the blood culture bottle during incubation. A recognized limitation is that polymicrobial cultures are subject to false-negative results and thus lower sensitivity, due to the slower growth of some Gram-negative bacilli in mixed cultures [65, 66]. Consequently, the bacterial concentrations present in the sample may be lower than the limit of detection for species-specific and resistance gene targets.
The CarbDetect (Alere Technologies GmbH, Loebstedter, Jena, Germany) platform is a novel oligonucleotide microarray-based assay designed for bacterial cultures [67, 68]. RNA-free, unfragmented genomic DNA from pure and monoclonal culture material is amplified ~ 50-fold and labeled with biotin-11-dUTP using a linear amplification protocol, of which only one antisense primer per target is used to generate single-stranded DNA products in order to simultaneously label and amplify an essentially unlimited number of sequence-specific targets. However, the amplification sensitivity is lower than that of a standard PCR assay, and consequently the method is restricted to pure culture and cannot be performed on swabs or other primary specimens. The biotin-labeled ssDNA is transferred and hybridized to DNA oligonucleotide microarrays bearing 238 probes for 35 carbapenemase genes; 26 ESBLs, narrow-spectrum β-lactamases, and AmpC genes; as well as 48 other relevant antibiotic resistance genes (e.g., aminoglycoside resistance). Additionally, eight species markers are provided, including E. coli (including enteroinvasive strains), K. pneumoniae, P. aeruginosa, A. baumannii, C. freundii, Shigella spp., Salmonella spp., and Enterobacter spp. (CarbDetect AS-2 Kit, https://alere-technologies.com). This assay is currently the most comprehensive resistance gene detection test commercially available and includes additional resistance targets beyond β-lactamase genes. The assay was evaluated with DNA extracted from 117 clinical Enterobacteriaceae, P. aeruginosa, and A. baumannii strains collected from urinary, blood, and stool samples, which was then used to identify bacterial species, carbapenemases, ESBLs, and narrow-spectrum β-lactamase genes in a single reaction, with 98.2% and 97.4% sensitivity and specificity, respectively [68]. The newly developed assay thus provides an accurate and convenient tool to identify and discriminate the most clinically relevant carbapenemases.
Next-Generation Sequencing (NGS)
The “first generation” of DNA sequencing technology, commonly referred to as Sanger sequencing, was the primary sequencing technology from 1975 to 2005. Sanger sequencing produces relatively long (500–1000 bp) high-quality DNA sequences and has long been accepted as the gold standard for DNA sequencing. The introduction of pyrosequencing technology by 454 Life Sciences in 2005 began the “next-generation sequencing” (NGS) revolution [69]. This high-throughput technology allowed for the generation and detection of thousands to millions of short sequencing reads in a single run with no need for cloning. Since then, a number of NGS technologies have emerged, and the development of various high-throughput platforms has paved the way for the application of whole-genome sequencing to the study of bacterial pathogens and antimicrobial resistance.
Using NGS technology, a large amount of sequence data can be generated in a relatively short time, with read lengths ranging from 100 to 300 bp (e.g., Illumina HiSeq/MiSeq platforms) to >20,000 bp (e.g., PacBio and MinION platforms) [70]. The primary steps in this process include DNA isolation from bacterial culture, library preparation, DNA fragment sequencing, and data analysis. Depending on the type of information needed, NGS can include either DNA or RNA (in the form of cDNA) as sequencing material. DNA NGS captures the entire genomic content and can be used to identify the presence of antimicrobial genes or genetic mutations, while RNA NGS (or RNA sequencing) can detect global gene expression, including that of genes indirectly contributing to antimicrobial resistance (e.g., porin or efflux pump genes). Coupled with appropriate bioinformatics pipelines for identification of antimicrobial resistance genes, NGS offers the unprecedented advantage of rapidly providing genetic information at the whole-genome level, thus making it ideal for identifying all possible genetic determinants of antimicrobial resistance within a microbial genome [71]. In addition, NGS can be directly performed on primary/uncultured clinical samples, which allows for detection of all pathogens and mining of the resistance information (resistome) in a bacterial community, i.e., metagenomics analysis.
NGS facilitates molecular characterization of bacterial pathogens on many levels, without the need for a priori selection of targets as is required for PCR. The sequence data generated can be used for many purposes, including speciation and strain discrimination; resistome, plasmidome, and virulome identification; outbreak investigations and source tracking; and transmission and evolution studies. With regard to antibiotic resistance monitoring, NGS offers especially great promise, as the full repertoire of resistance genes, the sequences of plasmids that bear them, and the chromosomal background of the host strains can all be deduced from the same data. In the case of CRE, NGS also allows for differentiation of carbapenemase producers from isolates that are resistant by virtue of other mechanisms, which can be important for infection control. Furthermore, with the advancement of long-read sequencing, the sequences of complete genomes and plasmids can be readily obtained without the need for transformation, conjugation, or PCR gap closure, thereby facilitating downstream bioinformatics analysis and accurate prediction of antimicrobial resistance.
Currently, real-time integration of NGS into clinical laboratories has been hampered by processing speed, financial costs, and automated data analysis [72]. In addition, the complexities of NGS require an evolving set of standards in order to ensure testing quality. Regulatory and accreditation requirements, professional guidelines, and best practices that help ensure the quality of NGS-based tests are needed [73]. However, with decreasing cost and turnaround time, improved sample preparation workflow, and development of user-friendly bioinformatics tools, it is only a matter of time until NGS becomes a key tool for antimicrobial surveillance and infection control, with widespread implementation in clinical microbiology settings.
Rapid Non-nucleic Acid-Based Tests: MALDI-TOF MS
Matrix-assisted laser desorption ionization-time of flight mass spectroscopy (MALDI-TOF MS) is increasingly utilized in clinical microbiology laboratories for identification of bacteria and yeasts. Several MALDI-TOF MS assays for detection of β-lactamase activity, including carbapenemases, have been developed in recent years [74,75,76,77,78,79,80,81,82,83,84]. The methodology utilized typically involves the following steps: a fresh bacterial culture, usually grown overnight, is suspended in a buffer and centrifuged; the pellet is then resuspended in a reaction buffer containing the β-lactam molecule; after incubation at 35 °C for 1 to 3 h, the reaction mixture is centrifuged, and the supernatant is mixed with an appropriate matrix and measured by MALDI-TOF MS. The resulting spectra displaying peaks representing the β-lactam molecule, its salts, and/or its degradation products are then analyzed [85].
A MALDI-TOF MS assay for detection of carbapenemases was published in 2011 by Hrabak et al. [74]. This method allows for detection of resistance to carbapenems in Enterobacteriaceae carbapenemase-mediated hydrolysis, without false-positive results. Several modified MALDI-TOF MS assays were subsequently published, with varying carbapenem targets and methodological details [75,76,77,78,79,80,81,82,83,84]. The MALDI-TOF MS carbapenemase detection assays usually yield high sensitivity (~95%–100%) and specificity (~95%–100%) in comparison with phenotypic detection methods. In addition, this approach can be directly used for clinical specimens, such as blood culture and urine samples [84, 86,87,88,89,90,91]. Detection of carbapenemases by MALDI-TOF MS can be a powerful, quick, and cost-effective method for microbiological laboratories, without false-positive or false-negative results [76]. In addition, MALDI-TOF MS has been used to characterize porin function in relation to carbapenem resistance, with one study suggesting that compared with SDS-PAGE, MALDI-TOF MS is able to rapidly identify porin-deficient strains within half an hour with greater sensitivity and less cost [92].
In summary, MALDI-TOF MS is becoming an essential tool in clinical microbiology laboratories, not only for rapid identification of bacterial pathogens but also for resistance detection. However, while various methods and models have been described in the literature, a notable limitation associated with detection of enzymatic carbapenem degradation by MALDI-TOF MS is the lack of well-standardized protocols. Moreover, whereas MALDI-TOF MS can detect production of carbapenemases and carbapenemase activity, it cannot differentiate specific carbapenemase types and is consequently less informative for molecular studies.
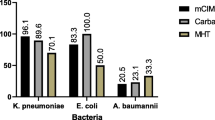
Conclusion
Carbapenemase-producing Enterobacteriaceae have now spread worldwide and become a major public health issue, challenging not only treatment solutions but also detection methods. However, appropriate treatment and infection control rely largely on efficient and timely identification of carbapenem-resistant bacteria. Therefore, there is an urgent need for rapid and accurate detection of carbapenemases, and it is necessary to introduce molecular methods into clinical diagnostic workflows. Clinical susceptibility testing provides valuable phenotypic resistance information for therapeutic decision-making but usually takes more than 48 h, which may delay appropriate treatment. Culture-based methods for carbapenemase detection, such as the MHT, Carba NP test, and mCIM, can provide rapid carbapenem resistance information but are unable to differentiate specific carbapenemases, which may be important for infection control and epidemiological investigations of CRE transmission. Currently, there are a variety of molecular-based methods able to detect most of the major carbapenemase gene families in global circulation, but they are largely limited to known carbapenemase sequence targets and can potentially miss novel variants or carbapenem resistance mechanisms. NGS, which can detect the entire genomic content or expression profile of a bacterial strain, is currently the most promising platform in antimicrobial resistance detection; however, further work is required to improve the workflow, including shortening turnaround times, reducing costs further, and improving automatic data-analyses pipelines. In summary, while no single detection platform can encompass all possible genes or resistance mechanisms, one can envision that future testing might incorporate rapid methods for both molecular detection of common carbapenemases and rapid non-nucleic acid-based determination of antimicrobial susceptibility, thus enabling timely identification of CRE and facilitating effective antibiotic therapy and infection control measures to prevent further CRE dissemination.
References
CDC. Facility guidance for control of carbapenem-resistant Enterobacteriaceae (CRE) – November 2015 update CRE toolkit. 2015. http://www.cdc.gov/hai/organisms/cre/cre-toolkit/.
Chen L, et al. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96. https://doi.org/10.1016/j.tim.2014.09.003.
Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–58, table of contents. https://doi.org/10.1128/cmr.00001-07.
Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20:821–30. https://doi.org/10.1111/1469-0691.12719.
Bialvaei AZ, Kafil HS, Asgharzadeh M, Yousef Memar M, Yousefi M. Current methods for the identification of carbapenemases. J Chemother. 2016;28:1–19. https://doi.org/10.1179/1973947815Y.0000000063.
Miller S, Humphries RM. Clinical laboratory detection of carbapenem-resistant and carbapenemase-producing Enterobacteriaceae. Expert Rev Anti Infect Ther. 2016;14:705–17. https://doi.org/10.1080/14787210.2016.1206815.
Osei Sekyere J, Govinden U, Essack SY. Review of established and innovative detection methods for carbapenemase-producing Gram-negative bacteria. J Appl Microbiol. 2015;119:1219–33. https://doi.org/10.1111/jam.12918.
Lutgring JD, Limbago BM. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol. 2016;54:529–34. https://doi.org/10.1128/jcm.02771-15.
Banerjee R, Humphries R. Clinical and laboratory considerations for the rapid detection of carbapenem-resistant Enterobacteriaceae. Virulence. 2017;8:427–39. https://doi.org/10.1080/21505594.2016.1185577.
Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother. 2012;67:906–9. https://doi.org/10.1093/jac/dkr563.
Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59:321–2. https://doi.org/10.1093/jac/dkl481.
Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–23. https://doi.org/10.1016/j.diagmicrobio.2010.12.002.
Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490–5. https://doi.org/10.1093/jac/dkp498.
Voets GM, Fluit AC, Scharringa J, Cohen Stuart J, Leverstein-van Hall MA. A set of multiplex PCRs for genotypic detection of extended-spectrum beta-lactamases, carbapenemases, plasmid-mediated AmpC beta-lactamases and OXA beta-lactamases. Int J Antimicrob Agents. 2011;37:356–9. https://doi.org/10.1016/j.ijantimicag.2011.01.005.
Lee JJ, et al. Fast and accurate large-scale detection of beta-lactamase genes conferring antibiotic resistance. Antimicrob Agents Chemother. 2015;59:5967–75. https://doi.org/10.1128/aac.04634-14.
Wang L, Gu H, Lu X. A rapid low-cost real-time PCR for the detection of Klebsiella pneumonia carbapenemase genes. Ann Clin Microbiol Antimicrob. 2012;11:9. https://doi.org/10.1186/1476-0711-11-9.
van der Zee A, et al. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect Dis. 2014;14:27. https://doi.org/10.1186/1471-2334-14-27.
Teo JW, La MV, Lin RT. Development and evaluation of a multiplex real-time PCR for the detection of IMP, VIM, and OXA-23 carbapenemase gene families on the BD MAX open system. Diagn Microbiol Infect Dis. 2016;86:358–61. https://doi.org/10.1016/j.diagmicrobio.2016.08.019.
Swayne RL, Ludlam HA, Shet VG, Woodford N, Curran MD. Real-time TaqMan PCR for rapid detection of genes encoding five types of non-metallo- (class A and D) carbapenemases in Enterobacteriaceae. Int J Antimicrob Agents. 2011;38:35–8. https://doi.org/10.1016/j.ijantimicag.2011.03.010.
Subirats J, Royo E, Balcazar JL, Borrego CM. Real-time PCR assays for the detection and quantification of carbapenemase genes (bla KPC, bla NDM, and bla OXA-48) in environmental samples. Environ Sci Pollut Res Int. 2017;24:6710–4. https://doi.org/10.1007/s11356-017-8426-6.
Smith M, et al. Rapid and accurate detection of carbapenemase genes in Enterobacteriaceae with the Cepheid Xpert Carba-R assay. J Med Microbiol. 2016;65:951–3. https://doi.org/10.1099/jmm.0.000310.
Singh P, Pfeifer Y, Mustapha A. Multiplex real-time PCR assay for the detection of extended-spectrum beta-lactamase and carbapenemase genes using melting curve analysis. J Microbiol Methods. 2016;124:72–8. https://doi.org/10.1016/j.mimet.2016.03.014.
Roth AL, Hanson ND. Rapid detection and statistical differentiation of KPC gene variants in Gram-negative pathogens by use of high-resolution melting and ScreenClust analyses. J Clin Microbiol. 2013;51:61–5. https://doi.org/10.1128/jcm.02193-12.
Nijhuis R, Samuelsen O, Savelkoul P, van Zwet A. Evaluation of a new real-time PCR assay (Check-Direct CPE) for rapid detection of KPC, OXA-48, VIM, and NDM carbapenemases using spiked rectal swabs. Diagn Microbiol Infect Dis. 2013;77:316–20. https://doi.org/10.1016/j.diagmicrobio.2013.09.007.
Naas T, Ergani A, Carrer A, Nordmann P. Real-time PCR for detection of NDM-1 carbapenemase genes from spiked stool samples. Antimicrob Agents Chemother. 2011;55:4038–43. https://doi.org/10.1128/aac.01734-10.
Naas T, Cotellon G, Ergani A, Nordmann P. Real-time PCR for detection of blaOXA-48 genes from stools. J Antimicrob Chemother. 2013;68:101–4. https://doi.org/10.1093/jac/dks340.
Mosca A, et al. Rapid and sensitive detection of bla KPC gene in clinical isolates of Klebsiella pneumoniae by a molecular real-time assay. SpringerPlus. 2013;2:31. https://doi.org/10.1186/2193-1801-2-31.
Milillo M, et al. Rapid and simultaneous detection of blaKPC and blaNDM by use of multiplex real-time PCR. J Clin Microbiol. 2013;51:1247–9. https://doi.org/10.1128/jcm.03316-12.
Mangold KA, et al. Real-time detection of blaKPC in clinical samples and surveillance specimens. J Clin Microbiol. 2011;49:3338–9. https://doi.org/10.1128/jcm.00268-11.
Kruttgen A, Razavi S, Imohl M, Ritter K. Real-time PCR assay and a synthetic positive control for the rapid and sensitive detection of the emerging resistance gene New Delhi Metallo-beta-lactamase-1 (bla(NDM-1)). Med Microbiol Immunol. 2011;200:137–41. https://doi.org/10.1007/s00430-011-0189-y.
Hindiyeh M, et al. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J Clin Microbiol. 2008;46:2879–83. https://doi.org/10.1128/jcm.00661-08.
Hindiyeh M, et al. Rapid detection of blaKPC carbapenemase genes by internally controlled real-time PCR assay using bactec blood culture bottles. J Clin Microbiol. 2011;49:2480–4. https://doi.org/10.1128/jcm.00149-11.
Hemarajata P, Yang S, Hindler JA, Humphries RM. Development of a novel real-time PCR assay with high-resolution melt analysis to detect and differentiate OXA-48-Like beta-lactamases in carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2015;59:5574–80. https://doi.org/10.1128/aac.00425-15.
Frasson I, et al. Rapid detection of blaVIM-1-37 and blaKPC1/2-12 alleles from clinical samples by multiplex PCR-based assays. Int J Antimicrob Agents. 2013;42:68–71. https://doi.org/10.1016/j.ijantimicag.2013.03.006.
Francis RO, Wu F, Della-Latta P, Shi J, Whittier S. Rapid detection of Klebsiella pneumoniae carbapenemase genes in Enterobacteriaceae directly from blood culture bottles by real-time PCR. Am J Clin Pathol. 2012;137:627–32. https://doi.org/10.1309/ajcp9snhjg2qglwu.
Favaro M, Sarti M, Fontana C. Multiplex real-time PCR probe-based for identification of strains producing: OXA48, VIM, KPC and NDM. World J Microbiol Biotechnol. 2014;30:2995–3001. https://doi.org/10.1007/s11274-014-1727-8.
Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Nordmann P. Probe ligation and real-time detection of KPC, OXA-48, VIM, IMP, and NDM carbapenemase genes. Diagn Microbiol Infect Dis. 2013;76:502–5. https://doi.org/10.1016/j.diagmicrobio.2013.05.004.
Cheng C, Zheng F, Rui Y. Rapid detection of blaNDM, blaKPC, blaIMP, and blaVIM carbapenemase genes in bacteria by loop-mediated isothermal amplification. Microb Drug Resist. 2014;20:533–8. https://doi.org/10.1089/mdr.2014.0040.
Chen L, et al. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J Clin Microbiol. 2011;49:579–85. https://doi.org/10.1128/jcm.01588-10.
Bogaerts P, et al. Analytical validation of a novel high multiplexing real-time PCR array for the identification of key pathogens causative of bacterial ventilator-associated pneumonia and their associated resistance genes. J Antimicrob Chemother. 2013;68:340–7. https://doi.org/10.1093/jac/dks392.
Mendes RE, et al. Rapid detection and identification of metallo-beta-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol. 2007;45:544–7. https://doi.org/10.1128/jcm.01728-06.
Bisiklis A, Papageorgiou F, Frantzidou F, Alexiou-Daniel S. Specific detection of blaVIM and blaIMP metallo-beta-lactamase genes in a single real-time PCR. Clin Microbiol Infect. 2007;13:1201–3. https://doi.org/10.1111/j.1469-0691.2007.01832.x.
Chen L, et al. Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob Agents Chemother. 2012;56:3444–7. https://doi.org/10.1128/AAC.00316-12.
Chavda KD, et al. Evaluation of a multiplex PCR assay to rapidly detect Enterobacteriaceae with a broad range of beta-lactamases directly from perianal swabs. Antimicrob Agents Chemother. 2016;60:6957–61. https://doi.org/10.1128/AAC.01458-16.
Poritz MA, et al. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6:e26047. https://doi.org/10.1371/journal.pone.0026047.
Blaschke AJ, et al. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn Microbiol Infect Dis. 2012;74:349–55. https://doi.org/10.1016/j.diagmicrobio.2012.08.013.
Salimnia H, et al. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol. 2016;54:687–98. https://doi.org/10.1128/jcm.01679-15.
Montgomery J, Draper N, Hemmert A, Crisp R. Rapid detection and genotyping of antimicrobial resistance determinants with the BioFire FilmArray® System. Open Forum Infect Dis. 2016;3:1999. https://doi.org/10.1093/ofid/ofw172.1547.
Tenover FC, et al. Detection of colonization by carbapenemase-producing Gram-negative Bacilli in patients by use of the Xpert MDRO assay. J Clin Microbiol. 2013;51:3780–7. https://doi.org/10.1128/jcm.01092-13.
Decousser JW, et al. Failure to detect carbapenem-resistant Escherichia coli producing OXA-48-like using the Xpert Carba-R assay(R). Clin Microbiol Infect. 2015;21:e9–10. https://doi.org/10.1016/j.cmi.2014.09.006.
Dortet L, Fusaro M, Naas T. Improvement of the Xpert Carba-R Kit for the detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2016;60:3832–7. https://doi.org/10.1128/aac.00517-16.
Hoyos-Mallecot Y, Ouzani S, Dortet L, Fortineau N, Naas T. Performance of the Xpert((R)) Carba-R v2 in the daily workflow of a hygiene unit in a country with a low prevalence of carbapenemase-producing Enterobacteriaceae. Int J Antimicrob Agents. 2017;49:774–7. https://doi.org/10.1016/j.ijantimicag.2017.01.025.
Findlay J, Hopkins KL, Meunier D, Woodford N. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J Antimicrob Chemother. 2015;70:1338–42. https://doi.org/10.1093/jac/dku571.
Huang TD, et al. Multicentre evaluation of the Check-Direct CPE(R) assay for direct screening of carbapenemase-producing Enterobacteriaceae from rectal swabs. J Antimicrob Chemother. 2015;70:1669–73. https://doi.org/10.1093/jac/dkv009.
Lau AF, et al. Clinical performance of Check-Direct CPE, a multiplex PCR for direct detection of bla(KPC), bla(NDM) and/or bla(VIM), and bla(OXA)-48 from perirectal swabs. J Clin Microbiol. 2015;53:3729–37. https://doi.org/10.1128/jcm.01921-15.
Endimiani A, et al. Rapid identification of bla KPC-possessing Enterobacteriaceae by PCR/electrospray ionization-mass spectrometry. J Antimicrob Chemother. 2010;65:1833–4. https://doi.org/10.1093/jac/dkq207.
Liu W, et al. Sensitive and rapid detection of the new Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification. J Clin Microbiol. 2012;50:1580–5. https://doi.org/10.1128/jcm.06647-11.
Solanki R, et al. Evaluation of LAMP assay using phenotypic tests and conventional PCR for detection of blaNDM-1 and blaKPC genes among carbapenem-resistant clinical Gram-negative isolates. J Med Microbiol. 2013;62:1540–4. https://doi.org/10.1099/jmm.0.059907-0.
Srisrattakarn A, et al. Rapid and simple identification of carbapenemase genes, bla NDM, bla OXA-48, bla VIM, bla IMP-14 and bla KPC groups, in Gram-negative bacilli by in-house loop-mediated isothermal amplification with hydroxynaphthol blue dye. World J Microbiol Biotechnol. 2017;33:130. https://doi.org/10.1007/s11274-017-2295-5.
Nakano R, et al. Rapid detection of the Klebsiella pneumoniae carbapenemase (KPC) gene by loop-mediated isothermal amplification (LAMP). J Infect Chemother. 2015;21:202–6. https://doi.org/10.1016/j.jiac.2014.11.010.
Garcia-Fernandez S, et al. Evaluation of the eazyplex(R) SuperBug CRE system for rapid detection of carbapenemases and ESBLs in clinical Enterobacteriaceae isolates recovered at two Spanish hospitals. J Antimicrob Chemother. 2015;70:1047–50. https://doi.org/10.1093/jac/dku476.
Cunningham SA, Vasoo S, Patel R. Evaluation of the check-points check MDR CT103 and CT103 XL microarray kits by use of preparatory rapid cell lysis. J Clin Microbiol. 2016;54:1368–71. https://doi.org/10.1128/jcm.03302-15.
Bogaerts P, et al. Evaluation of a DNA microarray for rapid detection of the most prevalent extended-spectrum beta-lactamases, plasmid-mediated cephalosporinases and carbapenemases in Enterobacteriaceae, Pseudomonas and Acinetobacter. Int J Antimicrob Agents. 2016;48:189–93. https://doi.org/10.1016/j.ijantimicag.2016.05.006.
Walker T, et al. Clinical impact of laboratory implementation of Verigene BC-GN Microarray-based assay for detection of Gram-negative bacteria in positive blood cultures. J Clin Microbiol. 2016;54:1789–96. https://doi.org/10.1128/jcm.00376-16.
Hill JT, Tran KD, Barton KL, Labreche MJ, Sharp SE. Evaluation of the nanosphere Verigene BC-GN assay for direct identification of gram-negative bacilli and antibiotic resistance markers from positive blood cultures and potential impact for more-rapid antibiotic interventions. J Clin Microbiol. 2014;52:3805–7. https://doi.org/10.1128/jcm.01537-14.
Siu GK, et al. Performance evaluation of the Verigene Gram-positive and Gram-negative blood culture test for direct identification of bacteria and their resistance determinants from positive blood cultures in Hong Kong. PLoS One. 2015;10:e0139728. https://doi.org/10.1371/journal.pone.0139728.
Braun SD, et al. Surveillance of extended-spectrum beta-lactamase-producing Escherichia coli in dairy cattle farms in the Nile Delta, Egypt. Front Microbiol. 2016;7:1020. https://doi.org/10.3389/fmicb.2016.01020.
Braun SD, et al. Rapid identification of carbapenemase genes in gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS One. 2014;9:e102232. https://doi.org/10.1371/journal.pone.0102232.
Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005;15:376–80.
Deurenberg RH, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. 2017;243:16–24. https://doi.org/10.1016/j.jbiotec.2016.12.022.
Chan KG. Whole-genome sequencing in the prediction of antimicrobial resistance. Expert Rev Anti Infect Ther. 2016;14:617–9. https://doi.org/10.1080/14787210.2016.1193005.
Bootsma HJ, Schouls LM. Next-generation sequencing of carbapenem-resistant Gram-negative microorganisms: a key tool for surveillance and infection control. Future Microbiol. 2015;10:299–302. https://doi.org/10.2217/fmb.14.134.
Gargis AS, Kalman L, Lubin IM. assuring the quality of next-generation sequencing in clinical microbiology and public health laboratories. J Clin Microbiol. 2016;54:2857–65. https://doi.org/10.1128/jcm.00949-16.
Hrabak J, Walkova R, Studentova V, Chudackova E, Bergerova T. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49:3222–7. https://doi.org/10.1128/jcm.00984-11.
Burckhardt I, Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol. 2011;49:3321–4. https://doi.org/10.1128/jcm.00287-11.
Hrabak J, et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:2441–3. https://doi.org/10.1128/jcm.01002-12.
Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J Clin Microbiol. 2012;50:927–37. https://doi.org/10.1128/jcm.05737-11.
Lee W, et al. Comparison of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry assay with conventional methods for detection of IMP-6, VIM-2, NDM-1, SIM-1, KPC-1, OXA-23, and OXA-51 carbapenemase-producing Acinetobacter spp., Pseudomonas aeruginosa, and Klebsiella pneumoniae. Diagn Microbiol Infect Dis. 2013;77:227–30. https://doi.org/10.1016/j.diagmicrobio.2013.07.005.
Hoyos-Mallecot Y, et al. MALDI-TOF MS, a useful instrument for differentiating metallo-beta-lactamases in Enterobacteriaceae and Pseudomonas spp. Lett Appl Microbiol. 2014;58:325–9. https://doi.org/10.1111/lam.12203.
Johansson A, Ekelof J, Giske CG, Sundqvist M. The detection and verification of carbapenemases using ertapenem and Matrix Assisted Laser Desorption Ionization-Time of Flight. BMC Microbiol. 2014;14:89. https://doi.org/10.1186/1471-2180-14-89.
Mirande C, et al. Rapid detection of carbapenemase activity: benefits and weaknesses of MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2015;34:2225–34. https://doi.org/10.1007/s10096-015-2473-z.
Vogne C, Prod'hom G, Jaton K, Decosterd LA, Greub G. A simple, robust and rapid approach to detect carbapenemases in Gram-negative isolates by MALDI-TOF mass spectrometry: validation with triple quadripole tandem mass spectrometry, microarray and PCR. Clin Microbiol Infect. 2014;20:O1106–12. https://doi.org/10.1111/1469-0691.12715.
Papagiannitsis CC, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol. 2015;53:1731–5. https://doi.org/10.1128/jcm.03094-14.
Oviano M, Sparbier K, Barba MJ, Kostrzewa M, Bou G. Universal protocol for the rapid automated detection of carbapenem-resistant Gram-negative bacilli directly from blood cultures by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF/MS). Int J Antimicrob Agents. 2016;48:655–60. https://doi.org/10.1016/j.ijantimicag.2016.08.024.
Hrabak J, Chudackova E, Walkova R. Matrix-assisted laser desorption ionization-time of flight (maldi-tof) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev. 2013;26:103–14. https://doi.org/10.1128/cmr.00058-12.
Sakarikou C, Ciotti M, Dolfa C, Angeletti S, Favalli C. Rapid detection of carbapenemase-producing Klebsiella pneumoniae strains derived from blood cultures by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS). BMC Microbiol. 2017;17:54. https://doi.org/10.1186/s12866-017-0952-3.
Johansson A, Nagy E, Soki J. Instant screening and verification of carbapenemase activity in Bacteroides fragilis in positive blood culture, using matrix-assisted laser desorption ionization--time of flight mass spectrometry. J Med Microbiol. 2014;63:1105–10. https://doi.org/10.1099/jmm.0.075465-0.
Hoyos-Mallecot Y, et al. Rapid detection and identification of strains carrying carbapenemases directly from positive blood cultures using MALDI-TOF MS. J Microbiol Methods. 2014;105:98–101. https://doi.org/10.1016/j.mimet.2014.07.016.
Foschi C, et al. Ease-of-use protocol for the rapid detection of third-generation cephalosporin resistance in Enterobacteriaceae isolated from blood cultures using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. J Hosp Infect. 2016;93:206–10. https://doi.org/10.1016/j.jhin.2016.02.020.
Fernandez J, Rodriguez-Lucas C, Fernandez-Suarez J, Vazquez F, Rodicio MR. Identification of Enterobacteriaceae and detection of carbapenemases from positive blood cultures by combination of MALDI-TOF MS and Carba NP performed after four hour subculture in Mueller Hinton. J Microbiol Methods. 2016;129:133–5. https://doi.org/10.1016/j.mimet.2016.08.014.
Oviano M, Ramirez CL, Barbeyto LP, Bou G. Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J Antimicrob Chemother. 2017;72:1350–4. https://doi.org/10.1093/jac/dkw579.
Hu YY, Cai JC, Zhou HW, Zhang R, Chen GX. Rapid detection of porins by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Front Microbiol. 2015;6:784. https://doi.org/10.3389/fmicb.2015.00784.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Niu, S., Chen, L. (2018). Molecular Detection and Characterization of Carbapenem-Resistant Enterobacteriaceae. In: Tang, YW., Stratton, C. (eds) Advanced Techniques in Diagnostic Microbiology. Springer, Cham. https://doi.org/10.1007/978-3-319-95111-9_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-95111-9_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-95110-2
Online ISBN: 978-3-319-95111-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)