Abstract
Magnetic resonance imaging (MRI) is routinely employed to assess muscular response and presence of inflammatory reactions in patients treated with metal-on-metal hip arthroplasty, driving the decision for revision surgery. However, MRI is lacking contrast for bony structures and as a result orthopaedic surgical planning is mostly performed on computed tomography images. In this paper, we combine the complementary information of both modalities into a novel framework for the joint segmentation of healthy and pathological musculoskeletal structures as well as implants on all images. Our processing pipeline is fully automated and was designed to handle the highly anisotropic resolution of clinical MR images by means of super resolution reconstruction. The accuracy of the intra-subject multimodal registration was improved by employing a non-linear registration algorithm with hard constraints on the deformation of bony structures, while a multi-atlas segmentation propagation approach provided robustness to the large shape variability in the population. The suggested framework was evaluated in a leave-one-out cross-validation study on 20 hip sides. The proposed pipeline has potential for the extraction of clinically relevant imaging biomarkers for implant failure detection.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In the past 20 years, metal-on-metal (MoM) hip arthroplasty has been one of the most effective surgical interventions for improving life quality. However, this implant type is associated with a non-negligible rate of failure (8% at 12 years from primary surgery [1]), due to adverse tissue inflammatory reactions and increased muscle atrophy [2]. Routine assessment of periprosthetic muscle response to the implant is performed on magnetic resonance (MR) images [3], whereas computed tomography (CT) imaging is preferred for surgical planning and post-operative follow-up, thanks to its improved contrast for bone and implant [4]. The two modalities provide complementary skeletal and muscular information, which are presently assessed independently in clinical practice. In this context, a single framework merging this information by means of joint automated segmentation could be beneficial for both early detection of implant failure and planning of revision surgery. By providing spatial relationship between muscle, bone and implant simultaneously, the combination of the two imaging modalities could help link implant position (not MR visible) with muscle damage (estimated on MR) to better characterise pain origin. Moreover, it could favour a patient-specific planning of surgical approach to minimise damage to healthy bone and muscular tissue.
In the musculoskeletal clinical field, manual segmentation is still the most frequently adopted solution in clinical routine for delineating regions of interest [5], despite the variety of image-based anatomical models and segmentation techniques presented in the literature. Methods for automated segmentation of hip bony structures in CT images are typically based on statistical shape models [6, 7], atlas-based segmentation propagation [8] or, more recently, hybrid approaches [9]. Segmentation of muscles on MR images is more problematic, because of their large inter-subject shape variability and the lack of image contrast between different muscular structures. A common approach for thigh muscles is the incorporation of atlases as priors into conventional segmentation techniques such as active contours or level-set algorithms [10, 11]. Remarkable results were also presented by Gilles et al. [12], who introduced a method to automatically segment hip muscles and bones on MR images by means of deformable multi-resolution simplex meshes. The performances of all discussed methods are strongly reliant on the variability encompassed in the training data set and they are often not suitable for pathological conditions. Klemt et al. [13] addressed this issue by developing a robust automated segmentation framework for abductor muscles on MR in both healthy subjects and patients with MoM prostheses. However, little work combining multimodal imaging for the segmentation of musculoskeletal structures has been proposed so far and it is often limited only to spine applications. An example is the method presented by Castro-Mateos et al. [14], which is based on a fast mesh-to-image registration to extract a surface model of CT-derived vertebrae and MR-derived intervertebral discs. Whilst being very suitable for bony structures, the applicability of this method to patients with hip arthroplasty would be hindered by the presence of metal artefact in the images and by the greater morphological and textural variability of muscles.
Taking advantage of the complementary information derived from CT and MR, we propose a fully automated joint segmentation framework of both modalities from patients treated with MoM arthroplasty. Our processing pipeline was designed to handle clinical data, characterised by highly anisotropic resolution and presence of severe metal artefact induced noise, and allows for a three-dimensional representation of patient-specific musculoskeletal hip anatomy. Key contributions of this work include the use of super resolution reconstruction (SRR) to improve clinical MR image quality; moreover, the development of a robust intra-subject multimodal registration allowed preservation of the rigid structure of bones, while deforming the muscles. Finally, a multi-channel multi-atlas based segmentation propagation guaranteed robustness to the large shape variability in the population.
2 Materials and Methods
2.1 Dataset and Templates Creation
Our dataset includes retrospectively collected images of 11 MoM hip implanted patients (7 females and 4 males, 10 unilateral and 1 bilateral replacement) who had both a CT and an MR scan acquired on the same day. For the MR acquisitions, a Siemens MAGNETON Avanto 1.5T scanner was employed for all patients, using the MARS MRI protocol proposed in [15], which is characterised by rapid 2D MRI acquisition but high voxel resolution anisotropy. This includes the collection of two T1-weighted Turbo Spin Echo (TSE) images: a high-resolution axial acquisition (TE\(\,{=}\,8\) ms, TR\(\,{=}\,509\) ms, typical imaging resolution\(\,{=}\,0.78 \times 0.78 \times 7.02\) mm\(^3\)) and a high-resolution coronal acquisition (TE\(\,{=}\,7.1\) ms, TR\(\,{=}\,627\) ms, typical imaging resolution\(\,{=}\,1.25 \times 1.25 \times 6.00\) mm\(^3\)). Eight CT images were acquired on a Siemens SOMATOM Sensation 16 CT Scanner, while three on a Siemens SOMATOM Definition AS machine (tube voltage in [80, 120] kVp). The images were processed (see Sect. 2.2), split along the left-right axis of symmetry and separated according to the presence of implant. Manual segmentation of pelvic bones, femora and implant were performed on CT, while Gluteus Maximus (GMAX), Gluteus Medius (GMED), Gluteus Minimus (GMIN) and Tensor Fasciae Latae (TFL) were individually manually delineated on the MR. As a result of these processes, we built two template data sets, composed of 10 implanted and 10 non-implanted hip sides respectively - for the sake of simplicity we will refer to the latter as the healthy data set despite the presence of metal artefact generated by the implanted side. Each template includes a CT image, a registered super-resolution reconstructed MR image and the respective joint manual segmentation of bones, muscles and implant. Within each dataset, the templates were robustly aligned onto the average space based on the method proposed in [13].
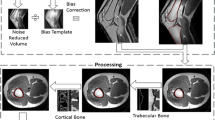
Proposed pipeline for joint automated segmentation of computed tomography (CT) and magnetic resonance (MR) pelvic images. The two modalities are first processed independently to enhance the image quality. Intra-subject multimodal registration is then performed to align them through a non-linear deformation with rigid constraints in bony structures. The registered CT and MR are split along the axis of symmetry and a multi-atlas based segmentation propagation approach is applied to obtain the automated segmentations of each side, which are finally recombined into the full field of view.
2.2 Pipeline for Automated Segmentation
A schematic representation of our processing framework is presented in Fig. 1. The pipeline was implemented in NiPype [16], combining registration and segmentation utilities of NiftyRegFootnote 1, NiftySegFootnote 2 and FSLFootnote 3 software packages with super-resolution reconstruction and the proposed novel multimodal registration framework. Our method is composed of three main blocks which are performed sequentially: image quality enhancement of each modality, intra-subject MR-to-CT registration, and atlas-based segmentation.
Image Quality Enhancement. In the first block, we aim at improving the quality of the clinical images for improved registration steps. The axial and the coronal MR images are first corrected for bias field effects [17]. In order to compensate for the highly anisotropic resolution of clinical MR images (up to a factor of 10), we combine both MR acquisitions into a \(1 \times 1 \times 1\) mm\(^3\) resolution image using the SRR algorithm presented in [18]. To ease the subsequent registration, the CT is also resampled to the same resolution using a cubic interpolation scheme. An initial estimate of bones segmentation on the CT is extracted by registering the templates to the target space and consequently propagating and fusing their segmentation, allowing the creation of masks for femur, pelvis and implant to be used in the intra-subject registration.
Intra-Subject MR-CT Registration with Bone Rigid Constraints. The subsequent step in our processing pipeline is the registration of the SRR MR image to the respective CT. Multimodal registration for hip musculoskeletal structures is challenging and no standard method has been proposed yet. A simple affine transformation is not sufficient to guarantee an accurate alignment of the images, due to differences in the patient’s pose in the scanners. On the other hand, high frequency deformations should be curbed when dealing with intra-subject registration to prevent non-physiological deformation. The applied transformation should embed a rigid behaviour for bones to preserve their shape, while allowing non-linear deformation of fat and muscular tissue. To tackle the discussed issues, we designed a registration pipeline composed of two steps. Firstly, the two images are affinely registered using a symmetric block-matching algorithm [19], in order to provide an initial global alignment. Subsequently, the non-linear registration is performed by imposing locally rigid hard constraints directly on the transformation through the following method, which we developed from the mathematical formulation proposed in [20]. Given a reference space \( X \) with the associated intensities \( R(X) \) (i.e. a reference image \( R \)), a set of masks \(M_j\) defined in the reference space labelling the rigid structures, and a floating image F defined in the floating space \( Y \), we defined our registration problem as the optimisation of the transformation \(\phi : X \rightarrow Y \) such that:
\(\mathcal {D}\) is a measure of similarity between the reference and the warped floating image, while \( P_{L} \) and \( P_{B} \) represent the linear elasticity and the bending energy penalty terms [21], whose contribution to the total cost function is weighted by \(\alpha \) and \(\beta \) respectively; \( A _{j}\) refers to a rigid transformation applied within the j-th mask. In order to guarantee inverse-consistency and symmetry of the registration [22], we exploit a scaling-and-squaring exponentiation of a stationary velocity field encoded by a cubic B-spline parametrisation defined over a set of control points \(\{\mu \}\). The transformation is optimised within a conjugate gradient scheme, and the rigid behaviour in the mask areas is ensured through the following steps:

Differently to current approaches such as [23] where a locally rigid behaviour can be promoted by the addition of a penalty term to the cost function (soft constraint), in our approach the rigid constraints are strictly embedded into the transformation model, not in the optimisation scheme (hard constraint). Thus, chain rule provides an analytical formulation of the conjugate gradient thereby avoiding constrained optimisation. Using the proposed method on a coarse-to-fine pyramidal approach, smooth transitions in the deformation field are maintained by the cubic B-spline parametrisation and the stationary velocity field exponentiation, while forcing rigid behaviours within the masks. To reduce the effect of undesired high-frequency components in the transformation, we set one control point every five voxels, and the masks are dilated at each pyramidal level to account for the local support of the control points. We underline that we extract the robust range of the intensity distributions for both the reference and the floating image, and we perform all the registration steps by flooring or ceiling all intensities outside this range, so as to decrease the influence of metal artefact induced noise.
Once registered with the proposed method, the CT and the MR are merged into a single four-dimensional (4D) volume. In order to employ the appropriate template dataset for the atlas-based segmentation – i.e. healthy or implanted – we developed a symmetry and implant detection algorithm. Based on left-right axis flip and rigid registration, it extracts the sagittal axis of symmetry from the inertia tensor of the image intensities. The 4D volume is split along this axis and each hip side is automatically classified according to the presence of implants.
Atlas-Based Segmentation. Each split hip side is segmented by means of multi-atlas segmentation propagation and label fusion. All the templates are registered to the target 4D image in a three-step process (rigid, affine and non-linear registration as implemented in NiftyReg). The transformation of the affine and the non-linear steps is initialised as the least trimmed squares average affine from all the template transformations estimated at the previous step. Since our templates were previously aligned to their mid-space (Sect. 2.1), this initialisation provides robustness against global failed registration. Notably, the non-linear step is a multi-channel registration, where both modalities contribute jointly and equally to the optimisation of the transformation. Using the estimated transformation, the segmentation of each template is propagated onto the target space. The candidate segmentations are then fused into a consensus through the STEPS algorithm [24], specifically modified to manage a multi-channel local similarity measure. The final segmentation is obtained by merging back the two hip sides and their estimated segmentation, providing a multi-label image that highlights different bones, muscles and implants on both the CT and the MR.
3 Validation and Experiments
Example of qualitative registration assessment with default NiftyReg regularisation parameters. The same axial and coronal slices are reported for the reference computed tomography, the super-resolution reconstructed magnetic resonance (MR) after affine registration, the super resolution reconstruction (SRR) MR after rigidly constrained non rigid registration and the SRR MR after standard non-linear registration. For these latter cases, the transformation Jacobian determinant maps are also displayed, showing the effect of the rigid constraints. Yellow arrows indicate exemplary areas where the proposed approach visually recovers a better alignment than the standard fully non-linear registration (e.g. in the femoral head size). (Color figure online)
3.1 Intra-Subject Registration Evaluation
The first set of experiments we performed aimed at identifying the optimal set of regularisation parameters \(\alpha \) and \(\beta \) as shown in (1) for the intra-subject registration. Normalised mutual information (NMI) was used as measure of similarity, since it is best suited for multimodal registration. For the sake of comparison, we performed the same study using the standard non-linear registration without the application of the rigid constraints, while keeping all the other parameters unchanged. Although this variant would assume non-rigid deformation of the bones, which is neither anatomically nor clinically correct, such a comparison allows us to verify whether our implementation also improved the registration results compared to the classical approach.
The choice of the best parameters was based on both qualitative and quantitative analysis. The former included visual assessment of the alignment between the CT and the registered MR and of the transformation Jacobian maps. An example of this comparison is reported in Fig. 2, where the Jacobian determinant maps clearly show how the standard registration algorithm fails in recovering a rigid behaviour within the bones, as opposed to the proposed method.
Target registration error (TRE) analysis. Top figure: comparison of TRE root mean square error (RMSE) values obtained from the rigidly constrained non-linear registration and the standard one with varying regularisation parameters \(\alpha \) - linear elasticity weight - and \(\beta \) - bending energy weight. TRE RMSE from affine registration is shown as well. Table: highest RMSE for each set of parameters. Starred values indicate the sets for which the rigidly constrained registration provided significantly lower RMSE than the standard (Wilcoxon rank sum test, \(p < 0.05\)). Highlighted in red are the results for the selected best set of parameters. Bottom figure: manual selection reproducibility error for the 10 landmarks and for the two modalities. List of landmarks abbreviations: greater trochanter (GT), tensor fasciae latae (TFL), anterior-inferior iliac spine (AIS), gluteus maximus (GMAX), ischium (Isc). Each landmark is identified in each side and it is categorized as healthy (H) or implanted (I) side. (Color figure online)
A quantification of the registration accuracy was obtained through landmarks analysis. Specifically, we labeled 5 landmarks (3 in bone, 2 in muscles) per hip side which could be conveniently located in both modalities and which cover the full field of view. The target registration error (TRE) was computed as the distance between the CT and the respective warped MR landmark. In order to limit the bias from the manual landmark choice, we repeated the selection twice at different times, we estimated the TRE for each selection and then computed the average TRE for each landmark and for each subject (reproducibility errors for the manual selection are shown in the bottom-left panel of Fig. 3). For each subject we extracted the root mean square error (RMSE) of the TRE across the ten landmarks and we compared the distribution of the RMSE with respect to the registration parameters. A summary of the obtained results is presented in Fig. 3. Overall the proposed method not only provided clinically plausible registration results, but also produced a more accurate alignment of the considered landmarks compared to a standard non-linear registration algorithm. The best set of parameters was identified as the one minimising the highest TRE RMSE among all the landmarks, so as to guarantee a reasonably good alignment across the whole field of view. We therefore concluded that the optimal results for the intra-subject registration resulted from the use of normalised mutual information with \(\alpha = 0.2\) and \({\beta = 0.01}\).
Example of automated segmentation obtained with the proposed automated pipeline. Top row shows the central axial and coronal slices for one of the subjects for both computed tomography and magnetic resonance, while the second row reports the same images overlaid with the segmentation result. A three-dimensional rendering of the full segmentation is also displayed on the bottom left for the same subject.
3.2 Leave-One-Out Cross Validation
The proposed pipeline was validated through a leave-one-out cross-validation (LOOCV) experiment on the template datasets, by calculating the Dice score between the automated segmentation result and the corresponding manual ground truth for each label and for each subject. The goal of the LOOCV was to compare the achieved results using both modalities jointly to those obtained using only the CT or only the MR images. We recall that the manual segmentation of muscles was not available for the CT, and similarly bones and implant labelling on the MR. Therefore only the available labels were considered in the single-modality experiments. For each analysed type – i.e. only CT, only MR, combined modalities for healthy and for implanted sides – the segmentation propagation and label fusion parameters were tuned to maximise the lowest average Dice Score across subjects and across labels. An example of the obtained automated segmentation is shown in Fig. 4.
The median Dice score for bones, muscles and implant extracted from the three LOOCV experiments are reported in Table 1. It can be observed that the multimodal and the single-modality approaches perform similarly with comparable Dice score values. Overall bone structures were better segmented than muscular ones, due to their lower shape and texture variability. Although the obtained results appear slightly lower with the proposed approach, the observed differences were statistically significant only in one case – i.e. for muscular structures in the healthy side – while for all the other cases the null hypothesis of same underlying distribution was accepted (Wilcoxon rank sum test with 5% significance level). This difference could arise from the need of finding a trade-off in the segmentation propagation and label fusion parameters to achieve a good accuracy in both skeletal and muscular structures for the 4D case. This might go at the expense of a slight reduction of performances with respect to the single-modality case, where the parameters are tuned only for the bones and implant (CT) or for the muscles (MR). Nonetheless, only the proposed framework is able to provide consistent and unified solution to the segmentation of both the CT and the MRI. The use of independent approaches to segment the muscles in the MR images and the bones and implants in the CT image would indeed not guarantee non-overlapping regions of interest. As an example, on our dataset we evaluated that on average 2% of the voxels labeled as muscle on the MR overlapped with CT-labeled bone voxels in our manual segmentation, while the proposed method guarantees no overlap by design. Without the use of a registration framework able to deal with the rigid nature of the bones while non-linearly deforming the surrounding soft tissue, it would be more challenging to accurately highlight the muscles in the CT space or the bones in the MR space, due to their poor contrast for these structures.
4 Conclusion
We presented a fully automated processing pipeline for the joint segmentation of bones, abductor muscles and implant on CT and MR images from hip arthroplasty patients. The combination of the two modalities enables accurate joint delineation of healthy and pathological musculoskeletal structures and of their spatial relationship.
As for other atlas-based approaches, the performance of our method could be improved by enlarging the template data sets to better encompass the population variability. Moreover, the presence of metal artefact-induced noise strongly affects the accuracy of both intra- and inter-subject registration; hence future developments of the processing pipeline will introduce novel metal artefact reduction techniques as an image quality enhancement step for the CT. In conclusion, the proposed pipeline is a promising tool towards patient-specific 3D visualisation of musculoskeletal structures, and towards the extraction of clinically relevant imaging biomarkers to detect implant failure. Thanks to our processing steps, the implant can be outlined also on the MR image, where it is typically obscured by the metal artefact. This could help identify the muscles that are at greater risk of developing atrophy due to the presence of the implant, and therefore inform the decision-making process for revision surgery.
References
UK National Joint Registry: 13th Annual Report (December 2015) (2016)
Berber, R., Khoo, M., Cook, E., Guppy, A., Hua, J., Miles, J., Carrington, R., Skinner, J., Hart, A.: Muscle atrophy and metal-on-metal hip implants: a serial MRI study of 74 hips. Acta Orthop. 86(3), 351–357 (2015)
MHRA of the Department of Health: Medical Device Alert. MHRA Database, pp. 1–7 (2012). http://www.mhra.gov.uk
Bogner, E., Sofka, C.: CT evaluation of total hip arthroplasty complication: Dissociation of acetabular component. HSS J. 3(1), 112–114 (2007)
Pedoia, V., Majumdar, S., Link, T.: Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn. Reson. Mater. Phy. 29(2), 207–221 (2016)
Seim, H., Kainmüller, D., Heller, M., Lamecker, H., Zachow, S., Hege, H.C.: Automatic segmentation of the pelvic bones from CT data based on a statistical shape model. In: Proceedings of the 1st Eurographics Conference on Visual Computing for Biomedicine - EG VCBM 2008, pp. 93–100 (2008)
Yokota, F., Okada, T., Takao, M., Sugano, N., Tada, Y., Tomiyama, N., Sato, Y.: Automated CT segmentation of diseased hip using hierarchical and conditional statistical shape models. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013. LNCS, vol. 8150, pp. 190–197. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-40763-5_24
Pettersson, J., Knutsson, H., Borga, M.: Automatic hip bone segmentation using non-rigid registration. In: Proceedings of the 18th International Conference on Pattern Recognition - ICPR 2006, vol. 3, pp. 946–949. IEEE (2006)
Chu, C., Chen, C., Liu, L., Zheng, G.: FACTS: fully automatic CT segmentation of a hip joint. Ann. Biomed. Eng. 43(5), 1247–1259 (2015)
Prescott, J., Best, T., Swanson, M., Haq, F., Jackson, R., Gurcan, M.: Anatomically anchored template-based level set segmentation: application to quadriceps muscles in MR images from the osteoarthritis initiative. J. Digit. Imaging 24(1), 28–43 (2011)
Ahmad, E., Yap, M., Degens, H., McPhee, J.: Atlas-registration based image segmentation of MRI human thigh muscles in 3D space. In: Mello-Thoms, C.R., Kupinski, M. (eds.) Proceedings of the SPIE Medical Imaging 2014: Image Perception, Observer Performance, and Technology Assessment, vol. 9037, p. 90371L. SPIE (2014)
Gilles, B., Magnenat-Thalmann, N.: Musculoskeletal MRI segmentation using multi-resolution simplex meshes with medial representations. Med. Image Anal. 14(3), 291–302 (2010)
Klemt, C., Modat, M., Pichat, J., Cardoso, M., Henckel, J., Hart, A., Ourselin, S.: Automatic assessment of volume asymmetries applied to hip abductor muscles in patients with hip arthroplasty. In: Ourselin, S., Styner, M. (eds.) Proceedings of the SPIE Medical Imaging 2015: Image Processing, vol. 9413, p. 94131M. SPIE (2015)
Castro-Mateos, I., Pozo, J., Lazary, A., Frangi, A.: Automatic construction of patient-specific finite-element mesh of the spine from IVDs and vertebra segmentations. In: Gimi, B., Krol, A. (eds.) Proceedings of the SPIE Medical Imaging 2016: Biomedical Applications in Molecular, Structural, and Functional Imaging, vol. 9788, p. 97881U. SPIE (2016)
Sabah, S., Mitchell, A., Henckel, J., Sandison, A., Skinner, J., Hart, A.: Magnetic resonance imaging findings in painful metal-on-metal hips: a prospective study. J. Arthroplasty 26(1), 71–76 (2011)
Gorgolewski, K., Burns, C., Madison, C., Clark, D., Halchenko, Y., Waskom, M., Ghosh, S.: Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front. Neuroinf. 5, 13 (2011)
Van Leemput, K., Maes, F., Vandermeulen, D., Suetens, P.: Automated model-based bias field correction of MR images of the brain. IEEE Trans. Med. Imaging 18(10), 885–896 (1999)
Ebner, M., Chouhan, M., Patel, P.A., Atkinson, D., Amin, Z., Read, S., Punwani, S., Taylor, S., Vercauteren, T., Ourselin, S.: Point-spread-function-aware slice-to-volume registration: application to upper abdominal MRI super-resolution. In: Zuluaga, M.A., Bhatia, K., Kainz, B., Moghari, M.H., Pace, D.F. (eds.) RAMBO/HVSMR -2016. LNCS, vol. 10129, pp. 3–13. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-52280-7_1
Modat, M., Cash, D., Daga, P., Winston, G., Duncan, J., Ourselin, S.: Global image registration using a symmetric block-matching approach. J. Med. Imaging 1(2), 024003 (2014)
Haber, E., Heldmann, S., Modersitzki, J.: A computational framework for image-based constrained registration. Linear Algebra Appl. 431(3–4), 459–470 (2009)
Ashburner, J., Ridgway, G.: Symmetric diffeomorphic modeling of longitudinal structural MRI. Front. Neurosci. 6, 197 (2013)
Modat, M., Daga, P., Cardoso, M., Ourselin, S., Ridgway, G., Ashburner, J.: Parametric non-rigid registration using a stationary velocity field. In: Proceedings of the IEEE Workshop on Mathematical Methods in Biomedical Image Analysis - MMBIA 2012, pp. 145–150 (2012)
Staring, M., Klein, S., Pluim, J.: A rigidity penalty term for nonrigid registration. Med. Phys. 34(11), 4098–4108 (2007)
Cardoso, J., Leung, K., Modat, M., Keihaninejad, S., Cash, D., Barnes, J., Fox, N., Ourselin, S.: STEPS: similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcelation. Med. Image Anal. 17(6), 671–684 (2013)
Acknowledgements
This work is supported by the EPSRC-funded UCL Centre for Doctoral Training in Medical Imaging [EP/L016478/1], the Royal National Orthopaedic Hospital NHS Trust, the Department of Healths NIHR-funded Biomedical Research Centre at University College London Hospitals and Innovative Engineering for Health award by the Wellcome Trust [WT101957] and EPSRC [NS/A000027/1], and by Wellcome/EPSRC [203145Z/16/Z].
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this paper
Cite this paper
Ranzini, M.B.M. et al. (2018). Joint Multimodal Segmentation of Clinical CT and MR from Hip Arthroplasty Patients. In: Glocker, B., Yao, J., Vrtovec, T., Frangi, A., Zheng, G. (eds) Computational Methods and Clinical Applications in Musculoskeletal Imaging. MSKI 2017. Lecture Notes in Computer Science(), vol 10734. Springer, Cham. https://doi.org/10.1007/978-3-319-74113-0_7
Download citation
DOI: https://doi.org/10.1007/978-3-319-74113-0_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-74112-3
Online ISBN: 978-3-319-74113-0
eBook Packages: Computer ScienceComputer Science (R0)