Summary
Leaf architecture is determined by cell shape, size, and density. As plant cells are enclosed by a rigid cell wall, changes to leaf architecture have to occur through downstream genetic systems that induce alterations in (1) cell wall composition, (2) synthesis, assembly, and orientation of cytoskeletal elements and/or (3) the degree of cross-linkage between wall components in response to upstream developmental and environmental cues. This chapter reviews how leaf architecture is influenced by molecular mechanisms that modulate the above wall modification processes. Upstream signaling systems such as salicylic (SA), jasmonic (JA), and gibberellic (GA) acid have significant effects on leaf architecture. GA promotes and JA and SA suppress growth. Leaf architectural changes are brought about by these upstream systems in concert or in an interactive manner, and the associated downstream molecular systems that are involved in executing changes to cell wall properties will be discussed. Evidence will be provided to show that xyloglucan endotransglucosylase/hydrolase and pectin methyltransferase/pectin methylesterase/pectin methylesterase inhibitor systems are key downstream execution points of leaf architectural changes common to different upstream molecular systems. Optimization of leaf architecture maximizes light interception, gas exchange properties, and photosynthesis. In addition, plant growth has been shown to be more sensitive to leaf area than to area-based photosynthesis rate. Therefore, understanding genes and molecular mechanisms that affect cell wall properties and leaf architecture has broader implications in terms of crop improvement, and candidate genes that can be manipulated to optimize leaf architecture in order to maximize net carbon assimilation and plant growth will be proposed.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
I. Introduction
A. Leaf Growth and Architecture
In general, a leaf is composed of upper and lower epidermes and layers of mesophyll cells usually organized into palisade and spongy tissue (Graham et al. 2006; Lambers et al. 2008). Traversing through the leaf mesophyll is a network of vasculature composed of two groups of specialized cells: xylem and phloem (Graham et al. 2006). Leaf growth occurs in three phases: (1) leaf initiation through leaf primordia formation in the apical meristem, (2) establishment of polar axes of the leaf (leaf-length, leaf-width, and leaf-depth directions), and (3) leaf expansion (Sinha 1999; Bowman et al. 2002; Kim and Cho 2006). Leaf architecture is determined by a large number of characteristics such as the size, shape, symmetry, venation, organization, and petiole characteristics (Ellis et al. 2009) that define leaf morphology as well as anatomical features such as cell types and their size, shape, density, and the size and distribution of intercellular air spaces. Leaf morphology and leaf cell anatomy can have large influences on photosynthetic rate per unit area and, even more, on whole-plant photosynthetic rate. In this chapter we will focus on genes and associated molecular mechanisms that affect leaf size/area, shape, and epidermal cell and mesophyll characteristics, with special reference to how these affect photosynthesis.
As plant cells are encircled by a rigid cell wall, cell wall biosynthesis and modification is required in order for the proper execution of all three growth phases and to establish specific leaf architecture (Sinha 1999; Buchanan et al. 2000; Kim et al. 2002; Baskin 2005; Cosgrove 2005; Caffall and Mohnen 2009; Guerriero et al. 2014; Ochoa-Villarreal et al. 2012; Tenhaken 2015). These modifications include: (1) alteration in cell wall composition, (2) alterations in the synthesis, assembly, and orientation of cytoskeletal elements such as microtubules and actin filaments, and/or (3) alterations in the degree of cross-linking within and between cell wall components. For example, initiation of leaf primordia has been shown to depend on cell wall composition while establishment of the polar axes of the leaf requires synthesis and proper arrangement of the cortical cytoskeleton (Sinha 1999; Buchanan et al. 2000; Bowman et al. 2002; Kim et al. 2002; Baskin 2005; Cosgrove 2005; Kim and Cho 2006; Caffall and Mohnen 2009; Ochoa-Villarreal et al. 2012; Guerriero et al. 2014; Tenhaken 2015). The extent to which cells can expand depends on both turgor pressure and the physical properties of the cell wall (Kim et al. 2002; Baskin 2005; Guerriero et al. 2014). During growth of a cell, the cell wall has to be sufficiently ductile to submit to the internal force of turgor and to allow expansion (Kim et al. 2002; Baskin 2005; Guerriero et al. 2014). It also has to synthesize new cell wall material to effectively encapsulate and reinforce the growing cell surface (Buchanan et al. 2000; Graham et al. 2006). Therefore, constant synthesis and modification of cell wall material, arrangement of cytoskeletons and the formation, disruption, and reformation of cross-linkages need to take place and these processes are under strict genetic regulation. Changes in cell wall architecture can occur in response to both external and internal signals. Developmental cues such as altered rates of cell division in leaf primordia have been shown to affect the extent to which cells can expand in a process known as “compensated cell enlargement” (Fujikura et al. 2007). In general, upstream signals perceived from developmental and environmental cues will need to affect downstream targets that are directly involved in modulating cell wall properties to direct changes in leaf architecture.
A large number of genes that code for enzymes and transcription factors involved in directly modulating cell wall properties have been characterized through genetic manipulations. This chapter will summarize how altered expression of some of these genes affects leaf architecture. Some of the key genes and molecular mechanisms specifically involved in modulating the three cell wall modification processes mentioned above will also be discussed.
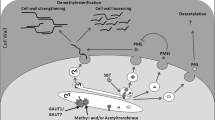
B. Alterations in Leaf Growth and Architecture Mediated by CAMTA/SA, PHYB/GA/PIF, and JAZ/JA Upstream Molecular Signaling Pathways
Three key upstream molecular systems namely the salicylic (SA), jasmonic (JA), and gibberellic (GA) acid signalling pathways and their influence on cell wall properties and leaf architecture will also be discussed in this chapter. We will look closely at the roles of CALMODULIN BINDING TRANSCRIPTION ACTIVATOR (CAMTA), PHYTOCHROME-B (PHYB), and JASMONATE ZIM-domain (JAZ) repressor proteins and their associated mechanisms in regulating leaf architecture; these genes are associated with SA, GA, and JA signalling pathways, respectively. CAMTA, PHYB, and JAZ genes were specifically selected owing to the significant alterations in leaf growth observed in the corresponding mutant plants (Figs. 8.1, 8.2 and 8.3). Some evidence for altered leaf growth upon altering expression of PHYB, JAZ, and CAMTA genes has been presented (Reed et al. 1993; Tsukaya et al. 2002; Foo et al. 2006; Finlayson et al. 2007; Doherty et al. 2009; Karve et al. 2012; Yang et al. 2012; Kim et al. 2013; Campos et al. 2016). Here we will look at how mesophyll architecture is altered in these mutant lines, and will discuss in the following sections the downstream molecular systems involved in altering cell wall properties in response to the above upstream systems.
Modulation of growth by CAMTA /SA, PHYB /GA/PIF, and JAZ/JA upstream molecular signalling pathways and their interactions. A schematic diagram is presented summarizing the interactions between SA, GA and JA signalling pathways which result in growth-defense trade-offs in plants. GA promotes shoot cell elongation and growth (Jaillais and Chory 2010; Chapman et al. 2012; Karve et al. 2012; Gommers et al. 2013; Leduc et al. 2014; Mazzella et al. 2014; Behringer and Schwechheimer 2015; Chaiwanon et al. 2016). Under light, a decrease in GA occurs as a result of both a reduction in the transcription of genes involved in GA biosynthesis, and an increase in gibberellin-2-oxidase which increases GA catabolism (Folta et al. 2003; Hisamatsu et al. 2005; Foo et al. 2006; Achard et al. 2007; Weller et al. 2009; Pierik et al. 2011; Hirose et al. 2012; Colebrook et al. 2014; Mazzella et al. 2014). Genes involved in GA biosynthesis may be downregulated by PHYB (Hisamatsu et al. 2005; Pierik et al. 2011; Hirose et al. 2012; Colebrook et al. 2014). Under shade or a lower red to far red (low R:FR) light ratio, PHYB is converted to its inactive form (Pr) which promotes the degradation of DELLA proteins (negative regulators of phytochrome-interacting factors, PIF). CAMTA mediated SA signalling under warm temperature induces SA-mediated defense responses (Kim et al. 2013). Activation of the CRT/DRE binding factor (CBF) pathway in low temperature by CAMTA genes, improves freezing tolerance in plants (Doherty et al. 2009; Kim et al. 2013) (Fig. 8.1). Rapid cold induction of CBF genes triggers the transcription of a large number of transcription factors that induce transcription of genes involved in freezing tolerance (Lee and Thomashow 2012). Herbivory-triggered jasmonic acid (JA) synthesis leads to the degradation of JAZ proteins, relieving the inhibition of several transcription factors, including group IIIe bHLHs (e.g., MYC2), and enhancing defence related processes (Hou et al. 2010; Havko et al. 2016; Campos et al. 2016). Antagonistic interactions between JAZ and DELLA proteins play a part in regulating the growth-defense trade-off mediated by GA and JA (Hou et al. 2010; Yang et al. 2012; Havko et al. 2016; Campos et al. 2016). Both CAMTA and JAZ can influence GA through CBF proteins. (Lee and Thomashow 2012) (Colour figure online)
The effect of altered CAMTA2 and CAMTA3 gene expression on leaf architecture and CO 2 diffusion through the leaf mesophyll. (a) Photographs comparing rosettes sizes, (b) representative micrographs of leaf cross sections, (c) total number of cells, and (d) the δ13CVPDB value calculated as the ratio of 13C to 12C isotopes in leaf tissue relative to a Vienna-Pee-Dee Belemnite standard (VPDB) (left panel) and the CO2 partial pressure at rubisco calculated using the δ13CVPDB values (right panel), are presented for A. thaliana Col-0 wild-type, and sid2–1, camta2/3, and camta2/3sid2–1 mutant lines (unpublished data by Y-S.K., S.M.W., T.D.S., M.F.T.). The δ13CVPDB value is a measure of discrimination against 13CO2 by a leaf. A smaller negative δ13CVPDB value indicates lower discrimination against 13CO2 and lower CO2 partial pressure at rubisco. Plants were grown hydroponically in 1/2-strength Hoagland’s solution under a light intensity of 120 μmol m−2 s−1, an 8-h photoperiod, day- and night-time temperatures of 22 °C and 20 °C, respectively, and 60% relative humidity. In (a), rosettes were photographed 41 days after seeding. In (b), leaf thickness is denoted by red double arrows. In (b-d), data are from 44-day old leaves. In (c) and (d) n = 3–4 plants per line. In (c) values represent the mean ± SE. In (d) box plots display the full range of variation and the line that divides the box in half marks the median. The mean is denoted by the small box in the middle of each box plot. The upper and lower whiskers represent scores outside the middle 50%. Statistical differences at α = 0.05 are marked with lower case letters (Colour figure online)
The effect of altered PHYB and JAZ g ene expression on leaf architecture and CO 2 diffusion through the leaf mesophyll. (a) Photographs comparing rosettes sizes, (b) representative micrographs of leaf cross sections, and (c) the δ13CVPDB value calculated as the ratio of 13C to 12C isotopes in leaf tissue relative to a Vienna-Pee-Dee Belemnite standard (VPDB) (left panel), and the CO2 partial pressure at rubisco calculated using the δ13CVPDB values (right panel), are presented for A. thaliana Col-0 wild-type and phyb, jazQ, and jazQphyB mutant lines. δ13CVPDB is described in the Fig. 8.2 legend. Plants were grown hydroponically in 1/2-strength Hoagland’s solution under a light intensity of 120 μmol m−2 s−1, 8-h photoperiod, day- and night-time temperatures of 22 °C and 20 °C, respectively, and 60% relative humidity. In (a), rosettes were photographed 55 days after seeding. In (b), leaf thickness is denoted by red double arrows. In (b–c), data are from 22-day old leaves. In (c) box plots display the full range of variation and the line that divides the box in half marks the median. The mean is denoted by the small box in the middle of each box plot. The upper and lower whiskers represent scores outside the middle 50%; n = 6 plants per line. Statistical differences at α = 0.05 are marked with lower case letters. (a–b – Campos et al. 2016; c – unpublished data by M.L.C., Y.Y., I.T.M., S.M.W., T.D.S., and G.A.H.) (Colour figure online)
Recently, CAMTA1, 2, and 3 genes were shown to suppress genes of the isochorismate synthase (ICS1) pathway of SA biosynthesis under warm temperature (Fig. 8.1) (Kim et al. 2013). SA biosynthesis is upregulated in the camta2/3 while the sid2–1 mutant line contains a loss-of function allele of ICS1 and is incapable of producing SA (Kim et al. 2013). Leaf growth in terms of both projected and total leaf area was significantly reduced in the camta2/3 double mutant compared to wild-type, while it was partially rescued in camta2/3sid2–1 (Fig. 8.2a) (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.). Downregulation of CAMTA expression caused marked changes in the mesophyll architecture that included the production of thin leaves carrying a large number of small, densely packed mesophyll cells and a reduction of intercellular air spaces (Fig. 8.2b–c) (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.). These data show that changes in leaf architecture in camta2/3 occurs in an SA dependent manner.
Under shade or a lower red to far red (FR) light ratio, PHYB is converted to its inactive form (Pr) which promotes the degradation of DELLA proteins (negative regulators of PIF) (Fig. 8.1) (Kozuka et al. 2005; Jaillais and Chory 2010; Colebrook et al. 2014; Mazzella et al. 2014; Havko et al. 2016). The degradation of DELLA proteins relieves inhibition of PIF transcription factors leading to growth (Kozuka et al. 2005; Jaillais and Chory 2010; Colebrook et al. 2014; Mazzella et al. 2014; Campos et al. 2016; Havko et al. 2016). Under unshaded light or higher red to FR light, PHYB remains in its active form (Pfr) which leads to suppression of PIF mediated growth promotion (Jaillais and Chory 2010; Karve et al. 2012; Colebrook et al. 2014; Mazzella et al. 2014; Havko et al. 2016). Herbivory-triggered JA synthesis leads to the degradation of JAZ proteins, relieving the inhibition of several transcription factors, including group IIIe bHLHs (e.g., MYC2), and enhancing defence related processes (Fig. 8.1) (Hou et al. 2010; Campos et al. 2016; Havko et al. 2016). Antagonistic interactions between JAZ and DELLA proteins play a part in regulating the growth-defense trade-off mediated by GA and JA (Hou et al. 2010; Yang et al. 2012; Campos et al. 2016; Havko et al. 2016). Leaf growth in the phyB mutant line has been examined (Tsukaya et al. 2002; Kozuka et al. 2005; Jaillais and Chory 2010; Colebrook et al. 2014; Mazzella et al. 2014; Campos et al. 2016; Havko et al. 2016). However, new data from jazQ and jazQphyB mutant lines reveal leaf area to be smaller in jazQ (Fig. 8.3a) (Campos et al. 2016). In contrast, a significant increase in both petiole length and projected and total leaf area, as well as flattened leaves were seen in phyB; these leaf characteristics were also evident in jazQphyB (Fig. 8.3a). Examination of the leaf cross sections revealed wider and shorter palisade tissue cells in the transverse sections, a reduced number of cell layers, a slight reduction in intercellular air spaces, and thinner leaves in both phyB and jazQphyB; such changes were not observed in jazQ (Fig. 8.3b) (Campos et al. 2016). In summary, the above studies provide evidence that CAMTA/SA, JAZ/JA, and PHYB/GA/PIF effects on leaf growth are accompanied by significant effects on mesophyll architecture as well as a role of underlying downstream molecular systems that modulate cell wall properties.
Use of a variety of different techniques such as microscopy, leaf gas exchange measurements, 13C discrimination analyses, and growth modeling enables greater understanding of the impact of leaf architecture on photosynthesis and plant growth. In addition, combination of physiological measurements gathered from the above techniques with gene expression data from RNA sequencing (RNA-seq) can help unravel molecular mechanisms affecting cell wall properties and leaf architecture. This chapter discusses downstream genetic mechanisms through which upstream molecular systems execute their effects on leaf architecture. In addition, key common downstream genes and molecular mechanisms that alter cell wall properties and consequently leaf architecture in response to SA, GA, and JA upstream signaling systems and the resulting effects on photosynthesis and overall plant growth are also reviewed. Candidate genes that may help to optimize leaf architecture in order to maximize net C assimilation and plant growth will also be presented.
II. Regulation of Cell Wall Composition
The development of the cell wall includes the formation of a middle lamella and the primary wall during initial growth, which in some cells is followed by formation of a secondary wall for further strength (Buchanan et al. 2000; Caffall and Mohnen 2009). The major constituents of the plant cell wall are cellulose (30%), hemicelluloses (30%), and pectins (35%) (Buchanan et al. 2000; Cosgrove 2005; Caffall and Mohnen 2009; Ochoa-Villarreal et al. 2012; Tenhaken 2015). A large gene superfamily, CELLULOSE SYNTHASE (CESA)/CELLULOSE SYNTHASE LIKE (CSL), includes genes that share significant sequence similarity. These genes code for enzymes catalyzing cellulose and hemicellulose synthesis, respectively (Cosgrove 2005; Burton et al. 2006; Suzuki et al. 2006; Held et al. 2008; Doblin et al. 2009; Dwivany et al. 2009; Yoshikawa et al. 2013).
Cellulose is synthesized by isoforms of the CESA family of cellulose synthase enzymes. Based on studies on A. thaliana, Nicotiana benthamiana, Gossypium hirsutum, Hordeum vulgare, Oryza sativa, Sorghum bicolor, and Zea mays, 8–12 CESA genes have been found to exist in plants (Pear et al. 1996; Burton et al. 2000, 2004; Robert et al. 2004; Tan et al. 2015). These cellulose synthase proteins interact to form a hexameric complex (Burton et al. 2004; Robert et al. 2004; Cosgrove 2005). The constituent CESA in the cellulose synthase complex differs based on whether the complex is associated with the primary or secondary cell wall (Burton et al. 2004; Robert et al. 2004; Cosgrove 2005). CESA3 and CESA5 in Z. mays and CESA4 in A. thaliana have been shown to be highly expressed in leaf blades (Holland et al. 2000; Burton et al. 2004).
A major portion of hemicellulose is made of xyloglucans followed by xylans, mannans, and other types of polymers such as mixed linkage glucan. The type and abundance of hemicellulose varies depending on the plant species. For example, dicot cell walls contain xyloglucans, xylans, mannans, and glucomannans while β -(1,3;1,4)-glucans are only found in Poales and other monocot groups. Arabinoxylans are the most prominent hemicelluose in graminae. CSLA – CSLJ genes are responsible for hemicellulose synthesis as follows: CSLA – β-mannan and glucomannan synthases, CSLC – β-glucan synthases, and CSLF and CSLH – mixed linkage glucan synthases (Cosgrove 2005; Burton et al. 2006; Suzuki et al. 2006; Held et al. 2008; Doblin et al. 2009; Dwivany et al. 2009; Chou et al. 2012; Yoshikawa et al. 2013). Recent studies show that xyloglucan synthesis is catalyzed by a multiprotein complex of CSLC4 and xylosyltransferases (XXT) (Chou et al. 2012).
Pectin is a complex heteropolysaccharide rich in galacturonic acid. It is the most abundant group of polymers in the primary cell wall. While pectin comprises 35% of primary cell wall in diocts and non-graminaceous monocots, it is about 2–10% in grasses (Ochoa-Villarreal et al. 2012). Pectin has various structural types, primarily homogalacturonan, rhamnogalacturonan I, and rhamnogalacturonan II (Buchanan et al. 2000; Caffall and Mohnen 2009; Ochoa-Villarreal et al. 2012; Xiao and Anderson 2013; Tenhaken 2015). Pectin is also highly substituted by side-chain modifications, such as methylesterification of the carboxyl groups of the galacturonic acid (Mouille et al. 2007; Caffall and Mohnen 2009; Wolf et al. 2009; Ochoa-Villarreal et al. 2012; Xiao and Anderson 2013; Kim et al. 2015; Tenhaken 2015). Homogalacturonan is synthesized in the Golgi apparatus and secreted as a highly methylesterified polymer. Methylesterification is catalysed by pectin methyltransferases (PMT) in the Golgi lumen. Pectin methylesterases (PMEs) present in the cell wall then de-methylesterify homogalacturonan. The interplay of PME-inhibitors (PMEI) and PMEs defines the levels of methylesterification in the cell wall, which is critical for cell expansion and overall plant growth and development. This review addresses the molecular mechanisms that affect leaf architecture by regulating cellulose, hemicellulose, and pectin synthesis.
A. Alterations in Cellulose Synthase Gene Expression
Naturally occurring small interfering RNA (siRNA) in developing leaves can suppress CESA expression (Held et al. 2008). In H. vulgare, a significant negative correlation was found between the expression levels of CESA in primary cell walls and antisense siRNA for CESA and leaf length, whereas a significant positive relationship was seen between leaf length and antisense siRNA expression levels for CESA (Held et al. 2008). The decrease in CESA transcript levels corresponded with a decrease in the rate of cellulose synthesis. Virus-induced gene silencing (VIGS) specifically targeting CESA1 resulted in the suppression of CESA as well as CSLA, CSLF, and GLYCOSYL TRANSFERASE8 (GT8) genes, which are glycosyl transferases. Consequently, an overall reduction in cellulose synthesis and incorporation of mixed linkage glucans were observed in developing leaves. siRNA for CESA and CSL genes have also been found in A. thaliana and O. sativa (Held et al. 2008). These data indicate that antisense siRNA can regulate CESA/CSL and GT8 gene expression and alter both cellulose and hemicellulose biosynthesis during early stages of leaf growth. It is also thought that difficulty in overexpressing CESA1 may be because of the effects of siRNA (Held et al. 2008). Arabidopsis thaliana mutant lines that are defective in CESA1 expression showed reduced cellulose content accompanied by a significant decrease in leaf and cotyledon areas (Arioli et al. 1998; Williamson et al. 2001; Beeckman et al. 2002). However, leaf shape was not affected indicating that altered cellulose content does not affect direction of expansion (Williamson et al. 2001; Beeckman et al. 2002) although arrangement of cellulose microfibrils would.
Recent studies indicate that effects on cell expansion brought about by changes in cellulose content are likely due in part to altered methylation status of pectin and that the synthesis of cellulose is tightly coupled with the synthesis of pectin and the degree of pectin methylesterification and vice versa. For example, VIGS of CESA1 and CESA2 in N. benthamiana resulted in a significant reduction in CESA1 and a decrease in cellulose that was compensated for by an increase in pectin (Burton et al. 2000). Interestingly, the degree of pectin methylesterification also showed a marked decrease with a subsequent increase in Ca2+ mediated cross linkages that helped strengthen the cell wall weakened by the lack of cellulose. Plants with suppressed CESA1 showed a significant reduction in leaf area and alterations in mesophyll architecture (Fig. 8.4a–f), which were similar to that seen in mutants with altered PMT gene expression (see Section IVB). The study by Burton et al. (2000) indicates that expression of CESA can regulate expression of PMEI and/or PME; demethylesterification requires enhanced activity of PME and decreased expression or activity of its inhibitor, PMEI. Held et al. (2008) did not measure PME or PMEI transcripts. Recent studies provide compelling evidence that altered expression of PMTs, which catalyze methylesterification of pectin in the Golgi, can cause changes in cellulose content (Kim et al. 2015; Weraduwage et al. 2016). Interestingly, overexpression of a PMT COTTON GOLGI-RELATED 2 (CGR2) led to an enhancement in pectin, methylated pectin as well as the crystalline cellulose content; conversely, suppression PMTs (CGR2 and CGR3) led to a decrease in these components (Fig. 8.5) (Kim et al. 2015; Weraduwage et al. 2016). The above data show that not only does CESA regulate PME and PMEI expression, but also that the expression of PMTs can control the degree of pectin methylesterification and cellulose synthesis.
Effect of altered cellulose synthase gene expression on leaf architecture. CESA was suppressed in N. benthamiana (Nt) via virus-induced gene silencing via a potato virus X vector (PVX) containing a putative CESA cDNA (PVX–NtCESA-1b) (Burton et al. 2000) (a–f). Scanning electron micrographs of (a) the adaxial side of PVX control leaf with a smooth epidermal surface, and trichomes, and mesophyll with adequate air spaces, (b) abaxial surface of PVX–NtCESA-1b with surface distortions, thinner leaf mesophyll with significantly reduced air spaces, (c) and (d) abaxial surface views of PVX control and PVX–NtCESA-1b leaves, respectively, (e) and (f) higher magnification views of PVX control and PVX–NtCESA-1b leaves, respectively, are shown. Bars in (a) and (b) = 200 μm; bars in (c) and (d) = 1 mm; bars in (e) and (f) = 100 μm. (Reproduced from Burton et al. 2000)
The effect of altered expression of pectin methyltransferase (CGR) on cell wall composition. (a) Quantification of crystalline cellulose, (b) neutral sugar, (c) uronic acids, and (d) the degree of methylesterification, from the alcohol insoluble residue (AIR) of leaf tissue of A. thaliana transgenic lines showing suppressed (cgr2–1 cgr3–1, cgr2–1, cgr3–1), and enhanced (CGR2OX, CGR3OX) CGR2 or CGR3 gene expression. In (b), AIR from leaf tissue was analyzed for quantification of neutral sugars using alditol acetate derivatives. In (c), uronic acids from AIR were measured using a colorimetric method (Filisetti-Cozzi and Carpita 1991). D-galacturonic acid was used as a standard to calculate concentration. In (d), release of methanol from methyl esters in AIR was measured after saponification (Wood and Siddiqui, 1971). Methanol was used as a standard to calculate concentration. Values are means + SD (n = 3 for each genotype). Values indicated by letters are statistically significantly different from the wild type (a, P < 0.01,and b, P < 0.05) by Student’s t test. (Reproduced from Kim et al. 2015)
In summary, we see that CESA and PMT/PME/PMEI molecular systems work in coordination to support cell wall synthesis and modification during cell expansion and have significant effects on leaf architecture. High PMT expression supports cell expansion and growth, enhanced CESA expression, and cellulose production to support cell wall building. A decrease in CESA can trigger cell wall hardening mediated by PME.
B. Potential PIF Mediated Effects on CESA and CESL Expression
Evidence for enhanced cellulose synthesis in response to enhanced leaf growth was seen in phyB and jazQphyB mutant lines where CESA4 expression was enhanced (Fig. 8.6) (Campos et al. 2016). In addition, expression of a number of CSL genes was also enhanced in these lines (Fig. 8.6). Overall, we see that cellulose synthesis responds to alterations in the upstream PHYB/GA/PIF molecular systems while closely interacting with downstream molecular systems such as PMT/PME/PMEI in order to produce sufficient amounts of cellulose to meet the demand for new cell wall material.
PIF mediated effects on genes associated with cellulose and hemicellulose synthesis. Expression levels of Cellulose synthase (CESA) and Cellulose synthase like (CSL, involved in hemicellulose synthesis) genes are presented for A. thaliana Col-0 wild-type, and jazQ, phyB, and jazQphyB mutant lines as determined by leaf messenger RNA sequencing (Campos et al. 2016). Values represent the mean ± SE and n = 3 plants per line. Statistically different expression levels in comparisons to Col-0 found according to the DESeq algorithm (P < 0.1, using a Benjamini-Hochberg adjusted for multiple testing) are marked with asterisks
III. Regulation of Cortical Microtubule and Microfilament Organization
Anisotropic (polarity-dependent) expansion of the cell wall is a key factor that determines cell shape. The balance between isotropic (polarity-independent) and anisotropic expansion processes determines the shape of an organ such as the leaf (Kim et al. 2002; Baskin 2005; Guerriero et al. 2014). Both turgor pressure and the physical properties of the cell wall determine the extent to which a cell can expand. Although the internal force exerted by turgor pressure on the cell wall is isotropic, because of the localized differences in the physical properties of the cell wall, the net expansion of the cell can be anisotropic, which subsequently determines cell shape and the architecture of the leaf. Anisotropic expansion rates per unit area of cell wall have two components: direction and angle (Baskin 2005).
The primary reason for anisotropic expansion of leaf cell and other cell walls is the arrangement of the cellular cytoskeleton, which is formed by the cortical microtubules and actin microfilaments (F-actin). The cortical microtubules located just beneath the plasma membrane mediate the directionality of the cellulose microfibril alignment (Buchanan et al. 2000; Kim et al. 2002; Baskin 2005; Guerriero et al. 2014). Arrangement of cellulose microfibrils perpendicular to the axis of elongation allows the primary cell wall to maintain strength and extensibility and facilitates anisotropic growth (Baskin 2005; Tenhaken 2015). While the alignment between microfibrils in primary cell walls is somewhat parallel, a stricter organization is seen in secondary cell walls where they exist in parallel arrays. These parallel microfibrils have been shown to arrange in different angles within each layer of secondary wall, thus limiting cells’ ability to expand and grow (Baskin 2005; Tenhaken 2015). However, the effect of F-actin on anisotropic cell expansion is rather indirect and does not depend on the directionality of F-actin alignment in the cortex. A network of fine F-actin facilitates the transport of Golgi vesicles containing building material for growth, including cell wall growth (Buchanan et al. 2000; Mathur and Hülskamp 2002; Mathur 2006; Guerriero et al. 2014). In addition, the movement of mitochondria and peroxisomes also takes place along F-actin. Studies have shown the abundance of fine/diffuse F-actin networks to enhance at cell bulges/lobes/protrusions or locations of anisotropic growth; thus vesicle trafficking to the growing area is enhanced (Mathur and Hülskamp 2002; Mathur 2006; Guerriero et al. 2014). On the other hand, dense F-actin networks have been shown to block the movement of vesicles and thereby lead to growth retardation (Mathur and Hülskamp 2002; Mathur 2006; Guerriero et al. 2014). Thus, the resistance of the cell wall to the internal turgor force during anisotropic growth depends on the net effect of microtubule arrangement and microfilament type and abundance.
A large number of genes have been found to regulate synthesis and arrangement of cortical microtubules and F-actin. This section will summarize these molecular mechanisms, identify points of interaction, and present how these molecular mechanisms determine leaf architecture.
A. Genes That Regulate Microtubule Alignment
The lining of cellulose microfibrils mirrors the array of microtubules in the cell cortex because the movement of cellulose synthase and deposition of cellulose is directed by microtubules (Buchanan et al. 2000; Chan 2012). Therefore, genetic regulation of the direction and angle of cortical microtubule alignment can have a drastic effect on anisotropic cell growth and leaf architecture. As mentioned earlier, leaf cell expansion can occur in three directions: length, width and depth, which ultimately affects overall leaf architecture. The direction of leaf cell expansion seems to be regulated by three major genes: ANGUSTIFOLIA (AN), ROTUNDI-FOLIA (ROT3) and LONGIFOLIA (LNG1, LNG2) (Tsuge et al. 1996; Tsukaya 1998, 2002; Kim et al. 2002; Kalve et al. 2014).
Interestingly, AN has been shown to facilitate anisotropic growth in leaf-width direction and inhibit expansion in the depth direction, whereas ROT3 enhances growth in leaf-length direction and inhibits expansion in the depth direction (Fig. 8.7). LNG1 and LNG2 have been shown to promote cell expansion in the leaf length direction. AN codes for a carboxy terminal binding protein and an mutant lines have narrow and thick leaves with significantly altered mesophyll architecture (Fig. 8.7). (Kim et al. 2002). The authors showed that restricted growth in the width direction and enhanced growth in the depth direction in an mutant lines was due to: (1) the more regular arrangement of cortical microtubules parallel to the leaf width direction and (2) a reduction in the angle between cortical microtubules and the plane parallel to the epidermal plane in the transverse section (Kim et al. 2002; Tsukaya 2002). Upregulation of XTH24, a xyloglucan endotransglucosylase/hydrolase in an an mutant line suggest interactions between AN and XTH resulting in the above cell wall modifications (Kim et al. 2002).
The effect of suppressed AN and ROT3 gene expression on leaf architecture. (a) The morphology of leaves of A. thaliana wild-type (wt), the an mutant, and the rot3 mutant are presented. In (a), from the left, are the two cotyledons, eight rosette leaves and three cauline leaves. Leaf cross sections of the fifth leaves of the wild type, an, and the rot3 mutant showing cell development in the (b) leaf width direction and (c) leaf length direction. (d) Transverse and longitudinal sections of leaves of the an rot3 double mutant. In (a–d), leaves were collected when fully expanded. In (b–d), the transverse sections reveal a region between the midrib and the leaf margin; longitudinal sections reveal a region in the center of the lamina. The leaf cross sections in horizontal rows are from leaves at the same stage of growth: stage I – leaf length = 1.0 mm, stage II – 5.0 mm; stage III – 10.0 mm, and stage IV – 15.0 mm. Bars = 100 μm. (Reproduced from Tsuge et al. 1996) (Colour figure online)
ROT3 has been shown to encode a cytochrome P-450 family steroid hydrolase, CYP90C1 (Tsuge et al. 1996; Kim et al. 2002; Tsukaya 2002). CYP90C1 was shown to catalyze the conversion of typhasterol to castasterone, one of the last steps of brassinosteroid (BR) biosynthesis (Kim et al. 1998, 2005, 2015; Ohnishi et al. 2006). However, while rot3 mutant lines showed a significant reduction in growth in length and an increase in breadth (Fig. 8.7), changes in microtubule organization was not observed. BRs have been shown to positively regulate MICROTUBULE DESTABILIZING PROTEIN40 (MDP40) gene expression; MDP40 is highly expressed in hypocotyls and cotyledons (Wang et al. 2012). MDP40 has also been shown to co-localize with cortical microtubules and regulate their arrangement to promote hypocotyl cell elongation (Wang et al. 2012). Interestingly, although the rot3 small leaf phenotype is similar to mutant lines deficient in BRs (korrigan1, dwarf1, deetiolated2), in contrast to rot3 both cell expansion and cell number is affected in these mutant lines (Fujioka et al. 1997; Choe et al. 2000; Nakaya et al. 2002; Tsukaya 2002). Therefore, ROT3 mediated cell elongation occurs via mechanisms other than microtubule alignment.
In contrast to AN and ROT3 genes, LNG1 and LNG2 have been shown to promote cell expansion in the leaf-length direction independent of ROT3 expression (Lee et al. 2006). Cold shock proteins characterized as nucleic acid binding proteins were recently found to regulate LNG1 expression in A. thaliana (Yang and Karlson 2012). However, detailed molecular mechanisms through which LNG exerts its effects on leaf architecture remain to be found. New data on the role of PHYB/GA/PIF mediated regulation of ROT3, LNG1, and LNG2, and potential ROT3-regulated genes, are presented in Section IIIB.
B. Regulation of F-Actin Formation and Abundance
The interplay between microtubule arrangement, microfilament type, and abundance generates the interdigitated appearance of leaf epidermal pavement cells (normal epidermal cells) and the genes involved in this process have been studied extensively. It has been shown that anisotropic growth resulting in lobe formation is initiated soon after cell division is completed and is clearly seen after the cell expands several-fold (Frank and Smith 2002). A general pattern has been established for the cortical fine F-actin and microtubule distribution in epidermal pavement cells at various growth stages in the wild-type A. thaliana leaves (Fig. 8.8a) (Fu et al. 2002, 2005). During growth of epidermal pavement cells, fine F-actin is abundant and microtubules are scarce in protruding lobe areas (Fig. 8.8a); in contrast, microtubules and dense F-actin is abundant in indentation areas (Mathur and Hülskamp 2002; Mathur 2006). It is known that such coordinated changes in the cytoskeletal material in the cell cortex is regulated mainly through interactions between RhoGTPases (ROPs), ROP-INTERACTIVE CRIB MOTIF PROTEIN (RIC), SUPPRESSOR OF CYCLIC AMP RECEPTOR PROTEIN (SCAR), WISKOTT–ALDRICH SYNDROME PROTEIN-FAMILY VERPROLIN HOMOLOGOUS PROTEIN (WAVE), ACTIN RELATED PROTEIN (ARP2/3) and actin binding and stabilizing proteins (Fig. 8.9). The center point of this molecular mechanism is held by RHO-RELATED GTPase FROM PLANT (ROP), constituting a family of 11 genes in the A. thaliana genome with most of them showing expression in leaves (Fu et al. 2002, 2005; Qian et al. 2009; Craddock et al. 2012) (Fig. 8.9). Fu et al. (2002, 2005, 2009) showed that changes in ROP2, 4, and 6 expression leads to significant changes in leaf architecture in a development stage dependent manner (Fig. 8.8b–m). Genetic manipulation studies of ROP genes revealed that ROP2 and ROP6 determine epidermal architecture through regulation of the formation and orientation of fine F-actin whereas ROP6 affects bundling and organization of cortical microtubules (Fig. 8.9) (Fu et al. 2002, 2005, 2009).
ROP GTPase modulation of cytoskeleton and morphogenesis of leaf cells. (a) A schematic illustration of A. thaliana leaf pavement cell development and associated fine actin filaments (red patches = MFs) and cortical microtubules (green lines = MTs) is presented. ROP-independent actin bundles are not shown. Arrows indicate directions of expansion. Stage I – includes young developing cells at the leaf base prior to lobe formation having only isotropic expansion, with a network of fine F-actin in the cell cortex with greater abundance in the lobe initiation sites, and randomly oriented cortical microtubules. Stage II – expanded cells with developing lobes located between the leaf base and tip area having a network of fine F-actin only in the lobe tips, with transversely arranged cortical microtubules confined to the invaginated areas and lobe shoulders (x – region of active ROP2; y – region of active ROP6). Stage III – mature cells with completed lobe extension having only randomly arranged cortical microtubules (Fu et al. 2002, 2005). When the leaf transitions from early to late growth stages, the arrangement of cortical microtubules was shown to change from random to transverse, which is important for expansion along long axis but prevents expansion in the lobe necks (Fu et al. 2002). (b–d) Scanning electron microscopy images of leaf trichomes (Bar = 200 μm), (e–g) leaf cross-sections (Bar = 250 μm), (h–j) scanning electron microscopy images of leaf pavement cells (Bar = 20 μm), and (k–m) pavement cells in bleached leaves (Bar = 40 μm) are presented for A. thaliana wild-type (images at left), and mutant lines with enhanced (images in the middle) and suppressed (images in the right) ROP2 expression. (a is reproduced from Fu et al. 2005; b–m are reproduced from Fu et al. 2002) (Colour figure online)
A summary of gene interactions in the ROP mediated molecular system regulating leaf architecture through the assembly and orientation of cytoskeletal elements. Corresponding genes encoding key proteins: Rho-related GTPase from plant (ROPs), Rop-interactive crib motif proteins (RIC), Suppressor of cyclic AMP receptor (SCAR), Wiskott–Aldrich syndrome protein-family verprolin-homologous protein (WAVE), and Actin related protein (ARP2/3), regulate fine F-actin network formation and microtubule assembly and orientation. Two pathways, one that operates through ROP2/ROP4 and RIC4, and the other that operates through ROP2/ROP4, SCAR/WAVE and ARP2/3 complex promote fine F-actin network formation. Another system operating via ROP6 and RIC1 promotes assembly of microtubule bundles and their orientation. While regions with fine F-actin promotes growth (protrusions), microtubule bundling creates resistance to growth (invaginations). This interaction leads to anisotropic growth, as seen specifically in pavement cell interdigitation. RIC4 and Mirotubule organization (MOR1) proteins help in feedback regulation between fine F-actin network formation and microtubule bundle assembly. Auxin has been found to suppress Rho guanine nucleotide exchange factors (RhoGEFs) gene expression; RhoGEFs activate ROP. New evidence (Campos et al. 2016) supports a role for Phytochrome interacting factors (PIF) in promoting ROP activity through: 1. upregulation of Pin-formed auxin efflux carrier gene family protein (PIN) expression and 2. through suppression of the negative regulator of RhoGTPase (RhoGAP) gene expression; PIF also suppressed Microtubule associated protein18 (MAP18) gene expression required for microtubule bundle formation. Upregulation or downregulation of gene expression is denoted by pointed and blunt ended arrows, respectively. Other abbreviations: ABP1 – Putative auxin receptor auxin binding protein 1, TMK – Transmembrane kinase subfamily of receptor-like kinases, RhoGEF – Rho-Guanine nucleotide exchange factors (GEFs), SPIKE – a RhoGEF or DOCK180-type guanine nucleotide exchange factor, SRA1 – Rac1-associated protein-1, NAP1 – Nck-associated protein, BRICK1 – SCAR/WAVE Actin-Nucleating Complex Subunit
Fu et al. (2005) further characterized the ROP mediated molecular mechanism for polar cell expansion in epidermal cells and showed that indentations and lobe formation by pavement cells is regulated by the interactions between ROP-INTERACTIVE CRIB MOTIF PROTEINS (RIC) RIC1 and RIC4. RIC1 was found to co-localize with cortical microtubules and loss of ROP2 and ROP4 enhanced this association (Fig. 8.9) (Fu et al. 2005). In contrast, ROP6 was found to directly associate with RIC1 and enhance its interaction with cortical microtubules (Fu et al. 2002, 2005, 2009). RIC4 co-localized with cortical fine F-actin in the growing lobe regions (Fu et al. 2005, 2009). In summary, these studies showed that RIC1 and RIC4 are associated with promoting microtubule and F-actin assembly, respectively (Fig. 8.9). The abundance and assembly of these cytoskeletal components are further regulated via feedback effects on ROP2-RIC4 interaction executed by MICROTUBULE ORGANI-ZATION (MOR1) proteins (Fig. 8.9) (Whittington et al. 2001).
ROP proteins are activated by Rho guanine nucleotide exchange factors (RhoGEFs) in plants and a single RhoGEF (SPIKE1 or SPK1) is present in A. thaliana (Qiu et al. 2002; Basu et al. 2008). Studies showed that SPK1 associates with many ROP proteins including ROP2, 3, 4, and 6 and that it is also capable of interacting with WAVE complex proteins such as SRA1 and NAP1 (Fig. 8.9) (Basu et al. 2008). Activation of SRA1 occurs primarily via ROP activation through SPIKE (Basu et al. 2008). Enhanced expression of SRA1 and NAP1 WAVE complex genes also affected leaf morphology through activation of an ACTIN RELATED PROTEIN complex (ARP2/3) (Li et al. 2004; Basu et al. 2005). The ARP2/3 complex, composed of seven ARP subunits, activates the polymerization of branched F-actin and leads to the assembly of branched F-actin networks (Li et al. 2003; Qian et al. 2009). The ARP2/3 complex is activated by proteins coded by the SCAR/WAVE gene family (Fig. 8.9) (Frank and Smith 2002; Djakovic et al. 2006; Qian et al. 2009). BRK1 proteins have been shown to co-localize with WAVE proteins and to associate with SCAR proteins stabilizing and promoting the accumulation of SCAR proteins (Frank and Smith 2002; Djakovic et al. 2006; Qian et al. 2009). Alterations in ARP subunit and BRK1 expression led to marked changes in F-actin polymerization and distribution with subsequent effects on the pavement cell architecture.
It is likely that similar mechanisms exist to regulate anisotropic growth of mesophyll cells. However, only a few studies looked at how ROP mediated changes in pavement cell architecture affected other aspects of leaf architecture, including leaf and cotyledon size and mesophyll architecture (Frank and Smith 2002; Fu et al. 2002; Qiu et al. 2002; Djakovic et al. 2006; Basu et al. 2008). In these studies, a reduction in pavement cell expansion and lobe formation correlated with a reduction in the size of mesophyll cells and consequently leaf size (Frank and Smith 2002; Fu et al. 2002; Qiu et al. 2002; Djakovic et al. 2006; Basu et al. 2008). In addition, there is evidence to suggest that the rate of cell division and expansion of the epidermis can affect the same processes in the inner layers of tissue, but results seem to vary depending on the species examined (Savaldi-Goldstein and Chory 2008; Marcotrigiano 2010). For example, genetically different epidermal layers on either side of the midrib of graft leaf chimeras between Nicotiana glauca and Nicotiana tabacum were used to show that the rate of cell division in the epidermal cell layer can determine the rate of cell division in the inner layers of the leaf (Marcotrigiano 2010). In addition, expression of BR synthesizing enzymes in an epidermal cell specific manner in brassinosteroid deficient mutant lines enhanced expansion of epidermal cells and subsequently that of mesophyll cells (Savaldi-Goldstein and Chory 2008; Marcotrigiano 2010). These results show that hormonal signaling from the epidermis to the inner layers can coordinate growth in different cell layers in a leaf (Savaldi-Goldstein and Chory 2008; Marcotrigiano 2010). In addition, Kawade et al. (2013) showed that epidermal cell proliferation is dependent on the movement of AN3 protein to the epidermal cells from mesophyll cells where it is synthesized. The detection of reduced cell proliferation in both mesophyll and epidermal cells in an3 mutants revealed that normal leaf growth is also dependent on signals that travel from the inner mesophyll to the outer epidermal cell layer (Kawade et al. 2013). Thus, inter-cell-layer controls can occur in either direction to coordinate leaf growth. In addition, physical properties of the epidermal cell wall may also determine its capability to bear the force exerted by internal tissues and hence regulate the growth capability of internal tissues (Savaldi-Goldstein and Chory 2008; Marcotrigiano 2010).
C. Potential PIF Mediated Effects on Fine F-Actin Network and Microtubule Bundle Formation
Previous studies have shown that epidermal pavement cell interdigitation is promoted by auxin, PUTATIVE AUXIN RECEPTOR AUXIN BINDING PROTEIN (ABP1), TRANSMEMBRANE KINASE SUBFAMILY OF RECEPTOR-LIKE KINASES (TMK), RhoGEF, ROP, and PIN-FORMED AUXIN EFFLUX CARRIER GENE FAMILY PROTEIN (PIN1) mediated feedback effects (Xu et al. 2010; Craddock et al. 2012) (Fig. 8.9). RNA-seq data from phyB and jazQphyB provide supporting evidence for the hypothesis that PIF may promote growth by positively affecting fine F-actin network formation and negatively affecting microtubule bundle formation (Figs. 8.9 and 8.10) (Campos et al. 2016). MICROTUBULE ASSOCIATED PROTEIN18 (MAP18) gene expression, which negatively regulates formation of well-organized microtubule bundles, was suppressed in a phyB mutant line (Figs. 8.9 and 8.10a). Well-organized microtubule bundles restrict growth. In addition, the negative regulator of ROP (RhoGAP) expression was also suppressed (Figs. 8.9 and 8.10a). While Rho guanine nucleotide exchange factors (RhoGEFs) activate ROP, RhoGAP inactivates ROP (Moon and Zheng 2003; Xu et al. 2010; Craddock et al. 2012). Only nine RhoGAP genes have been found in A. thaliana and data on molecular mechanisms regulating RhoGAP expression are rare (Kost 2010). Data obtained from phyB mutant lines show that PIF negatively affects RhoGAP expression (Fig. 8.9). The expression of both PIN3 and PIN7 auxin transporters was elevated in phyB (Figs. 8.9 and 8.10b) (Campos et al. 2016). Upregulation of PIN and downregulation of RhoGAP may activate ROP, specifically ROP2, which induces fine F-actin network formation and promotes growth (Fig. 8.9). Therefore, it seems that growth enhancement by PIF (Fig. 8.3) may be mediated, at least in part, by downregulation of well-organized microtubule bundle formation and through promotion of F-actin network formation.
PIF mediated upregulation of genes associated with cytoskeleton assembly and orientation. Expression levels of (a) Microtubule-associated Protein18 (MAP18) and RhoGAP (AT5G12150, which catalyzes RhoGTPase inactivation), (b) PIN3 and PIN7 of the Pin-formed (PIN) auxin efflux carrier gene family, (c) genes of cytochrome P450 proteins (Rotundifolia3 (ROT3, CYP90C1) and brassinosteroid-6-oxidase (BRGOX1, CYP85A1), and (d) Longifolia genes (LNG1, LNG2) are presented for A. thaliana Col-0 wild-type, and jazQ, phyB, and jazQphyB mutant lines determined by leaf messenger RNA sequencing (Campos et al. 2016). Values represent the mean ± SE and n = 3 plants per line. Statistically different expression levels in comparisons to Col-0 found according to the DESeq algorithm (P < 0.05, using a Benjamini-Hochberg adjusted for multiple testing) are marked with asterisks
The ability of PIF to alter gene expression of ROT3 shows that another pathway through which PIF can influence changes in leaf architecture may be through BR synthesis to enhance MDP40 gene expression that leads to microtubule destabilization. In fact, the expression of ROT3 (CYP90C1) and brassinosteroid-6-oxidase (CYP85A1), important cytochrome P450 proteins catalyzing the last steps of BR synthesis, was upregulated in the phyB mutant line (Fig. 8.10c). Furthermore, expression of both LNG alleles was enhanced in phyB (Fig. 8.10d).
In summary, AN, ROT3, and LNG1 and LNG2 are three major genes that regulate microtubule alignment in mesophyll cells and subsequently the direction of leaf cell expansion. There are also two key molecular systems that tightly regulate anisotropic cell expansion to determine the interdigitating architecture of pavement cells. One system operates through (i) ROP2/ROP4 and RIC4 and (ii) ROP2/ROP4, SCAR/WAVE, and ARP2/3 complexes to promote fine F-actin formation and assembly (Fig. 8.9). The second system acts through ROP6 and RIC1 to promote microtubule assembly and orientation. There is clear evidence for coordinated antagonistic regulation of F-actin and cortical microtubule distribution in the protruding lobe and invaginated neck regions of pavement cells (Fig. 8.9). In addition, regulation of epidermal cell expansion by the above two systems also seems to have significant effects on mesophyll architecture and overall leaf growth in a direct or indirect manner. Upstream, the PHYB/GA/PIF molecular system seems to enhance growth at least in part via downregulation of well-organized microtubule bundle formation and through promotion of F-actin network formation.
IV. Cross-Linkages Between Different Cell Wall Constituents
In addition to composition, and the orientation of cellulose microfibrils and F-actin formation and abundance, cross-linkages between cell wall constituents also assert strength, and therefore, resistance to cell expansion and growth. Xyloglucans, which form a major portion of hemicelluloses, are cross linked with cellulose microfibrils and pectin, thereby adding rigidity and mechanical strength to the cell wall (Cosgrove 2005; Ochoa-Villarreal et al. 2012; Tenhaken 2015). This interaction between xyloglucan hemicellulosic polymers and cellulose fibers is modulated by expansin and xyloglucan endotransglucosylase/hydrolase (XTH) enzymes (Cosgrove 2005; Ochoa-Villarreal et al. 2012; Tenhaken 2015). Expansins are primarily involved in wall loosening whereas XTHs are more versatile in function. XTH catalyzes the endolytic cleavage of existing xyloglucan-xyloglucan or xyloglucan-other polymer chains, after which reformation of cross-linkages with different xyloglucans or polymers (xyloglucan endotransglucosylase, XET) or with water (xyloglucan endohydrolase, XEH) occurs. XTH’s ability to recruit new xyloglucan or other polymer chains to the existing cell wall likely leads to wall strengthening whereas hydrolysis of cross-linkages may lead to wall loosening (Rose et al. 2002; Becnel et al. 2006). Therefore, XTH can regulate plasticity of the cell wall and subsequently cell size and leaf architecture (Nishitani and Tominaga 1992; Rose et al. 2002; Jan et al. 2004; Becnel et al. 2006).
Pectin is synthesized in the Golgi apparatus and secreted to the cell wall and is considered to be a critical element that controls cell wall elasticity and expansion. As mentioned earlier, the degree of pectin methylesterification depends on the action of PMT, PME, and PMEI. A higher degree of demethylation frees carboxyl groups of galacturonic acids to form Ca2+ and Mg2+ intermolecular linkages that lead to hardening of pectin and reduce extensibility of the cell wall (Heldt and Piechulla 2010; Kim et al. 2015). The middle lamella, which is responsible for adhesion between adjacent cells, is composed mostly of pectin with a low degree of methylesterification (Caffall and Mohnen 2009; Wolf et al. 2009; Neumetzler et al. 2012). Therefore, as with XTHs, alterations in genes encoding PMT, PME, and PMEI can have a significant effect on cell size and leaf architecture. This section will look at how alterations in XTH, PME, and PMEI gene expression and that of PMTs affect leaf architecture. Expansins will not be discussed in this review.
A. Xyloglucan Endotransglucosylase/Hydrolase
The XTH gene family has 33 genes in A. thaliana and 29 in O. sativa encoding xyloglucan endotransglucosylase/hydrolase (Yokoyama and Nishitani 2001; Yokoyama et al. 2004). Developmental stage and organ-based expression patterns of XTH has been extensively studied and the following genes have been shown to be highly expressed in young to mature rosette leaves of A. thaliana: XTH4, 6–9, 16, 22–24, 27, 28, and 31, 32. Interestingly, not all XTHs positively affect cell expansion. For example, overexpression of XTH3, XTH17, and XTH24 resulted in the development of smaller leaves as a result of the production of a large number of small cells in the mesophyll or as a result of a reduction in cell number (Verica and Medford 1997; Matsui et al. 2005; Cho et al. 2006; Han et al. 2013). These results point to effects on cell proliferation. On the other hand, many XTH genes, when overexpressed, enhance cell expansion and leaf size indicating that these are involved in cell wall loosening (Ogawa et al. 1996; Itoh et al. 2002; Jan et al. 2004; Shin et al. 2006; Liu et al. 2007; Miura and Hasegawa 2010; Hara et al. 2014). Some XTHs, namely XTH27, seem to specifically and positively regulate growth of tracheids with no role in leaf expansion (Matsui et al. 2005). Many studies have shown XTH to be involved in stress responses to salinity. For example, salt stress induces XTH17 and XTH3 gene expression that results in alterations in cell wall properties, remodelling of stomata, and alterations in mesophyll architecture; the modified leaf architecture increases water retention and survival (Yokoyama and Nishitani 2001; Cho et al. 2006; Chan et al. 2011; Keuskamp et al. 2011; Han et al. 2013). Regulation of XTH expression by many growth regulators including SA, JA, and GA is also evident (Yokoyama and Nishitani 2001; Jan et al. 2004; Keuskamp et al., 2011; Campos et al. 2016; unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.).
There is evidence to support that not only is CESA expression coordinated with pectin methylesterification and demethylesterifcation, it is also coordinated with cell wall loosening by XTH expression, specifically with XTH21 expression (Liu et al. 2007). Suppression of XTH21 led to a reduction in CESA2 and 4 expression in A. thaliana (Liu et al. 2007). Leaf architecture reported for XTH21 suppressed A. thaliana lines are similar to the leaf characteristics reported for CGR2 and CGR3 suppressed lines (Kim et al. 2015; Weraduwage et al. 2016) or CESA1 silenced N. benthamina lines (Burton et al. 2000). The fact that XTH24 is upregulated in an mutants (Tsuge et al. 1996; Kim et al. 2002), and that XTHs are regulated in response to stress and growth regulators, show that XTHs form a key molecular system that modifies cell wall properties and leaf architecture in response to external stimuli; while doing so, it is capable of altering the action of CESA to support cell wall modifications and growth.
ERECTA (ER) is another gene family that has been found to affect leaf architecture. These are leucine-rich repeat receptor-like kinases known to control a variety of developmental processes including leaf initiation, stem elongation, and leaf elongation in the length direction (Shpak et al. 2004; Masle et al. 2005; Sánchez-Rodríguez et al. 2009; Villagarcia et al. 2012). Three ER family genes have been found in A. thaliana and suppression of these genes caused significant changes in leaf shape, size, and mesophyll anatomy (Shpak et al. 2004; Masle et al. 2005; Sánchez-Rodríguez et al. 2009; Villagarcia et al. 2012). In addition, genes of the ER family, through their effects on epidermal cell expansion, have been shown to reduce stomatal density and improve transpiration efficiency (Masle et al. 2005; Villagarcia et al. 2012). Interestingly, alterations in expression of ER genes have been found to alter cell wall composition while the degree of pectin methylesterification was unaffected (Sánchez-Rodríguez et al. 2009). Thus, it is hypothesized that ER regulates leaf architecture by its effects on cell proliferation (Shpak et al. 2004; Masle et al. 2005; Villagarcia et al. 2012). er mutants were shown to have fewer, loosely arranged large mesophyll cells in the spongy tissue (Masle et al. 2005; Ferjani et al. 2007). Analyses of cell proliferation and cell expansion rates revealed that the cell enlargement in er mutants was “compensated cell enlargement” triggered by reduced rates of cell proliferation (Ferjani et al. 2007). The molecular mechanisms involved in cell-to-cell communication that link cell proliferation and cell enlargement in determinate organs such as leaves is not clear (Ferjani et al. 2007). The exact mechanism through which ER genes affect cell wall properties also remains to be found. The effects on cell proliferation suggest a possible involvement of XTH, e.g. XTH24. Furthermore, ER modulation of epidermal cell expansion indicates potential impacts on genes involved in regulating epidermal cell interdigitation (Fig. 8.6).
In summary, there is strong evidence for the participation XTH genes in regulating cell expansion, cell proliferation, and mesophyll and leaf architecture. Coordinated expression of these genes in relation to other molecular systems will be discussed in the following section.
1.XTH as a Key Downstream Point of Execution of Leaf Architectural Changes, and Its Modulation by CAMTA/SA, JAZ/JA and PHYB/GA/PIF
As discussed previously, some XTH genes regulate wall loosening. Interestingly, expression of AN and PIF have opposite effects on XTH24 expression (Figs. 8.11a and 8.12) (Campos et al. 2016). An increase in XTH24 expression in an mutant lines led to narrow and thick leaves. Therefore, wider and shorter cells in the palisade tissue seen in phyB are unlikely a result of increased XTH24, but may occur through the action of different XTHs and other mechanisms such as enhanced PMT activity as described below or through alterations in LNG and ROT3 expression (Fig. 8.10c–d). LNG leads to formation of longer leaves and ROT3 leads to longer, thinner leaves as a result of shorter cells in palisade tissue. We hypothesize that ROT3 acts on anisotropic cell expansion through XTH4, 8, 9, 17, 23 because BRs have been shown to induce their expression (Yokoyama and Nishitani 2001) and because ROT3, BR2OX, and XTH4 are all induced in phyB (Fig. 8.10c) (Campos et al. 2016).
Comparison of altered expression of genes coding for XTH and EXP in A. thaliana mutant lines showing altered leaf architecture. Expression levels of Xyloglucan endotransglucosylase/hydrolase (XTH) in A. thaliana Col-0 wild-type and (a) jazQ, phyB, and jazQphyB mutant lines (Campos et al. 2016), and (b) camta1/2/3, and camta1/2/3sid2–1 mutant lines (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.), are shown. (c) Expression levels of Expansin (EXP) genes in A. thaliana Col-0 wild type and jazQ, phyB, and jazQphyB mutant lines are presented. Expression levels were determined by leaf messenger RNA sequencing. In (a) and (c), values represent the mean ± SE and n = 3 plants per line and statistically different expression levels in comparisons to Col-0 found according to the DESeq algorithm (P < 0.05, using a Benjamini-Hochberg adjusted for multiple testing) are marked with asterisks. In (b), values presented are Log2 fold differences in expression (transgenic/Col-0) of XTH genes as determined by RNA-seq analysis of leaves and negative values indicate indicated a lower level of expression relative to Col-0
CAMTA /SA, JAZ /JA, and PHYB /GA/PIF-mediated effects on XTH gene expression. A schematic diagram is presented summarizing the effects of salicylic acid (SA), JASMONATE ZIM-domain (JAZ) repressors, and phytochrome interacting transcription factor (PIF)-mediated effects on specific target xyloglucan endotransglucosylase/hydrolase (XTH) gene expression. Data was derived from messenger RNA sequencing obtained from null mutant lines of Phytochrome-B (PHYB) and/or JAZ gene expression (Campos et al. 2016), and from mutant lines having enhanced SA production as a result of suppressed Calmodulin binding transcription activator (CAMTA) gene expression (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.). Upregulation and downregulation of gene expression is denoted by pointed and blunt ended arrows, respectively. Other abbreviations: DELLA – PIF transcription factor repressors, CBF- CRT/DRE Binding Factor, ICS1 – Isochorismate synthase, CBP60G and SARD1 – transcription factors with CAMTA DNA-binding motifs in promoter regions that positively regulate ICS1
Interestingly, our data revealed a general PIF mediated upregulation of XTH genes (and expansins) in the phyB mutant with wider and shorter palisade tissue cells and larger leaf area whereas XTH31 gene expression was suppressed in jazQ with smaller leaves (Figs. 8.11a, c and 8.12) (Campos et al. 2016). Both XTH8 and XTH31 were downregulated in camta1/2/3 in an SA dependent manner; camta2/3 produced significantly small cells and leaves (Figs. 8.11b and 8.12) (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.). These data show that PIF may affect XTH in a manner opposite to that of JAZ and SA. In other words, PIF would mostly enhance the expression of XTH, and transcription factors suppressed by JAZ (e.g., MYC) would downregulate the expression of XTH, to enhance and suppress leaf growth, respectively (Fig. 8.12). In contrast, transcription factors suppressed by JAZ and SA may act synergistically on XTH to suppress leaf growth by suppressing XTH31 that is common to both pathways and may play an important role in growth suppression during defense or stress responses (Fig. 8.12). This also supports the hypothesis that changes to leaf architecture occurring in response to stress responses take place first at the genetic level at common action points such as changes to XTH. Overall, it is clear that XTH, which directly regulates cell wall loosening and the capability of cell expansion, is a key downstream execution point of leaf architecture changes common to AN, CAMTA/SA, JAZ/JA, and PHYB/GA/PIF mediated molecular mechanisms.
B. Regulation of Ca2+ Mediated Cross-Linking of Pectin
1. Pectin Methylesterase and Pectin Methylesterase Inhibitor
Pectin methylesterases (PME) catalyze removal of the methyl moiety from methylated galacturonic acid and release methanol (Pilling et al. 2004; Oikawa et al. 2011). This methanol is given off as a gas and methanol emissions from forests occur when leaves are developing (Hu et al. 2011). Pectin methylesterase is a large gene family constituting more than 67 genes in A. thaliana (Markovic and Janecek 2004; Lionetti et al. 2007). A similarly large family of more than 69 genes encoding for PME inhibitor (PMEI) proteins has also been discovered in A. thaliana and other plants (Giovane et al. 1995; Jiang et al. 2002; Raiola et al. 2004; Lionetti et al. 2007; Wolf et al. 2009; Volpi et al. 2011). While the action of PME promotes cell wall hardening as explained above, demethylesterified pectin has been found susceptible to fungal endopolygalacturonases and pectin lyase (de Vries and Visser 2001; Lionetti et al. 2007). In fact, PME activity plays a significant role in mediating plant-pathogen interactions (Chen et al. 2000; Wietholter et al. 2003; Lionetti et al. 2007; Raiola et al. 2011). For example, overexpression of PMEI1 reduces PME activity while enhancing the degree of methylesterified pectin in cell walls and resistance to pectin degrading fungal enzymes; suppression of PMEI1 enhanced susceptibility (Lionetti et al. 2007; An et al. 2008; Volpi et al. 2011).
Data on the effect of altered PME and PMEI expression and activity on leaf architecture are scarce. Only a few studies provide evidence to support the idea that enhanced PMEI and reduced PME activity promote cell wall extensibility and cell expansion in cotyledons and leaves leading to their increased size (Pilling et al. 2004; Neumetzler et al. 2012; Peaucelle et al. 2012; Müller et al. 2013a, b; Levesque-Tremblay et al. 2015). However, enhanced PMEI and reduced PME activity has the opposite effect on growth and differentiation of the shoot meristem, stems, and hypocotyls (Peaucelle et al. 2008, 2012). Recently, a small Golgi-localized protein, FRIABLE1, was found to be a negative regulator of PME expression in A. thaliana (Neumetzler et al. 2012). Overall, based on existing data, negative regulation of PME by PMEI promotes leaf cell expansion and leaf growth owing to reduced cell wall hardening. Although these studies did not observe PME mediated alterations of cell adhesion in leaf cells, enhanced PME expression was found to promote cell adhesion between cotyledon cells leading to a reduction in cotyledon size (Neumetzler et al. 2012). In addition, alterations in a SQUAMOSA PROMOTER BINDING PROTEIN–LIKE GENE (SBP-BOX) reduced pectin methylesterase activity, pectin-Ca2+ cross-linkages, and cell-to-cell adhesion resulting in large intercellular airspaces in the fruit pericarp in Solanum lycopersicon (Orfila et al. 2001; Eriksson et al. 2004; Manning et al. 2006; Caffall and Mohnen 2009). Therefore, it is likely that PME activity plays a role in leaf cell-to-cell adhesion. However, PMTs, such as CGR2 and CGR3, seem to have a stronger effect on cell-to-cell adhesion as discussed below.
2. Pectin Methyltransferase
Pectin methyltransferase (PMT) catalyzes methylesterification of pectin. Out of the 29 putative PMT genes in A. thaliana, only the effects of QUASIMODO1, 2, and 3 (QUA1, QUA2, QUA3) (Mouille et al. 2007; Miao et al. 2011), TUMOROUS SHOOT DEVELOPMENT2 (TSD2) (Krupkova et al. 2007), and COTTON GOLGI-RELATED (CGR2, 3) genes (Held et al. 2011; Kim et al. 2015) on cell expansion have been investigated. Interestingly, partial suppression of QUA1, QUA2, or TSD2 did not enhance cell-cell adhesion, but reduced it as evident by cell detachment in the hypocotyl (Bouton et al. 2002; Krupkova et al. 2007; Mouille et al. 2007). Even though suppression of PMT expression is predicted to result in a decrease in pectin methylesterification and an enhancement of cell-to-cell adhesion, this was not seen during the above studies. Therefore, the function of the above genes as PMTs needs to be further characterized. It may also be that adhesive and expansion capabilities in leaf cells were reduced due to the suppression of the above genes causing the reduced leaf size in the corresponding mutant lines. However, a detailed anatomical study has to be conducted to test this possibility.
Recent studies provide compelling evidence to support the role of CGR2 and CGR3 in regulating mesophyll cell expansion and overall leaf architecture in A. thaliana (Kim et al. 2015; Weraduwage et al. 2016). Pectin content and pectin methylesterification in leaves were reduced in a double knockout mutant of CGR2 and CGR3 genes (cgr2/3) and the opposite effect was verified in lines overexpressing CGR2 (CGR2OX) (Held et al. 2011; Kim et al. 2015; Weraduwage et al. 2016). cgr2/3 mutant lines produced thin but dense leaf mesophyll with enhanced cell number and reduced air spaces compared to the wild-type (Fig. 8.13a–c) (Kim et al. 2015; Weraduwage et al. 2016). CGR2OX produced thinner leaves compared to the wild-type, but thicker than cgr2/3. Cells and intercellular air spaces in CGR2OX leaves were also larger than in the wild-type (Kim et al. 2015; Weraduwage et al. 2016). Both projected and total leaf area were markedly reduced in cgr2/3 and enhanced in CGR2OX (Fig. 8.13f) (Weraduwage et al. 2016). Above phenotypes in cgr2/3 were partially resored in cgr2com by CGR2 complementation. However, despite the changes in leaf expansion, changes in overall leaf shape were not detected (Fig. 8.13f). These data show that CGR2 and CGR3 are involved in cell expansion and thereby play a crucial role in determining leaf architecture. The authors hypothesized a reduction in expression of CGR2 and CGR3 causes cell wall hardening as a result of reduced pectin methylesterification and a greater degree of Ca2+ mediated cross-linking of pectin (Kim et al. 2015; Weraduwage et al. 2016). An increase in cell-to-cell adhesion may have caused the increase in cell density and reduced intercellular airspaces in the cgr2/3 mutant while the promotion of mesophyll cell expansion observed with CGR2 overexpression is probably due to a reduction in cell wall hardening and cell adhesion brought about by an increase in pectin methylesterification. However, given that no change in leaf shape was apparent, expression of CGR2 and/or CGR3 does not seem to affect microtubule alignment, but rather have a role in general cell expansion independently from the cytoskeleton.
The effect of altered CGR2 and CGR3 gene expression on leaf architecture. (a) Representative micrographs of leaf cross sections, (b) leaf thickness, (c) the relationship between the number of mesophyll cells and size of the intercellular air spaces in the leaf mesophyll, (d) the surface area of mesophyll cells facing intercellular air spaces per unit leaf area (S mes), (e) the surface area of chloroplasts facing intercellular air spaces per unit leaf area (S c), (f) size comparison of rosettes, and (g) the δ13CVPDB value calculated as the ratio of 13C to 12C isotopes in leaf tissue relative to a Vienna-Pee-Dee Belemnite standard (VPDB) (top panel), and the CO2 partial pressure at rubisco calculated using the δ13CVPDB values (bottom panel), are presented for A. thaliana wild-type Col-0 and mutant lines: cgr2/3 (loss of function double mutant line of CGR2 and CGR3), cgr2com (cgr2/3 complemented by CGR2), and CGR2OX (CGR2 overexpression line). δ13CVPDB is described in the Fig. 8.2 legend. In (a), leaf thickness is denoted by red double arrows. In (a–e), data were obtained from 34-day old leaves. In (f), rosettes were photographed 45 days after seeding. In (b), (d), and (e) values represent the mean ± SE and n = 4 plants per line. In (c) n = 4 plants per line were used to obtain the mean values for the area of air spaces as a % of area of leaf cross section and the number of mesophyll cells per mm2 of leaf cross section. Differences between means were tested by carrying out a one-way ANOVA at a = 0.05, followed by a Fisher’s Least Significant Difference Test. Statistical differences at P < 0.05 are marked with lower case letters. (Reproduced from Weraduwage et al. 2016)
In summary, data presented above show that PMTs, PME, and PMEI form an effective molecular system to mediate the degree of pectin methylesterification in order to fine tune cell expansion and adhesion and, consequently, mesophyll and overall leaf architecture. Based on the data available so far, PMTs such as CGR2 and CGR3 seem to cause more controllable alterations in cell expansion and leaf architecture compared to PMEI, PME, and other identified putative PMTs. The following section summarizes recent evidence showing how the PMT/PME/PMEI system can act as a key downstream molecular system common to different upstream signaling pathways targeting changes in leaf architecture.
3. PMT/PME/PMEI System as a Key Downstream Execution Point of Leaf Architectural Changes and Its Modulation by CAMTA/SA, JAZ/JA, and PHYB/GA/PIF
In general, PIF positively affects PMEI expression whereas CAMTA/SA has a significant inhibitory effect (Figs. 8.14 and 8.15) (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T., and by M.L.C., Y.Y., I.T.M., S.M.W., T.D.S., and G.A.H.). This may lead to an inactivation of PMEs in phyB and activation in camta1/2/3 with a corresponding increase and decrease in pectin methylesterification, respectively (Fig. 8.15). This is further supported by the fact that the mesophyll architecture of cgr2/3 with reduced CGR2 and CGR3 expression and reduced pectin methylesterifiation was similar to camta2/3 (Figs. 8.2 and 8.13). Significant changes in PMEI expression could not be detected in jazQ mutant lines (Fig. 8.14). This may be why jazQ did not show any drastic changes in mesophyll architecture despite having smaller leaves (Fig. 8.3). Thus, the PMT/PME/PMEI system, which regulates the degree of methylation of pectin and subsequently cell wall extensibility and adhesive properties, is likely a key downstream execution point common to the PHYB/GA/PIF and CAMTA/SA genetic systems, which seem to have opposite effects on pectin methylesterification (Fig. 8.15). Changes to pectin methylesterification can have more drastic negative effects on leaf architecture than those observed by effects on XTH and expansins alone. These data also emphasize PMT/PME/PMEI as a key downstream molecular system through which changes to leaf architectural changes are executed in response to stress (Fig. 8.15).
Comparison of altered expression of genes coding for PMEI in Arabidopsis mutant lines showing altered leaf architecture. Expression levels of Pectin methylesterase inhibitor (PMEI) in A. thaliana Col-0 wild-type and (a) jazQ, phyB, and jazQphyB mutant lines (Campos et al. 2016), and (b) camta1/2/3 and camta1/2/3sid2–1 mutant lines (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.), are shown. Expression levels were determined by leaf messenger RNA sequencing. Values represent the mean ± SE and n = 3 plants per line. In (a), values represent the mean ± SE and n = 3 plants per line and statistically different expression levels in comparison to Col-0 found according to the DESeq algorithm (P < 0.05, using a Benjamini-Hochberg adjusted for multiple testing) are marked with asterisks. In (b), values presented are Log2 fold differences in expression (transgenic/Col-0) of PMEI genes as determined by RNA-seq analysis of leaves and positive and negative values indicate a higher or a lower level of expression relative to Col-0, respectively
PHYB /GA/PIF and CAMTA /SA mediated effects on PMEI gene expression and pectin methylesterification. A schematic diagram is presented summarizing the effects of Phytochrome interacting transcription factor (PIF) and salicylic acid (SA) on specific target Pectin methylesterase inhibitor (PMEI) gene expression. Data were derived from messenger RNA sequencing data obtained from null mutant lines of Phytochrome-B (PHYB) gene expression (Campos et al. 2016) and from mutant lines having enhanced SA production as a result of suppressed Calmodulin binding transcription activator (CAMTA) gene expression (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.). Different PMEI genes are denoted by their GenBank accession numbers. The degree of pectin methylation will depend upon: 1. the activity of pectin methylesterase (PME) that demethylates pectin, 2. the expression and activity of PMEI that inhibits PME, and 3. the expression and activity of pectin methyltransferase. A higher degree of methylesterification of pectin has been shown to reduce cell wall hardening and promote cell expansion, while a lower degree of methylesterification promotes cell wall hardening and cell-to-cell adhesion. New evidence was also found to support upregulation of Longifolia (enhances cell expansion in the leaf-length direction) and Rotundifolia3 (enhances cell expansion in the leaf length direction while limiting cell expansion in the leaf-depth direction) gene expression by PIF (Campos et al. 2016). Upregulation and down regulation of gene expression is denoted by pointed and blunt ended arrows, respectively. Other abbreviations: DELLA –PIF transcription factor repressors, CGR2 and CGR3 – Cotton Golgi-related 2 and 3, QUA2 and QUA3 – Quasimodo 2 and 3
V. Broader Implications of Understanding Genes and Molecular Mechanisms That Affect Cell Wall Properties and Leaf Architecture
The architecture of the leaf is designed to ultimately produce an organ that is optimized to act as a solar collector as well as an efficient gas exchanger to maximize photosynthesis. Although a large number of mutations affecting leaf architecture have been studied during the past two decades (Sections II-IV), these mutations have not been studied in the context of positive or negative effects on photosynthesis. In fact, alterations in leaf architecture have profound effects not only on photosynthesis, but also on respiration and overall plant growth (Lambers et al. 2008; Weraduwage et al. 2015, 2016). In the preceding sections, leaf architecture of many mutants including that of phyB, jazQ, camta and cgr mutants and the potential molecular mechanisms involved in bringing about such architectural changes were discussed. Here, we will discuss the impact of such architectural changes on photosynthesis, C partitioning, and plant growth and identify genetic candidates that can be used as tools for crop improvement.
A. Mesophyll Architecture and Its Impact on CO2 Availability at Rubisco and Area-Based Photosynthesis
CO2 diffusion into the chloroplast stroma through the cell wall, plasma membrane, and chloroplast envelope takes place along the route that poses the lowest resistance (Terashima et al. 2006). The active area through which CO2 diffuses in to the chloroplast stroma is the chloroplast surface area facing intercellular air spaces per unit leaf area (S c), and S c and internal conductance are positively correlated (Terashima et al. 2006). Lower S c leads to a reduction in CO2 concentration at rubisco with subsequent reductions in carboxylation/oxygenation ratio (Terashima et al. 2006). camta2/3 (Fig. 8.2d), and cgr2/3 (Fig. 8.13g) with densely packed cells in the leaf mesophyll, and phyB and jazQphyB with thin leaves (Fig. 8.3c) showed a reduction in the degree of 13CO2 discrimination during CO2 assimilation and a decrease in average CO2 partial pressure at rubisco (Weraduwage et al. 2016; unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T.; unpublished data by M.L.C., Y.Y., I.T.M., S.M.W., T.D.S., and G.A.H). A decrease in average CO2 partial pressure at rubisco was also accompanied by a reduction in area-based photosynthesis rates in camta2/3 (unpublished data by Y-S.K., S.M.W., T.D.S., and M.F.T), phyB and jazQphyB (Campos et al. 2016), and cgr2/3 (Weraduwage et al. 2016). The above effects on leaf gas exchange properties were greatest in camta2/3 and cgr2/3 where the mesophyll cell density, and hence leaf dry mass per unit leaf area (LMA), was greatest. Further examination of the leaf mesophyll of cgr2/3 revealed a significant reduction in mesophyll cell surface area facing intercellular air spaces per unit leaf area (S mes) and S c (Fig. 8.13d–e). Thus, the reduction in CO2 availability for photosynthesis in camta2/3 and cgr2/3 is likely a result of reduced S mes, S c, and intercellular air spaces.
LMA has been shown to both positively and negatively affect mesophyll conductance depending on the plant species (Flexas et al. 2008; Soolanayakanahally et al. 2009; Tosens et al. 2012). Milla-Moreno et al. (2016) showed that in P. balsamifera, even though an increase in the number of cell layers in palisade tissue can increase leaf thickness and subsequently decrease internal CO2 conductance, there was a greater positive effect on mesophyll conductance in terms of enhanced S mes and S c. Thus, in P. balsamifera, LMA and mesophyll conductance was positively correlated (Milla-Moreno et al. 2016). However, in camta2/3 and cgr2/3 a negative correlation is likely between LMA and mesophyll conductance. A reduction in GA biosynthesis or sensitivity to this growth regulator increases LMA in A. thaliana and S. lycopersicon; GA supplementation lowers LMA (Dijkstra et al. 1990; Nagel et al. 2001; Poorter et al. 2009). Under light, a decrease in GA occurs as a result of both a reduction in the transcription of genes involved in GA biosynthesis and an increase in gibberellin-2-oxidase that increases GA catabolism (Folta et al. 2003; Hisamatsu et al. 2005; Foo et al. 2006; Achard et al. 2007; Weller et al. 2009; Pierik et al. 2011; Hirose et al. 2012; Colebrook et al. 2014; Mazzella et al. 2014). phyB and jazQphyB maintained fairly large intercellular air spaces, but with shorter and wider cells in the palisade tissue and reduced cell layers and subsequently thinner leaves, which resulted in lower LMA and area-based photosynthesis in these lines (Campos et al. 2016). Reduced area-based photosynthesis rates were also reported in gigantea-2 (gi-2) mutant lines with thin leaves and lower LMA (Weraduwage et al. 2015), and in er mutants despite having larger intercellular airspaces in the leaf mesophyll (Masle et al. 2005). These observations are attributed to the lack of photosynthetic machinery on a leaf area basis in thinner leaves (Masle et al. 2005; Campos et al. 2016). One difference between sun and shade leaves is that the former produces thicker leaves (longer palisade cells or multiple cell layers, higher LMA) that enables the housing of more chloroplasts per unit leaf area and increased amounts of photosynthetic enzymes per unit leaf area, allowing for the maintenance of higher area-based photosynthesis rates; the opposite is seen in shade leaves (Lambers et al. 2008).
The advantages of producing leaves with a larger surface area (lower LMA) and the disadvantages of producing leaves with higher LMA is discussed in the following sections. In summary, molecular mechanisms affecting cell wall properties and leaf architecture play a significant role in modulating area-based photosynthesis rates through CO2 diffusion into cells as well as concentrating resources required for photosynthesis per unit leaf area.
B. Mesophyll Architecture and Its Impact on Area-Based Respiration and Daily C Gain
An increase in leaf cell density enhances leaf mass density (LMD, dry mass of leaf per unit volume of leaf tissue) and both leaf thickness and LMD can affect LMA (Lambers et al. 2008; Poorter et al. 2009; Weraduwage et al. 2016). For example, despite having thinner leaves, an increase in leaf cell density resulted in higher LMA in cgr2/3 (Weraduwage et al. 2016). Analysis of anatomical components that affect LMA has revealed palisade tissue cell properties to be the major contributor to variations in LMA in mature leaves of Populus balsamifera (Milla-Moreno et al. 2016).
As mentioned before, thicker leaves usually possess larger LMA as a result of a greater number of cell layers (Lambers et al. 2008; Poorter et al. 2009; Villar et al. 2013; Weraduwage et al. 2015). Changes in leaf architecture such as a larger number of smaller cells with thicker cell walls can also lead to (1) changes in chemical composition such as a higher proportion of lignin and other cell wall polysaccharides per unit leaf area that can result in higher LMD and LMA and (2) cellular changes such as increased organelle numbers (Cunningham et al. 1999; Lambers et al. 2008). For example, cgr2/3 cells possess a large number of small chloroplasts per unit leaf area that could also lead to higher LMA; chloroplast thickness was not reported in this study. However, this only translated to higher area-based photosynthesis rates during early growth stages of cgr2/3 and at latter stages area-based photosynthesis rates were significantly lower as a result of lower S c (Weraduwage et al. 2016).
Leaves with larger LMA require more energy to maintain a greater number of cells and organelles per unit leaf area and hence incur higher maintenance respiratory costs (Lambers et al. 2008). This was observed in cgr2/3 in which higher leaf cell density and higher LMA positively correlated with enhanced area-based respiration (Weraduwage et al. 2016). Consequently, photosynthesis to respiration ratios were lower, and coupled with smaller projected leaf area, a significant reduction in daily carbon gain, net assimilated C for growth and overall plant growth was seen in cgr2/3 (Fig. 8.16). In contrast, in mutant lines such as gi-2 (Weraduwage et al. 2015) and CGR2OX (Weraduwage et al. 2016), which produced larger, thinner leaves with smaller LMA, area-based respiration was smaller and photosynthesis to respiration ratios were larger and coupled with larger projected leaf area, a significant increase in daily carbon gain, net assimilated C for growth, and overall plant growth was seen (Fig. 8.16). It is also assumed that phyB and jazQphyB incur lower construction costs to build their thinner leaves compared to that in jazQ (Campos et al. 2016). Overall, the above data indicate that molecular mechanisms affecting cell wall properties and leaf architecture can have a significant impact on area-based respiration in leaves and daily C gain.
CGR2 and CGR3 mediated pectin methylesterification regulates the relationship between photosynthesis and plant growth. A schematic diagram outlining how (a) suppression or (b) over-expression of CGR2 and CGR3 gene expression affects the relationship between photosynthesis and plant growth in A. thaliana is presented. Suppression of CGR2 and CGR3 reduces the degree of pectin methylation, which leads to an increase in cell-to-cell adhesion, cation-mediated cross linking of galacturonic acid, and consequent hardening of cell walls. CGR2 over-expression increases the degree of pectin methylation, which allows cell expansion through reduced cell-to-cell adhesion and cation-mediated cross linking. It has been proposed that pectin methyltransferase enzyme, through its ability to directly alter cell expansion, determines the amount of C partitioned to leaf area growth versus growth in terms of LMA. For example, while more C is partitioned to growth in terms of LMA in the CGR2 and CGR3 double knockout mutant, in the CGR2 over-expression line more C is partitioned to leaf area growth. An increase in LMA leads to enhanced area-based respiration and a reduction in leaf area contribute to a reduction in whole plant photosynthesis. Collectively, this results in a reduction in C available for growth and consequently a decrease in overall plant growth; an opposite trend is seen in CGR2OX. Thus, CGR2 and CGR3, through their ability to alter the degree of methylated pectin in the cell wall of the mesophyll cells, determine how photosynthate is utilized to grow the plant. In other words, CGR2 and CGR3 mediated pectin methylesterification affects the relationship between photosynthesis and plant growth by regulating the proportions of C that are partitioned to leaf area growth and LMA. Final overall growth mainly depends on the expression patterns of CGR2 and CGR3 and how much C is partitioned to area growth and LMA and not on area-based photosynthesis. PME: pectin methylesterase; PMEI: PME inhibitor; PMT: pectin methyltransferase. CH3OH is methanol. (Reproduced from Weraduwage et al. 2016)
C. Leaf Architecture and Its Impact on Light Capture, Whole-Plant Photosynthesis, and Growth
Light capture is optimized by having a larger leaf area, and this is seen especially in plants grown under low light (Lambers et al. 2008). Shade plants often produce leaves with smaller LMA (Niinemets 2001; Lambers et al. 2008). This reduces area-based respiration rates, maximizing daily C gain, and allows compensation for lower area-based photosynthesis rates (because of lower LMA as described in Section VA) under low light (Lambers et al. 2008). Although area-based photosynthesis rates were lower, whole plant photosynthesis was enhanced in A. thaliana mutants capable of producing large rosettes, e.g., gi-2 (Weraduwage et al. 2015), phyB, and jazQphyB (Campos et al. 2016) and CGR2OX (Fig. 8.16) (Weraduwage et al. 2016) as a result of larger projected leaf area. Projected leaf area represents the effective leaf surface area capable of intercepting light (Honda and Fisher 1978). Model based analyses revealed that in CGR2OX more C is partitioned to leaf area growth (Fig. 8.16). Greater whole plant photosynthesis as a result of larger leaf area coupled with lower area-based respiration as a result of lower LMA led to an enhancement in C available for growth and consequently an increase in overall plant growth in CGR2OX (Fig. 8.16). This was also seen in jazQphyB (Campos et al. 2016).
LMA or 1/specific leaf area has been shown to be a key trait that determines variation in relative growth rates between plants growing in nutrient rich and nutrient poor conditions (Poorter and Remkes 1990; Garnier 1992; Lambers et al. 2008). Also, LMA often increases under water stress as a result of an increase in cell wall thickness and a reduction in cell size (Cutler et al. 1977; Utrillas and Alegre 1997; Van Volkenburgh and Boyer 1985; Fredeen et al. 1991; Niinemets 2001; Lambers et al. 2008). Thus, under drought stress, enhanced LMA may in part contribute to growth reductions based on factors mentioned above (Niinemets 2001).
D. Genes Such as CGR2 and CGR3 That Alter Cell Wall Properties Can Modulate the Relationship Between Photosynthesis and Growth
It has been shown that, while the correlation between area-based photosynthesis and plant growth is not clear, relative growth rate and leaf growth parameters such as leaf area per unit leaf dry mass (inverse of LMA), leaf area per unit plant dry mass, and investment of C in leaf growth are strongly and positively correlated (Shipley 2002; Lambers et al. 2008; Poorter et al. 2009). In dicots, there is a negative correlation between relative growth rate and the root:shoot ratio, highlighting the importance leaf expansion and overall growth (Garnier 1991; Lambers et al. 2008). Similarly, model based analyses of A. thaliana leaf growth revealed that while photosynthetic C is required for growth, the magnitude of plant growth depends on the proportions of C partitioned to leaf area growth and LMA (Weraduwage et al. 2015, 2016).
The Arabidopsis leaf area growth model was developed to simulate the C flow from the beginning to end of the A. thaliana life cycle while also simulating the utilization of assimilated C in respiration and the partitioning of the remaining C to grow leaves in the form of area and LMA, root growth, and reproduction (Weraduwage et al. 2015). The model can be fitted with measured data to determine partitioning coefficients of C that give rise to the growth patterns of different plants (Weraduwage et al. 2015). The responses of overall plant growth (plant dry weight) to varying magnitude of model inputs such as photosynthesis and partitioning coefficients can also be determined (Weraduwage et al. 2015). For example, the model revealed that enhanced leaf and plant growth in gi-2 (Weraduwage et al. 2015), and differences in leaf and plant growth and architecture in cgr2/3 and CGR2OX (Weraduwage et al. 2016), were not a result of differences in area-based photosynthesis rates, but mainly a result of altered C partitioning to leaf area growth and growth in terms of LMA (Figs. 8.16 and 8.17) (Weraduwage et al. 2016). C partitioning to leaf area growth was greater in CGR2OX and gi-2 and significantly smaller in cgr2/3; the opposite trend was seen in terms of C partitioning to LMA (Figs. 8.16 and 8.17). These findings obtained using the Arabidopsis leaf area growth model agree with inferences derived from classical growth models developed by Monsi (1960), Poorter and Lambers (1991), and Tillman (1991) where enhancements in growth rates were shown to positively correlate with biomass allocation to leaves. Therefore, “photosynthesis drives growth through alterations in carbon partitioning to new leaf area growth and leaf mass per unit leaf area” (Weraduwage et al. 2016). It was found that CGR2 and CGR3 genes can directly affect the relationship between photosynthesis and growth by directly altering the capacity of cell expansion and cellular organization in the leaf mesophyll, thus creating varying carbon demands for leaf area growth and LMA that will in turn drive C partitioning for these processes (Fig. 8.16). It can be hypothesized that other genes such as XTH8, XTH21, and XTH31, which have a profound effect on leaf architecture (Fig. 8.12) (Ogawa et al. 1996; Itoh et al. 2002; Jan et al. 2004; Liu et al. 2007), may also be able to affect C partitioning between leaf area growth and LMA.
Comparison of how changes in area-based photosynthesis and C partitioning to leaf growth affects plant growth in A. thaliana . The Arabidopsis Leaf Area Growth Model was used to test how changes in area-based photosynthesis and C partitioning to leaf growth affects plant growth. The model simulates plant growth based on the use of assimilated C in respiration and partitioning of the remaining C to leaf area growth and LMA, root growth, and reproduction. The model is capable of simulating plant growth (blue, black, and orange lines) based on the magnitude of a particular model input, e.g., area-based photosynthesis, C partitioning to different growth processes etc. Measured data from Col-0 wild-type, CGR2 overexpressed (CGR2OX) or CGR2, and CGR3 suppressed (cgr2/3) A. thaliana lines were used to test whether the enhancement in growth in CGR2OX was as a result of alterations in area-based photosynthesis or as a result of alterations in C partitioning to leaf area growth (with corresponding reductions of C partitioning to LMA); CGR2OX showed enhanced growth and cgr2/3 showed suppressed growth (Kim et al. 2015; Weraduwage et al. 2016). In order to do so, area-based photosynthesis (a) or partitioning coefficients to leaf area growth (b) for Col-0 were replaced by that of CGR2OX (orange line) or cgr2/3 (black lines). In (a) and (b), blue lines represent modeled growth for Col-0 and red squares represent measured data points (mean ± SD) for Col-0 at 29, 49, 63, and 82 days after seeding. Black lines represent modeled data for Col-0 when its area-based photosynthesis rates (a) or C partitioning to leaf area growth (b) are replaced by that of cgr2/3. Orange lines represent modeled data for Col-0 when its area-based photosynthesis rates (a) or C partitioning to leaf area growth (b) are replaced by that of CGR2OX. Asterisks represent measured data for CGR2OX at 63 and 82 days after seeding and dotted lines indicates the upper limit of measured data for cgr2/3. (Reproduced from Weraduwage et al. 2016). Based on the model, the observed enhancement in growth in CGR2OX and reduced growth in cgr2/3 occurs as a result of small changes in C partitioning to leaf area growth and LMA (Weraduwage et al. 2016). For more information on the Arabidopsis Leaf Area Growth Model, please read Weraduwage et al. (2015, 2016) (Colour figure online)
In summary, alterations of leaf and mesophyll architecture through cell wall modifications can have a marked influence on net C assimilation as a result of effects on: 1. CO2 availability at rubisco, 2. light interception, and 3. respiratory costs. Thus, cell wall plasticity is a key factor influencing photosynthetic processes in plants. Therefore, genes and molecular systems that modulate cell wall properties can be utilized to optimize leaf architecture to maximize photosynthesis. Genes that code for PMTs, specifically CGR2 and CGR3, and XTH through their role in regulating cell wall plasticity and mesophyll architecture can direct C partitioning between leaf area growth and LMA and thereby, directly affect the relationship between photosynthesis and plant growth. Thus, understanding genes and corresponding molecular mechanisms that affect cell wall properties and leaf architecture is of utmost importance to select candidate genes for crop improvement.
VI. Conclusions
Based on results from key studies carried out during the past two and a half decades, several major molecular systems that interact to regulate cell wall composition (CESA/CSL, PMT/PME/PMEI, XTH), anisotropic cell expansion (ROP and associated genes, LNG, ROT3, AN, XTH), and isotropic cell expansion (XTH, PMT/PME/PMEI) to cause profound changes in leaf architecture were identified. Upstream signaling systems such as CAMTA/SA, JAZ/JA, and PHYB/GA/PIF can cause significant changes in leaf architecture by interacting with a large number of midstream and downstream molecular pathways. PHYB/GA/PIF can affect leaf architecture by altering expression of genes belonging to a variety of molecular pathways including: ROPs, LNG and ROT3, CESA/CSL, XTH, EXP, and PMEI. AN interacted with PHYB/GA/PIF at downstream XTH genes. Recent evidence suggests that the effects of CAMTA/SA on leaf architecture may be occurring through XTH and PMEI and that of JAZ/JA through XTH and EXP. The major downstream execution points of leaf architecture changes common to CAMTA/SA, JAZ/JA, and PHYB/GA/PIF mediated signaling pathways were the XTH and PMT/PME/PMEI systems. XTH expression is also affected by both ROT3 and AN. Overall, growth promotion by PHYB/GA/PIF and growth suppression by JAZ/JA and CAMTA/SA seems to occur via modulation of XTH and PMEI of the PMT/PME/PMEI systems in opposite directions. It is also clear that changes in leaf architecture that occur in response to stress responses take place first at the genetic level at common action points such as XTH and PMT/PME/PMEI.
C for growth for most organisms on earth is supplied through photosynthesis and the leaf is the primary photosynthetic organ in plants. Alterations in leaf architecture have a profound impact on (1) CO2 availability at rubisco and area-based photosynthesis, (2) area-based respiration, and (3) light capture and whole-plant photosynthesis, with ultimate effects on daily C gain growth. In addition, molecular systems such as the PMT/PME/PMEI, and specifically CGR2 and CGR3 genes (PMTs) that alter cell wall properties, can modulate the relationship between photosynthesis and growth by directly altering the capacity of cell expansion and cellular organization in the leaf mesophyll thereby creating differential carbon demands for leaf area growth and LMA that can in turn drive C partitioning for these processes. Therefore, our understanding of genes and molecular mechanisms that affect cell wall properties and leaf architecture will facilitate the identification of novel genetic model systems that can be utilized to improve photosynthesis and growth of crop plants.
Abbreviations
- ABA:
-
abscisic acid
- ABP1 :
-
PUTATIVE AUXIN RECEPTOR AUXIN BINDING PROTEIN
- AIR:
-
alcohol insoluble residue
- AN :
-
ANGUSTIFOLIA
- ARP2/3 :
-
ACTIN RELATED PROTEIN
- BR:
-
brassinosteroids
- BRICK1 :
-
SCAR/WAVE ACTIN-NUCLEATING COMPLEX SUBUNIT
- C:
-
carbon
- CAMTA :
-
CALMODULIN BINDING TRANSCRIPTION ACTIVATOR
- CBF :
-
CRT/DRE BINDING FACTOR
- CBP60G :
-
CALMODULIN BINDING PROTEIN 60G
- CESA :
-
CELLULOSE SYNTHASE
- CGR :
-
COTTON GOLGI-RELATED
- cgr2com:
-
cgr2/3 complemented by CGR2
- CGR2OX:
-
CGR2 overexpression line of Arabidopsis thaliana
- CO2 :
-
carbon dioxide
- CRT:
-
C-repeat
- CSL :
-
CELLULOSE SYNTHASE LIKE
- CYP85A1 :
-
BRASSINOSTEROID-6-OXIDASE
- CYP90C1 :
-
CYTOCHROME P-450 FAMILY STEROID HYDROLASE
- DELLA:
-
PIF transcription factor repressors
- DRE:
-
dehydration responsive element
- ER :
-
ERECTA
- F-actin:
-
actin microfilaments
- FR:
-
far-red
- GA:
-
gibberellic acid
- GT8 :
-
GLYCOSYL TRANSFERASE8
- IAA:
-
auxin
- ICS1 :
-
ISOCHORISMATE SYNTHASE
- IIIe bHLHs:
-
basic helix-loop-helix subgroup IIIe transcription factors
- JA:
-
jasmonic acid
- JAZ:
-
jasmonate ZIM-domain repressors
- jazQ:
-
JAZ quintuple mutation
- LMA:
-
leaf dry mass per unit leaf area
- LMD:
-
leaf mass density
- LNG :
-
LONGIFOLIA
- MAP18 :
-
MICROTUBULE ASSOCIATED PROTEIN18
- MDP40 :
-
MICROTUBULE DESTABILIZING PROTEIN40
- MF:
-
fine actin filament
- MOR1 :
-
MICROTUBULE ORGANIZATION PROTEIN
- MT:
-
microtubule
- MYC2:
-
a basic helix-loop-helix transcription factor
- NAP1 :
-
NCK-ASSOCIATED PROTEIN
- nt:
-
nucleotide
- PCR:
-
polymerase chain reaction
- Pfr:
-
active form of phytochrome that absorbs far-red light
- PHYB :
-
PHYTOCHROME-B
- PIF:
-
phytochrome-interacting factors
- PIN :
-
PIN-FORMED AUXIN EFFLUX CARRIER GENE FAMILY PROTEIN
- PME :
-
PECTIN METHYLESTERASE
- PMEI :
-
PECTIN METHYLESTERASE INHIBITOR
- PMT:
-
pectin methyltransferase
- Pr:
-
inactive form of phytochrome that absorbs red light
- PVX:
-
potato virus X vector
- QUA :
-
QUASIMODO
- R:
-
red
- RhoGAP:
-
negative regulator of ROP
- RhoGEF:
-
Rho-guanine nucleotide exchange factors
- RIC :
-
ROP-INTERACTIVE CRIB MOTIF PROTEIN
- RNA-seq:
-
RNA sequencing
- ROP :
-
RHO-RELATED GTPASE FROM PLANT (RhoGTPase)
- ROT3 :
-
ROTUNDIFOLIA3
- rubisco:
-
ribulose bisphosphate carboxylase oxygenase
- SA:
-
salicylic acid
- SARD1 :
-
SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1
- SBP-BOX :
-
SQUAMOSA PROMOTER BINDING PROTEIN–LIKE GENE
- S c :
-
chloroplast surface area facing intercellular air spaces per unit leaf area
- SCAR :
-
SUPPRESSOR OF CYCLIC AMP RECEPTOR
- sid2-1 :
-
salicylic acid induction-deficient 2-1
- siRNA:
-
small interfering RNA
- S mes :
-
mesophyll cell surface area facing intercellular air spaces per unit leaf area
- SPIKE :
-
RHOGEF OR DOCK180-TYPE GUANINE NUCLEOTIDE EXCHANGE FACTOR
- SRA1 :
-
RAC1-ASSOCIATED PROTEIN-1
- TMK :
-
TRANSMEMBRANE KINASE SUBFAMILY OF RECEPTOR-LIKE KINASES
- TSD2 :
-
TUMOROUS SHOOT DEVELOPMENT2
- VIGS:
-
virus-induced gene silencing
- VPDB:
-
Vienna-Pee-Dee Belemnite standard
- WAVE :
-
WISKOTT–ALDRICH SYNDROME PROTEIN-FAMILY VERPROLIN HOMOLOGOUS PROTEIN
- XEH :
-
XYLOGLUCAN ENDOHYDROLASE
- XET :
-
XYLOGLUCAN ENDOTRANSGLUCOSYLASE
- XTH :
-
XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE
- XXT :
-
XYLOSYLTRANSFERASE
- δ13CVPDB :
-
ratio of 13C to 12C isotopes in leaf tissue relative to a Vienna-Pee-Dee Belemnite standard
References
Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP (2007) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143:1163–1172
An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228:61–78
Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W et al (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279:717–720
Baskin TI (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21:203–222
Basu D, Le J, El-Essal SE, Huang S, Zhang C, Mallery EL et al (2005) DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17:502–524
Basu D, Le J, Zakharova T, Mallery EL, Szymanski DB (2008) A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc Natl Acad Sci U S A 105:4044–4049
Becnel J, Natarajan M, Kipp A, Braam J (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61:451–467
Beeckman T, Przemeck GKH, Stamatiou G, Lau R, Terryn N, De Rycke R et al (2002) Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol 130:1883–1893
Behringer C, Schwechheimer C (2015) B-GATA transcription factors – insights into their structure, regulation and role in plant development. Front Plant Sci 6:90
Bouton S, Leboeuf E, Mouille G, Leydecker MT, Talbotec J, Granier F et al (2002) QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14:2577–2590
Bowman JL, Eshed Y, Baum SF (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18:134–141
Buchanan BB, Gruissem W, Jones RL (2000) Biochemistry and Molecular Biology of Plants. Wiley, Somerset
Burton RA, Gibeaut DM, Bacic A, Findlay K, Roberts K, Hamilton A et al (2000) Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12:691–705
Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134:224–236
Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A et al (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-d-glucans. Science 311:1940–1942
Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344:1879–1900
Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM et al (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 7:12570
Chaiwanon J, Wang W, Zhu J-Y, Oh E, Wang Z-Y (2016) Information integration and communication in plant growth regulation. Cell 164:1257–1268
Chan J (2012) Microtubule and cellulose microfibril orientation during plant cell and organ growth. J Microsc 247:23–32
Chan Z, Grumet R, Loescher W (2011) Global gene expression analysis of transgenic, mannitol-producing, and salt-tolerant Arabidopsis thaliana indicates widespread changes in abiotic and biotic stress-related genes. J Exp Bot 62:4787–4803
Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M (2012) Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One 7:9
Chen M-H, Sheng J, Hind G, Handa AK, Citovsky V (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J 19:913–920
Cho SK, Kim JE, Park JA, Eom TJ, Kim WT (2006) Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580:3136–3144
Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS et al (2000) Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J 21:431–443
Chou Y-H, Pogorelko G, Zabotina OA (2012) Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol 159:1355–1366
Colebrook EH, Thomas SG, Phillips AL, Hedden P (2014) The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol 217:67–75
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6:850–861
Craddock C, Lavagi I, Yang Z (2012) New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol 22:492–501
Cunningham SA, Summerhayes B, Westoby M (1999) Evolutionary divergences in leaf structure and chemistry, comparing rainfall and soil nutrient gradients. Ecol Monogr 69:569–588
Cutler JM, Rains DW, Loomis RS (1977) The importance of cell size in the water relations of plants. Physiol Plant 40:255–260
de Vries RP, Visser J (2001) Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev 65:497–522
Dijkstra P, Reegen H, Kuiper PJ (1990) Relation between relative growth rate, endogenous gibberellins, and the response to applied gibberellic acid for Plantago major. Physiol Plant 79:629–634
Djakovic S, Dyachok J, Burke M, Frank MJ, Smith LG (2006) BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development 133:1091–1100
Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB et al (2009) A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci U S A 106:5996–6001
Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21:972–984
Dwivany FM, Yulia D, Burton RA, Shirley NJ, Wilson SM, Fincher GB et al (2009) The CELLULOSE-SYNTHASE LIKE C (CSLC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Mol Plant 2:1025–1039
Ellis B, Daly DC, Hickey LJ, Mitchell JV, Johnson KR, Wilf P, Wing SL (2009) Manual of Leaf Architecture. Cornell University Press, Ithaca
Eriksson EM, Bovy A, Manning K, Harrison L, Andrews J, De Silva J et al (2004) Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol 136:4184–4197
Ferjani A, Horiguchi G, Yano S, Tsukaya H (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144:988–999
Filisetti-Cozzi TM, Carpita NC (1991) Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197:157–162
Finlayson SA, Hays DB, Morgan PW (2007) phyB-1 sorghum maintains responsiveness to simulated shade, irradiance and red light : far-red light. Plant Cell Environ 30:952–962
Flexas J, Ribas-Carbó M, Diaz-Espejo A, Galmés J, Medrano H (2008) Mesophyll conductance to CO2: current knowledge and future prospects. Plant Cell Environ 31:602–621
Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J 36:203–214
Foo E, Ross JJ, Davies NW, Reid JB, Weller JL (2006) A role for ethylene in the phytochrome-mediated control of vegetative development. Plant J 46:911–921
Frank MJ, Smith LG (2002) A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr Biol 12:849–853
Fredeen AL, Gamon JA, Field CB (1991) Responses of photosynthesis and carbohydrate-partitioning to limitations in nitrogen and water availability in field-grown sunflower. Plant Cell Environ 14:963–970
Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14:777–794
Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120:687–700
Fu Y, Xu T, Zhu L, Wen M, Yang Z (2009) A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr Biol 19:1827–1832
Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T et al (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9:1951–1962
Fujikura U, Horiguchi G, Tsukaya H (2007) Dissection of enhanced cell expansion processes in leaves triggered by a defect in cell proliferation, with reference to roles of endoreduplication. Plant Cell Physiol 48:278–286
Garnier E (1991) Resource capture, biomass allocation and growth in herbaceous plants. Trends Ecol Evol 6:126–131
Garnier E (1992) Growth analysis of congeneric annual and perennial grass species. J Ecol 80:665–675
Giovane A, Balestrieri C, Quagliuolo L, Castaldo D, Servillo L (1995) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit. Purification by affinity chromatography and evidence of a ripening-related precursor. Eur J Biochem 233:926–929
Gommers CMM, Visser EJW, Onge KRS, Voesenek LACJ, Pierik R (2013) Shade tolerance: when growing tall is not an option. Trends Plant Sci 18:65–71
Graham LE, Graham JM, Wilcox LW (2006) Plant biology. Pearson Prentice Hall, New Jersey
Guerriero G, Hausman JF, Cai G (2014) No stress! Relax! Mechanisms governing growth and shape in plant cells. Int J Mol Sci 15:5094–5114
Han Y, Wang W, Sun J, Ding M, Zhao R, Deng S et al (2013) Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants. J Exp Bot 64:4225–4238
Hara Y, Yokoyama R, Osakabe K, Toki S, Nishitani K (2014) Function of xyloglucan endotransglucosylase/hydrolases in rice. Ann Bot 114:1309–1318
Havko N, Major I, Jewell J, Attaran E, Browse J, Howe G (2016) Carbon assimilation and partitioning by jasmonate: an accounting of growth–defense tradeoffs. Plants 5:7
Heldt H-W, Piechulla B (2010) Plant Biochemistry. Academic, London
Held MA, Penning B, Brandt AS, Kessans SA, Yong W, Scofield SR, Carpita NC (2008) Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc Natl Acad Sci U S A 105:20534–20539
Held MA, Be E, Zemelis S, Withers S, Wilkerson C, Brandizzi F (2011) CGR3: a Golgi-localized protein influencing homogalacturonan methylesterification. Mol Plant 4:832–844
Hirose F, Inagaki N, Hanada A, Yamaguchi S, Kamiya Y, Miyao A et al (2012) Cryptochrome and phytochrome cooperatively but independently reduce active gibberellin content in rice seedlings under light irradiation. Plant Cell Physiol 53:1570–1582
Hisamatsu T, King RW, Helliwell CA, Koshioka M (2005) The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol 138:1106–1116
Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP (2000) A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol 123:1313–1324
Honda H, Fisher JB (1978) Tree branch angle: maximizing effective leaf area. Science 199:888–890
Hou X, Lee LY, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19:884–894
Hu L, Millet DB, Mohr MJ, Wells KC, Griffis TJ, Helmig D (2011) Sources and seasonality of atmospheric methanol based on tall tower measurements in the US Upper Midwest. Atmos Chem Phys 11:11145–11156
Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14:57–70
Jaillais Y, Chory J (2010) Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol 17:642–645
Jan A, Yang G, Nakamura H, Ichikawa H, Kitano H, Matsuoka M et al (2004) Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol 136:3670–3681
Jiang CM, Li CP, Chang JC, Chang HM (2002) Characterization of pectinesterase inhibitor in jelly fig (Ficus awkeotsang Makino) achenes. J Agric Food Chem 50:4890–4894
Kalve S, Fotschki J, Beeckman T, Vissenberg K, Beemster GTS (2014) Three-dimensional patterns of cell division and expansion throughout the development of Arabidopsis thaliana leaves. J Exp Bot 65(22):6385–6397
Karve AA, Jawdy SS, Gunter LE, Allen SM, Yang X, Tuskan GA et al (2012) Initial characterization of shade avoidance response suggests functional diversity between Populus phytochrome B genes. New Phytol 196:726–737
Kawade K, Horiguchi G, Usami T, Hirai Masami Y, Tsukaya H (2013) ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr Biol 23:788–792
Keuskamp DH, Sasidharan R, Vos I, Peeters AJ, Voesenek LA, Pierik R (2011) Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J 67:208–217
Kim G-T, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12:2381–2391
Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R et al (2002) The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J 21:1267–1279
Kim G-T, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H (2005) CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 41:710–721
Kim G-T, Cho K-H (2006) Recent advances in the genetic regulation of the shape of simple leaves. Physiol Plant 126:494–502
Kim Y, Park S, Gilmour SJ, Thomashow MF (2013) Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J 75:364–376
Kim S-J, Held MA, Zemelis S, Wilkerson C, Brandizzi F (2015) CGR2 and CGR3 have critical overlapping roles in pectin methylesterification and plant growth in Arabidopsis thaliana. Plant J 82:208–220
Kost B (2010) Regulatory and cellular functions of plant RhoGAPs and RhoGDIs. In: Yalovsky S, Baluška F, Jones A (eds) Integrated G Proteins Signaling in Plants. Springer, Berlin/Heidelberg, pp 27–48
Kozuka T, Horiguchi G, Kim GT, Ohgishi M, Sakai T, Tsukaya H (2005) The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by photoreceptors and sugar. Plant Cell Physiol 46:213–223
Krupkova E, Immerzeel P, Pauly M, Schmulling T (2007) The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant DEVELOPMENT. Plant J 50:735–750
Lambers H, Chapin F, Pons T (2008) Plant physiological ecology. Springer, NewYork
Leduc N, Roman H, Barbier F, Péron T, Huché-Thélier L, Lothier J et al (2014) Light signaling in bud outgrowth and branching in plants. Plants 3:223
Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C–repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci U S A 109:15054–15059
Lee YK, Kim GT, Kim IJ, Park J, Kwak SS, Choi G, Chung WI (2006) LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133:4305–4314
Levesque-Tremblay G, Muller K, Mansfield SD, Haughn GW (2015) HIGHLY METHYL ESTERIFIED SEEDS is a pectin methyl esterase involved in embryo development. Plant Physiol 167:725–737
Li S, Blanchoin L, Yang Z, Lord EM (2003) The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiol 132:2034–2044
Li Y, Sorefan K, Hemmann G, Bevan MW (2004) Arabidopsis NAP and PIR regulate actin-based cell morphogenesis and multiple developmental processes. Plant Physiol 136:3616–3627
Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M et al (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143:1871–1880
Liu YB, Lu SM, Zhang JF, Liu S, Lu YT (2007) A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 226:1547–1560
Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ et al (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952
Marcotrigiano M (2010) A role for leaf epidermis in the control of leaf size and the rate and extent of mesophyll cell division. Am J Bot 97:224–233
Markovic O, Janecek S (2004) Pectin methylesterases: sequence-structural features and phylogenetic relationships. Carbohydr Res 339:2281–2295
Masle J, Gilmore SR, Farquhar GD (2005) The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436:866–870
Mathur J (2006) Local interactions shape plant cells. Curr Opin Cell Biol 18:40–46
Mathur J, Hülskamp M (2002) Microtubules and microfilaments in cell morphogenesis in higher plants. Curr Biol 12:R669–R676
Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T et al (2005) AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42:525–534
Mazzella MA, Casal JJ, Muschietti JP, Fox AR (2014) Hormonal networks involved in apical hook development in darkness and their response to light. Front Plant Sci 5:52
Miao Y, Li HY, Shen J, Wang J, Jiang L (2011) QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J Exp Bot 62:5063–5078
Milla-Moreno EA, McKown AD, Guy RD, Soolanayakanahally RY (2016) Leaf mass per area predicts palisade structural properties linked to mesophyll conductance in balsam poplar (Populus balsamifera L.). Botany 94:225–239
Miura K, Hasegawa PM (2010) Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol 20:223–232
Monsi M (1960) Dry-matter reproduction in plants 1. Schemata of dry-matter reproduction. Bot Mag Tokyo 73:81–90
Moon SY, Zheng Y (2003) Rho GTPase-activating proteins in cell regulation. Trends Cell Biol 13:13–22
Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hematy K et al (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50:605–614
Müller K, Levesque-Tremblay G, Bartels S, Weitbrecht K, Wormit A, Usadel B et al (2013a) Demethylesterification of cell wall pectins in Arabidopsis plays a role in seed germination. Plant Physiol 161:305–316
Müller K, Levesque-Tremblay G, Fernandes A, Wormit A, Bartels S, Usadel B, Kermode A (2013b) Overexpression of a pectin methylesterase inhibitor in Arabidopsis thaliana leads to altered growth morphology of the stem and defective organ separation. Plant Signal Behav 8:e26464
Nagel OW, Konings H, Lambers H (2001) Growth rate and biomass partitioning of wildtype and low-gibberellin tomato (Solanum lycopersicum) plants growing at a high and low nitrogen supply. Physiol Plant 111:33–39
Nakaya M, Tsukaya H, Murakami N, Kato M (2002) Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol 43:239–244
Neumetzler L, Humphrey T, Lumba S, Snyder S, Yeats TH, Usadel B et al (2012) The FRIABLE1 gene product affects cell adhesion in Arabidopsis. PLoS One 7:14
Niinemets Ü (2001) Global-scale climatic controls of leaf dry mass per area, density, and thickness in trees and shrubs. Ecology 82:453–469
Nishitani K, Tominaga R (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267:21058–21064
Ochoa-Villarreal M, Aispuro-Hernández E, Vargas-Arispuro I, Martínez-Téllez MÁ (2012) Plant cell wall polymers: function, structure and biological activity of their derivatives. In: De Souza Gomes A (ed) Polymerization. InTech, Rijeka. https://doi.org/10.5772/46094 Available from: https://www.intechopen.com/books/polymerization/plant-cell-wall-polymers-function-structure-and-biological-activity-of-their-derivatives
Ogawa S, Toyomasu T, Yamane H, Murofushi N, Ikeda R, Morimoto Y et al (1996) A step in the biosynthesis of gibberellins that is controlled by the mutation in the semi-dwarf rice cultivar tan-ginbozu. Plant Cell Physiol 37:363–368
Ohnishi T, Szatmari AM, Watanabe B, Fujita S, Bancos S, Koncz C et al (2006) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18:3275–3288
Oikawa PY, Giebel BM, Sternberg Lda S, Li L, Timko MP, Swart PK et al (2011) Leaf and root pectin methylesterase activity and 13C/12C stable isotopic ratio measurements of methanol emissions give insight into methanol production in Lycopersicon esculentum. New Phytol 191:1031–1040
Orfila C, Seymour GB, Willats WG, Huxham IM, Jarvis MC, Dover CJ et al (2001) Altered middle lamella homogalacturonan and disrupted deposition of (1-->5)-α-L-arabinan in the pericarp of Cnr, a ripening mutant of tomato. Plant Physiol 126:210–221
Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci U S A 93:12637–12642
Peaucelle A, Louvet R, Johansen JN, Hofte H, Laufs P, Pelloux J, Mouille G (2008) Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol 18:1943–1948
Peaucelle A, Braybrook S, Hofte H (2012) Cell wall mechanics and growth control in plants: the role of pectins revisited. Front Plant Sci 3:121
Pierik R, De Wit M, Voesenek LA (2011) Growth-mediated stress escape: convergence of signal transduction pathways activated upon exposure to two different environmental stresses. New Phytol 189:122–134
Pilling J, Willmitzer L, Bucking H, Fisahn J (2004) Inhibition of a ubiquitously expressed pectin methyl esterase in Solanum tuberosum L. affects plant growth, leaf growth polarity, and ion partitioning. Planta 219:32–40
Poorter H, Lambers H (1991) Is interspecific variation in relative growth rate positively correlated with biomass allocation to the leaves? Am Nat 138:1264–1268
Poorter H, Remkes C (1990) Leaf area ratio and net assimilation rate of 24 wild species differing in relative growth rate. Oecologia 83:553–559
Poorter H, Niinemets Ü, Poorter L, Wright IJ, Villar R (2009) Causes and consequences of variation in leaf mass per area (LMA): a meta-analysis. New Phytol 182:565–588
Qian P, Hou S, Guo G (2009) Molecular mechanisms controlling pavement cell shape in Arabidopsis leaves. Plant Cell Rep 28:1147–1157
Qiu JL, Jilk R, Marks MD, Szymanski DB (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14:101–118
Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett 557:199–203
Raiola A, Lionetti V, Elmaghraby I, Immerzeel P, Mellerowicz EJ, Salvi G et al (2011) Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol Plant-Microbe Interact 24:432–440
Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5:147–157
Robert S, Mouille G, Höfte H (2004) The mechanism and regulation of cellulose synthesis in primary walls: lessons from cellulose-deficient Arabidopsis mutants. Cellulose 11:351–364
Rose JK, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43:1421–1435
Sánchez-Rodríguez C, Estévez JM, Llorente F, Hernández-Blanco C, Jordá L, Pagán I et al (2009) The ERECTA receptor-like kinase regulates cell wall-mediated resistance to pathogens in Arabidopsis thaliana. Mol Plant-Microbe Interact 22:953–963
Savaldi-Goldstein S, Chory J (2008) Growth coordination and the shoot epidermis. Curr Opin Plant Biol 11:42–48
Shin YK, Yum H, Kim ES, Cho H, Gothandam KM, Hyun J, Chung YY (2006) BcXTH1, a Brassica campestris homologue of Arabidopsis XTH9, is associated with cell expansion. Planta 224:32–41
Shipley B (2002) Trade-offs between net assimilation rate and specific leaf area in determining relative growth rate: relationship with daily irradiance. Funct Ecol 16:682–689
Shpak ED, Berthiaume CT, Hill EJ, Torii KU (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131:1491–1501
Sinha N (1999) Leaf development in angiosperms. Annu Rev Plant Physiol Plant Mol Biol 50:419–446
Soolanayakanahally RY, Guy RD, Silim SN, Drewes EC, Schroeder WR (2009) Enhanced assimilation rate and water use efficiency with latitude through increased photosynthetic capacity and internal conductance in balsam poplar (Populus balsamifera L.). Plant Cell Environ 32:1821–1832
Suzuki S, Li L, Sun YH, Chiang VL (2006) The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142:1233–1245
Tan H-T, Shirley NJ, Singh RR, Henderson M, Dhugga KS, Mayo GM et al (2015) Powerful regulatory systems and post-transcriptional gene silencing resist increases in cellulose content in cell walls of barley. BMC Plant Biol 15:62
Tenhaken R (2015) Cell wall remodeling under abiotic stress. Front Plant Sci 5:771
Terashima I, Hanba YT, Tazoe Y, Vyas P, Yano S (2006) Irradiance and phenotype: comparative eco-development of sun and shade leaves in relation to photosynthetic CO2 diffusion. J Exp Bot 57:343–354
Tillman D (1991) Relative growth rates and plant allocation patterns. Am Nat 138:1269–1275
Tosens T, Niinemets Ü, Vislap V, Eichelmann H, Castro Díez P (2012) Developmental changes in mesophyll diffusion conductance and photosynthetic capacity under different light and water availabilities in Populus tremula: how structure constrains function. Plant Cell Environ 35:839–856
Tsuge T, Tsukaya H, Uchimiya H (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L). Heynh Development 122:1589–1600
Tsukaya H (1998) Genetic evidence for polarities that regulate leaf morphogenesis. J Plant Res 111:113–119
Tsukaya H (2002) The leaf index: heteroblasty, natural variation, and the genetic control of polar processes of leaf expansion. Plant Cell Physiol 43:372–378
Tsukaya H, Kozuka T, Kim G-T (2002) Genetic control of petiole length in Arabidopsis thaliana. Plant Cell Physiol 43:1221–1228
Utrillas MJ, Alegre L (1997) Impact of water stress on leaf anatomy and ultrastructure in Cynodon dactylon (L.) Pers. under natural conditions. Int J Plant Sci 158:313–324
Van Volkenburgh E, Boyer JS (1985) Inhibitory effects of water deficit on maize leaf elongation. Plant Physiol 77:190–194
Verica JA, Medford JI (1997) Modified MER15 expression alters cell expansion in transgenic Arabidopsis plants. Plant Sci 125:201–210
Villagarcia H, Morin A-C, Shpak ED, Khodakovskaya MV (2012) Modification of tomato growth by expression of truncated ERECTA protein from Arabidopsis thaliana. J Exp Bot 63:6493–6504
Villar R, Ruiz-Robleto J, Ubera JL, Poorter H (2013) Exploring variation in leaf mass per area (LMA) from leaf to cell: an anatomical analysis of 26 woody species. Am J Bot 100:1969–1980
Volpi C, Janni M, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R (2011) The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol Plant-Microbe Interact 24:1012–1019
Wang X, Zhang J, Yuan M, Ehrhardt DW, Wang Z, Mao T (2012) Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24:4012–4025
Weller JL, Hecht V, Vander Schoor JK, Davidson SE, Ross JJ (2009) Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 21:800–813
Weraduwage S, Chen J, Anozie FC, Morales A, Weise SE, Sharkey TD (2015) The relationship between leaf area growth and biomass accumulation in Arabidopsis thaliana. Front Plant Sci 6:167
Weraduwage SM, Kim S-J, Renna L, Anozie FC, Sharkey TD, Brandizzi F (2016) Pectin methylesterification impacts the relationship between photosynthesis and plant growth in Arabidopsis thaliana. Plant Physiol 171:833–848
Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411:610–613
Wietholter N, Graessner B, Mierau M, Mort AJ, Moerschbacher BM (2003) Differences in the methyl ester distribution of homogalacturonans from near-isogenic wheat lines resistant and susceptible to the wheat stem rust fungus. Mol Plant-Microbe Interact 16:945–952
Williamson RE, Burn JE, Birch R, Baskin TI, Arioli T, Betzner AS, Cork A (2001) Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215:116–127
Wood PJ, Siddiqui IR (1971) Determination of methanol and its application to measurement of pectin ester content and pectin methylesterase activity. Anal Biochem 39:418–428
Wolf S, Mouille G, Pelloux J (2009) Homogalacturonan methyl-esterification and plant development. Mol Plant 2:851–860
Xiao C, Anderson CT (2013) Roles of pectin in biomass yield and processing for biofuels. Front Plant Sci 4:67
Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ et al (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143:99–110
Yang Y, Karlson D (2012) Effects of mutations in the Arabidopsis cold shock domain protein 3 (AtCSP3) gene on leaf cell expansion. J Exp Bot 63:4861–4873
Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q et al (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci U S A 109:E1192–E1200
Yokoyama R, Nishitani K (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42:1025–1033
Yokoyama R, Rose JK, Nishitani K (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol 134:1088–1099
Yoshikawa T, Eiguchi M, Hibara K-I, Ito J-I, Nagato Y (2013) Rice SLENDER LEAF 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J Exp Bot 64:2049–2061
Acknowledgments
We are grateful to Drs. Sean E. Weise, (Department of Biochemistry and Molecular Biology), Cliff Foster (the Cell Wall Facility, Great Lakes Bioenergy Research Center), Alicia Withrow and Melinda Frame (Center for Advanced Microscopy) of Michigan State University (East Lansing, MI), and to Dr. Suvankar Chakraborty (Stable Isotope Ratio Facility for Environmental Research) of the University of Utah (Salt Lake City, UT) for their support. We also wish to thank Jim Klug and Cody Keilen (Growth Chamber Facility) of Michigan State University for their assistance and all members of the Brandizzi, Thomashow, Howe, and Sharkey labs for their support. Funding for this research was provided by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U. S. Department of Energy (award number DE-FG02-91ER20021) and in part by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494). Partial salary support for MT, GH, TDS, and FB came from Michigan AgBioResearch.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Weraduwage, S.M. et al. (2018). Molecular Mechanisms Affecting Cell Wall Properties and Leaf Architecture. In: Adams III, W., Terashima, I. (eds) The Leaf: A Platform for Performing Photosynthesis. Advances in Photosynthesis and Respiration, vol 44. Springer, Cham. https://doi.org/10.1007/978-3-319-93594-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-93594-2_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-93592-8
Online ISBN: 978-3-319-93594-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)