Abstract
Nuclear pore complexes (NPCs) are dynamic structures embedded in double lipid layer of the nuclear envelope (NE), which act as guardians of nucleocytoplasmic transport, and contribute to genome organization, genome stability and gene expression regulation. Some of these cellular functions orchestrated by NPCs are usurped by viruses that replicate in the nucleus. Non-mitotic cells are one of the major targets of HIV-1, thus the passage of the virus through the NPC is a key step for viral replication. In recent years, research regarding multiple aspects of the early steps of HIV-1 life cycle highlights dynamic and concerted interactions between viral components, NPC and chromatin state. HIV-1 is a member of this host-pathogen activity which ensures favourable conditions for the production of its own progeny. This chapter aims to review the existing and emerging concepts showing how individual nucleoporins (Nups) may be a “cellular code” that dictates HIV-1 fate in infected cells.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
8.1 Introduction
Eukaryotic chromosomes are protected and enclosed by the NE, a double-lipid bilayer, which allows the communication between the inner and the outer sides of the nucleus. This link is mediated by dynamic windows, called NPCs, which are uniformly or unequally distributed depending on the cell cycle (Maeshima et al. 2006). Nuclear pore components play critical roles in some vital cellular functions and can be also hijacked by viruses. Some Nups are also directly involved in the nuclear entry of viruses that replicate in the nucleus. Recent studies highlighted the importance of Nups for viral nuclear import and viral replication of different viruses that replicate in the nucleus: Human Immunodeficiency Virus (HIV), Hepatitis B Virus (HBV), Herpes Simplex Virus (HSV), Influenza Virus, and Adenovirus (Brass et al. 2008; Copeland et al. 2009; Di Nunzio 2013; Di Nunzio et al. 2012, 2013; Konig et al. 2008; Lelek et al. 2015; Matreyek et al. 2013; Schmitz et al. 2010; Trotman et al. 2001). In particular, RanBP2/Nup358, main component of the cytoplasmic fibrils, and Nup153, the most dynamic Nup, are involved in HIV-1 docking at the pore and nuclear translocation respectively (Di Nunzio et al. 2012, 2013; Matreyek et al. 2013; Schaller et al. 2011). The main component of the nuclear basket of the NPC, Tpr, regulates the chromatin landscape around nuclear pores and is critical for HIV-1 replication (Lelek et al., 2015).
This manuscript provides a detailed view of the recent advances in the early steps of HIV-1 infection, taking into account results obtained by crystal structures for interactions between viral components and host factors, next generation sequencing methods to identify integration sites, as well as new cutting-edge microscopy to visualize the viral journey into the host cell.
8.2 Early Steps of HIV-1 Infection
HIV-1 has the peculiarity to infect non-dividing cells and, hence, it enters the nucleus through the NPC channel. The life cycle of HIV-1 begins with binding of the viral envelope glycoproteins to receptor CD4 and co-receptors CCR5 or CXCR4 on the target cell, followed by viral fusion to the cellular membrane and release of the viral core into the cytoplasm. The viral genome packaged into the capsid (CA) shell, the viral core, is formed by two positive strands of RNA that are retrotranscribed in DNA which forms the pre-integration complex (PIC) when coupled with other known, such as the integrase (IN), and unknown factors. The PIC has been estimated to have a size ~ 56 nm (Miller et al. 1997) which far exceeds the size of molecules that can pass through the NPC channel, thus, it is generally accepted that the nuclear import of the HIV-1 PIC is an energy-dependent active process that is governed by different viral and cellular factors. Once inside the nucleus, the PIC can integrate into the host chromatin. This step is considered to be essential for productive replication and ensure the stable insertion of the viral genome sequence in the host chromatin. HIV-1 inserts its genome in the active chromatin located near NPCs (Lelek et al. 2015; Marini et al. 2015). Viral and host factors directly or indirectly influence the selection of the target site. These include nucleoporins, chromatin structure, the viral integrase and chromatin tethering factors like lens-epithelium-derived growth factor/p75 (LEDGF/p75). One of the latest findings shows that chromatin organization around NPCs is determinant for the fate of viral progeny. In fact, the nuclear basket of the pore maintains an open chromatin that is favourable for viral replication (Lelek et al. 2015). Therefore, even if the majority of integrated proviruses are transcribed to ensure their progeny, a small fraction remains silent and forms viral reservoir, which is the most critical barrier for the eradication of HIV-1. The passage through the pore can determine directly or indirectly HIV-1 integration sites as it has been shown in infected cells depleted for some Nups, such as RanBP2/Nup358, Nup153, Nup98 and Tpr (Di Nunzio et al. 2013; Lelek et al. 2015; Ocwieja et al. 2011; Schaller et al. 2011). A particular euchromatin landmark, H3K36me3, associated to viral integration sites has been found in the chromatin near the nuclear basket. Its location near the pore channel is regulated by the presence of Tpr, in fact cells depleted for Tpr have less H3K36me3 near the NPC (Lelek et al. 2015). Interestingly, infected lymphocytes depleted for Tpr show a silent virus while the global profile of gene expression is not altered. Results obtained by super resolution imaging identified a peak density of H3K36me3 at approximately 500 nm from the NPC position (Lelek et al. 2015), this is in agreement with data obtained by DNA FISH showing that HIV-1 preferentially integrates at the nuclear periphery, within 1 μm from the nuclear envelope (NE) (Marini et al. 2015). In contrast, proviral integrations are disfavoured in the chromatin associated with the nuclear lamina (Marini et al. 2015). This chromatin located between pores is poor in marks of active genes, such as H3K36me3, and weakly associated with integration sites (Lelek et al. 2015). It is clear that the fate of HIV-1 integration is dictated by PIC-host nuclear factor interactions and the chromatin landscape near the NPC, in particular, the chromatin located in the vicinity of the nuclear pore complex creates a favourable environment for HIV-1 integration sites selection.
8.3 Role of NPC Components in HIV-1 Replication
8.3.1 Nups and HIV-1 Uncoating
After fusion to the cellular membrane, HIV-1 releases the viral core into the cytoplasm. The HIV-1 core then travels through the microtubule network of the host cell to traffic towards the nucleus (McDonald et al. 2002). This pathway is used only by HIV-1 carrying an envelope WT, while HIV-1 delta Env pseudoptyped with VSV-G, commonly used for the investigation of the early steps of HIV-1 life cycle, enters in the cytoplasm by endocytosis (Campbell and Hope, 2015). However, only the VSV-G pseudotyped virus is able to escape from endosomes and is highly infectious, giving the virus 20- to 130-fold higher infectivity (Aiken, 1997; Luo et al. 1998). Even if the two viruses enter in contact with different cytoplasmic components, both need the NPC to enter the nucleus (Di Nunzio et al. 2012). Once at the NPC, the viruses dock at the pore through RanBP2/Nup358 (Di Nunzio et al. 2012), spending several hours before translocating inside the nucleus. HIV-1 needs to uncoat (loss of the integrity of core) before integrating. This is one of the most intriguing steps of the HIV-1 life cycle. In the past years, the most accredited model defined the uncoating as the process by which the capsid core dissociates from the rest of the RTC immediately after viral infection (Aiken 2006). Furthermore, some studies suggest that CA remains associated with the RTC before to translocate into the nucleus (Arhel et al. 2007; Di Nunzio et al. 2012, 2013; Jacques et al. 2016; Lelek et al. 2012; Matreyek et al. 2013; Price et al. 2014). Uncoating may occur following three principal models that have been proposed (Campbell and Hope 2015): (1) immediately after viral fusion with the cellular membrane (Francis et al. 2016; Mamede et al., 2017); (2) during cytoplasmic trafficking (Francis et al. 2016; Hulme et al. 2011); (3) with the aid of nuclear pore factors (Di Nunzio et al. 2012, 2013; Lelek et al. 2015; Price et al. 2014). A recent study using an artificial system for viral imaging observed that the majority of HIV-1 cores uncoat soon after release into the cytoplasm. Less than 5% of viral particles uncoat gradually during their cytoplasmic journey and only a small fraction shows a late uncoating at the pore (Francis et al. 2016). Recent studies attributed the CA shell an important protective role versus the viral genome avoiding its exposure to cytosolic DNA sensors. This observation is not compatible with a model of premature uncoating (Rasaiyaah et al. 2013). Interestingly, results from different teams and techniques have shown that inhibition of reverse transcription delays uncoating (Hulme et al. 2011; Yang et al. 2013). The peak of reverse transcription is around 7–8 hrs from infection (Butler et al. 2001), suggesting a late uncoating. Other studies show that the CA is the viral determinant for HIV-1 nuclear import (Yamashita et al. 2007) through the interaction with cellular host factors (Brass et al. 2008; Di Nunzio et al. 2012, 2013; Konig et al. 2008; Lee et al. 2010; Matreyek et al. 2013; Schaller et al. 2011). Because of the large size (120 nm × 60 nm × 40 nm), the viral core cannot translocate through the NPC channel, which has an estimated diameter of ~ 39nm (Pante and Kann 2002). Thus, current models propose that uncoating begins before to enter in the nucleus (Ambrose and Aiken 2014; Arhel et al. 2007; Chen et al. 2016; Fassati 2012; Hilditch and Towers 2014; Burdick et al., 2017). Nups have been found to have an active role in PIC maturation and in the uncoating processes (Bichel et al., 2013). The peculiarity of lentiviruses with respect to retroviruses in usurping the NPC could be due to the specific interaction of the lentiviruses CA to particular Nups. The first Nup that the viral CA can meet is RanBP2/Nup358. The interaction between them has been well documented (Di Nunzio et al. 2012; Schaller et al. 2011). The C-terminus of RanBP2/Nup358 comprising a cyclophilin (cyp)-homology domain has a major contribution to the in vitro cores binding, probably helped by FG repeats dispersed along the entire protein (Di Nunzio et al. 2012). A recent study highlights the possibility that the CA isomerization mediated by the cyclophilin-homology domain of RanBP2/Nup358 may be preserved by HIV-1 to target the nuclear pore and synchronize nuclear entry with CA uncoating (Bichel et al. 2013). Interestingly, the cyclophilin A binding loop is an exclusive feature of lentiviruses capsid, while retroviruses do not have this loop in their CA preventing their docking at nuclear pores. Another critical Nup for HIV-1 infection is Nup153. This Nup plays multiple cellular functions and also participates in the replication of several viruses. HBV, for example, binds Nup153 to help the uncoating and the release of its DNA genome into the nucleus (Schmitz et al. 2010). HIV-1 CA also binds the human Nup153 (Di Nunzio et al. 2013). It is possible that Nup153 participates in HIV-1 uncoating, but the mechanism is still under investigation. Interestingly, the multifaceted role of viral CA can determine the differences between the two retroviral subfamilies, gamma retroviruses and lentiviruses. In the case of gamma retroviruses, like MLV, the uncoating step is better understood than for lentiviruses, such as HIV-1. MLV uncoating is triggered by an accessory p12 protein that interacts with MLV cores, preventing uncoating and ensuring completion of reverse transcription. The C-terminus tail of p12 binds condensed chromatin, which is typical during mitosis, and tethers the CA-associated PIC to the chromatin helping with the integration step (Elis et al. 2012; Wight et al. 2014). Consistent with this mechanism, when the mitosis ends and the NE reassembles, p12 is released and orchestrates CA uncoating (Schneider et al. 2013). In contrast to retroviruses, lentiviruses have a more unstable core, and in vitro studies have failed to show association of CA with PIC (Farnet and Haseltine 1991; Fassati and Goff 2001; Forshey et al. 2002; Miller et al. 1997). The high fragility of HIV-1 CA increases the difficulty of studying the fate of viral CA traveling into the host cell environment. Microscopy studies could overcome the problems correlated with the CA stability, but the limited number of cores that can mature in functional PICs poses a problem for the interpretation of the imaging results. Recent new advances in imaging approaches have led to a better comprehension of the molecular mechanism underlying the uncoating step (Chin et al. 2015; Lelek et al. 2012, 2015; Peng et al. 2014; Burdick et al., 2017) and might aid in the design of new compounds to specifically target this step.
8.3.2 Nups and HIV-1 Translocation
HIV-1 is a component of Retroviridae family. Contrary to other subclasses, the lentivirus category, which includes HIV-1, has the ability to infect non-dividing cells as well as dividing cells, likely sharing the same nuclear import mechanism (Katz et al. 2003). Larger complexes are more efficiently translocated in nuclei of actively dividing cells with respect to quiescent cells, probably due to the dependency of the nuclear import on the phosphorylation of some critical factors and on cell metabolism (Feldherr and Akin 1994). NPCs are the exclusive controllers of nucleocytoplasmic trafficking and regulate the passage of macromolecules in and out the nucleus. Recent experiments based on atomic force microscopy suggest that Nups containing FG repeats form condensates in the centre of NPC channel that may transiently dissolve to transport larger cargoes (Stanley et al. 2017). Viruses are also subjected to the nucleocytoplasmic transport rules. Herpesviruses and adenoviruses, possess a large and relatively stable icosahedral capsid (CA), which envelops the viral genome. Since their capsids are larger than the nuclear pore channel, these viruses need to uncoat before entering the nucleus. Viral capsids dock at the NPC cytoplasmic side, and the interaction with Nups is used as a cue to trigger genome nuclear release. Herpesviruses bind to the NPC via an importin β-dependent interaction with RanBP2/Nup358 and the interaction with Nup214 triggers DNA release in the nucleus of the target cell (Cohen et al. 2011). The CA of adenovirus recruits histone H1, importin β, importin 7 and hsp70 (Mercer et al. 2010) which trigger the disassembly/conformational changes required for the import of viral DNA into the nucleus through the NPC (Cohen et al. 2011; Hsieh et al. 2010). Different retroviruses have different behaviours during nuclear entry. MLV enters the nucleus during mitosis while HIV-1 reaches the chromatin during interphase. Both pathways are regulated by different cellular factors that play critical and indispensable roles in retroviruses nuclear import. The differences between these two retroviruses, MLV and HIV-1, have been one of the most intriguing observations in the field of viral nuclear import. Biochemical data suggest that the divergence between the two retroviruses could be attributed to a different stability of viral CA. In the case of HIV-1, the presence of CA as component of the PIC is still uncertain, however, increasing evidences suggest that this viral protein is a determinant of HIV-1 nuclear import. Low levels of HIV-1 CA has been observed associated with the reverse transcription (RTC) and the PIC (Farnet and Haseltine 1991; Fassati and Goff 2001; Forshey et al. 2002; Miller et al. 1997). For many years it was believed that the traffic through the pore was independent on the viral CA. Recently several teams showed the critical role of viral CA in HIV-1 translocation and integration, highlighting a new role for HIV-1 CA in the early steps of viral infection (Brass et al. 2008; Chen et al. 2016; Dharan et al. 2016; Di Nunzio et al. 2012, 2013; Jacques et al. 2016; Konig et al. 2008; Lelek et al. 2015; Matreyek et al. 2013; Ocwieja et al. 2011; Saito et al. 2016; Sowd et al. 2016). Direct interactions between viral cores and Nups have been shown to be essential for an efficient viral translocation (Di Nunzio et al. 2012, 2013; Lelek et al. 2015; Matreyek et al. 2013; Schaller et al. 2011). Some groups detected the presence of CA inside of the nucleus (Chin et al. 2015; Hulme et al. 2015; Peng et al. 2014) and its interaction with nuclear factors, such as the polyadenylation factor CPSF6 which is a mRNA processing protein that shuttles between the nucleus and the cytoplasm (Chin et al. 2015; Lee et al. 2010). This factor predominantly localizes inside the nucleus due to the presence of a serine/arginine (SR)-rich nuclear localization signal located at the C-terminus of the protein (Lee et al. 2010; Price et al. 2012). The first study showing the critical role of CPSF6 for HIV-1 infection was based on a mouse cDNA-expression screen that identified the truncated form of CPSF6 lacking the C-terminal SR rich domain, which accumulated in the cytoplasm and prevented HIV-1 nuclear import (Lee et al. 2010). The direct binding between CPSF6 and in vitro cores was later shown (Fricke et al. 2013). Overall these findings uncovered that HIV-1 usurps the nuclear pore machinery to dock, translocate and integrate into the host chromatin. These steps are concerted to aid the PIC to mature and allow the provirus to integrate into the host genome to replicate (Fig. 8.1). Nups also have a key role in HIV-1 translocation in fact, the depletion of particular Nups induces a block in HIV-1 nuclear entry. Several studies have determined the individual role of Nups in major viral steps, such as viral docking at the pore and nuclear translocation. The depletion of RanBP2/Nup358, which is exclusively located in the outer side of the pore, reduces HIV-1 nuclear entry by up to ten fold (Di Nunzio et al. 2012; Schaller et al. 2011). This is similar to the depletion of Nup153, a Nup predominantly located in the nuclear side of the NPC (Di Nunzio et al. 2013; Matreyek et al. 2013). Outwardly, these two Nups showing opposite locations at NPCs seem to participate at the same step of HIV-1 infection. But several differences in their roles have been identified. RanBP2/Nup358 is involved in the docking step of the virus at the pore (Di Nunzio et al. 2012), while Nup153 is required for nuclear translocation, which is mediated by its flexible C-terminus domain (Di Nunzio et al. 2013; Lelek et al. 2015; Matreyek et al. 2013). According to in vitro studies, the Cyp loop of CA determines the interaction with the Cyp-like domain of RanBP2/Nup358 (Lin et al. 2013), while a specific region of the C-terminus domain of Nup153 that interacts with the interphase CA (NTD)-CA (CTD) in the CA hexamer is responsible for the binding to the viral core (Matreyek et al. 2013). This CA pocket is present only in assembled or partially assembled core but not in monomers, suggesting that Nup153 interacts and favours HIV-1 nuclear entry through the binding of CA hexamers (Figs. 8.1 and 8.2). Similarly to HIV-1, the yeast ortholog of human Nup153, Nup124p binds gag of the retrotransposon Tf1 to help nuclear import (Varadarajan et al. 2005). This Nup contains 29 FG repeats at its terminal tail, which are responsible for this binding as it has been shown in vitro (Matreyek et al. 2013) and in vivo (Lelek et al. 2015). Even if Nup153 is predominantly located at the nuclear basket, it has also been observed at the cytoplasmic side of the pore aiding nuclear import of cargo (Lim et al. 2007). Nup153 can assume two conformations a “collapse” conformation for cargo transport and a “release” for cargo interaction in the cytoplasmic side of the NPC (Cardarelli et al. 2012). This motion of Nup153 could correlate with the motion of the molecules actively transported through the NPC. This cellular process could be usurped by HIV-1 to translocate inside the nucleus (Fig. 8.1). However, the mechanistic requirements underlying the in vivo viral nuclear translocation mediated by Nup153 or other factors and the state of the viral CA during translocation have yet to be formally demonstrated. In summary, the viral core is believed to dock onto the cytoplasmic side of the NPC through interactions with RanBP2/Nup358 (Di Nunzio et al. 2012; Schaller et al. 2011). HIV-1 subsequently interacts with some Nups, such as Nup153, Nup98-Nup96, Tpr, and factors like TNPO3 and CPSF6, directly or indirectly to release the PIC into the nucleoplasm (Di Nunzio et al. 2012, 2013; Krishnan et al. 2010; Lee et al. 2010; Lelek et al. 2015; Matreyek et al. 2013; Price et al. 2014; Valle-Casuso et al. 2012).
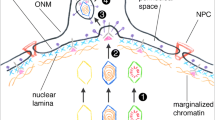
HIV-1 nuclear import and integration in target cells. An intact core is shown before/during interaction with the NPC (left) (docking), in the centre it is represented the PIC translocation, potentially piloted by Nup153 (blue), which is in a collapsed conformation compatible with nuclear import of cargo. The CA hexamer (orange) could be part of this latter complex. On the right, the proviral integration into the host euchromatin located underneath the pore (Lelek et al. 2015; Marini et al. 2015) enriched in open chromatin feature H3K36me3 and nuclear factors LEDGF/p75 (pink), CPSF6 (green)
HIV-1 nuclear import and integration in Tpr depleted cells. NPCs represented lack of Tpr, thus the chromatin underneath the nuclear basket is less de-condensed. An intact core is shown before/during interaction with the NPC (left) (docking), in the centre it is represented the PIC translocation, potentially piloted by Nup153 (blue), which is in a collapsed conformation compatible with nuclear import of cargo. The CA hexamer (orange) could be part of this latter complex. On the right, the proviral integration into the host chromatin less de-condensed due to the loss of Tpr fibrils located underneath the pore (Lelek et al. 2015; Marini et al. 2015)
8.3.3 Nups and HIV-1 Integration
The regulation of nucleocytoplasmic transport is the major known role of NPC. However, recent evidences show Nups as regulators of different processes in the cell. In particular, Nups can affect fundamental genome functions independently of their nuclear transport function (Ibarra and Hetzer 2015). The multifaceted role of Nups could be attributed to particular domains present in them. In particular, Nups are able to influence the chromatin landscape. The association of active decondensed chromatin to NPCs has been interpreted as the link between the transcription of active genes with mRNA export (Blobel 1985). The functional reason for the presence of active chromatin at NPCs could be the presence of the transcriptional activator SAGA and the mRNA export machinery at the nuclear basket (Cabal et al. 2006; Dieppois et al. 2006; Garcia-Oliver et al. 2012; Iglesias et al. 2010; Rodriguez-Navarro et al. 2004; Taddei and Gasser 2004). In yeast, gene regulation regulated by Nups-chromatin interactions is spatially confined at the nuclear periphery. In metazoans, Nups can exist in two populations, one associated with the pore and the other free in the nucleoplasm. Thus, gene regulation mediated by Nups-chromatin interactions is more complicated. In Drosophila, the silencing of nuclear basket Nups, Nup153 and Mtor/Tpr, reduces the expression of thousands of genes. Interestingly, these Nups are associated with active chromatin marks (Vaquerizas et al. 2010). It is possible that the same Nup can participate in different transcriptional regulation events depending on their location at the NPC or off-NPC. For example, in Drosophila Nup98 shows preferential binding for active genes only if located out of the pore (Kalverda et al. 2010). However, in human cells, Nup98 also participates in gene activation at the pore (Liang et al. 2013). Nups seem to organize chromatin topology, controlling the accessibility of transcription factors. The complex formed by NPCs and the underlying chromatin could act as a functional hub that recruits the enzymatic machinery that leads to epigenetic reformatting and the transcriptional regulation of particular genes. This model might explain the role of Nups in transcriptional memory, in which Nup98 favours specific chromatin structure changes, such as H2A.Z deposition and H3K4me2 (Ahmed et al. 2010; Brickner et al. 2007; Light et al. 2010, 2013). NPCs also play a critical role in chromatin organization, defining active and silent regions near the pore. During interphase the chromatin is well organized within the nucleus. In particular, condensed chromatin (heterochromatin) is associated with nuclear periphery, interrupted by stretches of less condensed chromatin (euchromatin) at NPCs (Krull et al. 2010; Ma et al. 2015; Raices and D’Angelo 2012) (Fig. 8.1). The chromatin near the pore represents actively transcribed regions and it is also the target of HIV-1 PIC (Lelek et al. 2015; Marini et al. 2015). This might ensure/promote efficient viral gene expression after integration. HIV-1 integrase (IN) binds to LEDGF/p75, which was the first cellular factor identified to help HIV-1 integration into active genes (Cherepanov et al. 2003; Ciuffi et al. 2005). LEDGF/p75 promotes efficient infection (Llano et al. 2006; Shun et al. 2007) and tethers IN to favour target sites (Fig. 8.1). The HIV-1 integration machinery must also interact with many additional host factors during infection, including nuclear trafficking and pore proteins during nuclear entry, histones during initial target capture, and DNA repair proteins during completion of the DNA joining steps. The NPC has been described as the link between HIV-1 translocation and integration into the host chromatin (Di Nunzio 2013; Lelek et al. 2015). It is possible that HIV-1 cores recruit cellular factors that promote or participate in these concerted steps. The viral-host complexes can favour the accessibility to viral components, usually hided in the inner core, or modify the conformation of the CA influencing the engagement of other host factors that mediate the nuclear translocation and integration of the PIC into the host chromosomal DNA. The viral CA shows some plasticity in the use alternative pathways to aid the PIC nuclear entry and HIV-1 CA mutants insensitive to certain Nups show different integration sites pattern than wild type viruses (Schaller et al. 2011). For example, HIV-1 CA carrying point mutations, like N74D, which is unable to recruit the cellular factors CPSF6 and RanBP2/Nup358, shows a different pattern of integration sites than wild type HIV-1 (Koh et al. 2013; Schaller et al. 2011; Sowd et al. 2016). Other HIV-1 CA mutants, such as N57A, which are more severely defective in arrested cells than dividing cells (Yamashita et al. 2007), also show a different integration pattern than the wild type virus (Schaller et al. 2011). It is possible that HIV-1 CA mutant engages different host factors determining a viral nuclear entry at a different cell cycle step than the wild type virus. The results underscore the plasticity of HIV-1 CA and its ability to recruit different host factors to reach and integrate in cellular chromosomes to guarantee new viral progeny. Beside the role in HIV-1 nuclear import, CPSF6 is also involved in defining the distribution of HIV-1 integration sites. Interestingly, the depletion of CPSF6 provokes changes in the kinetics of viral infectivity and reduces integration into transcriptionally active and spliced genes. CPSF6 binds CA and it has been proposed to drive the PIC to actively transcribed chromatin (Chin et al. 2015; Sowd et al. 2016). The integrase-LEDGF/p75 interaction, on the other hand, promotes integration into gene bodies where nucleosomes are remodelled for an efficient HIV-1 integration (Lesbats et al. 2011; Sowd et al. 2016). A double CPSF6 and LEDGF/p75 knockout diminishes integration into genes below the levels observed for each of them. These observations support a model in which NPC components act in concert with other cellular factor to favour HIV-1 replication.
Studies of different integrating genomic parasites show that their host chromatin targeting preferences have evolved to optimize their coexistence with the host and to favour the release of new viral progeny. One example comes from the yeast Ty retrotransposons, which must coexist with their hosts indefinitely so they integrate into host genomic locations that do not damage the survival of yeast cell (Bushman 2003; Craig and Marszalek 2002). Instead, HIV-1 infected T cells typically survive only a day or two before to be killed by the cellular immune system or by the toxicity of infection (Perelson et al. 1996). Thus, HIV-1 needs to push the release of the newborn viruses within few days from infection. Viral integration within transcription units is usually favourable for efficient transcription (Jordan et al. 2001; Lewinski et al. 2005) potentially explaining the targeting preference. But, HIV-1 can also coexist long time as chimera into the genome of infected patients as latent virus (Razooky et al. 2015). The mechanisms underlying the establishment of latency are under investigation and it is highly possible that chromatin factors associated with nuclear entry proteins can play a critical role on the persistence of HIV-1 in infected cells. Other retroviruses, such as gamma retroviruses favour integration near transcription start sites of cancer genes (Wu et al. 2003) to ensure a favourable environment for proviral transcription with the additional advantage to promote the proliferation and/or survival of the infected cells. Two recent studies (Lelek et al. 2015; Marini et al. 2015) have shown that the classical nuclear entry pathway adopted by HIV-1, which involves the interaction of the virus to canonical Nups, creates a favourable chromatin environment for viral replication underneath the NPC. These Nups, in particular Tpr, organizes the chromatin near the pore, which is the target of HIV-1 integration. In the NPC surrounding chromatin there is a high density of the epigenetic landmark H3K36me3 (Lelek et al. 2015), which is highly present in HIV-1 integration sites (Wang et al. 2007), and LEDGF/p75 (Fig. 8.1). Depletion of Tpr results in the loss of the nuclear basket and a concomitant loss of euchromatin containing the chromatin feature H3K36me3, into the vicinity of NPCs (Fig. 8.2). Interestingly, the nuclear import and integration of the viral DNA are not affected by Tpr depletion but the viral genes are under expressed. Thus, in the absence of Tpr, HIV-1 integration can occur but in silent chromatin. As a consequence the replication of the HIV-1 genome is attenuated (Lelek et al. 2015) (Fig. 8.2). Because NPC components have a critical role in chromatin organization around pores, depletion of one that changes the chromatin topology promotes the integration of the virus in other available regions dictated by the new NPC composition. The PIC is probably guided by CPSF6 and/or Nup153 to target active genes, spatially available near the pores and maintained by the nuclear basket protein Tpr. Additionally, LEDGF/p75 helps the virus to integrate into the body of active genes (Fig. 8.1). The integration sites are critical for the outcome of viral transcription. HIV-1 integrates within transcriptional units in both acutely and latently infected cells (Wang et al. 2007). The viral persistence is characterized by viral reservoirs where a replication competent virus persists for long time in a quiescent state (Van Lint et al. 2013). This observation increases the complexity of how viral reservoir are established. Interestingly, the loss of the nuclear basket due to the depletion of Tpr reproduces a phenotype similar to that of persistent viral infected cells, in which the virus integrates but does not replicate. According to this scenario, proviral transcription is influenced by chromatin structure and epigenetic features at the viral integration site. It is possible that the virus evolved the ability to establish a viral reservoir to co-exist with the host for a long term, especially in unfavourable environmental conditions (Lucic and Lusic 2016).
More extensive studies will help to elucidate the cellular mechanisms usurped by the virus to promote its own survival and persistence. In fact, persistent infected cells remain one of the most important limit to eradicate HIV-1.
8.4 Concluding Remarks
Studies based on the role of NPC components highlight the emerging role of Nups at the pore and off-pore as organizer of chromatin structure and nuclear topology. Furthermore, it is clear that the link between Nups and the transcriptional machinery is critical for gene expression. A clear challenge consists in a better comprehension of how, at a molecular level, Nups contribute to divergent genome-associated functions, such as transcriptional activation or repression. An important step will be the understanding of the physical interactions between Nups and chromatin-associated proteins. All these processes regulated by Nups seem to play a role in HIV-1 life cycle. In particular, the NPC is the exclusive pathway adopted by HIV-1 to reach the host chromatin in non-dividing cells and probably also in dividing cells. Nuclear basket Nups bind particular chromatin regions and regulate gene activity, however it is still under investigation how Nups, chromatin factors and genes are concerted to orchestrate HIV-1 replication. Nuclear basket Nups may be another “cellular code” for specifying HIV-1 fate through their contacts with the underlying chromatin. With a deeper understanding of the relationships between HIV-1 components, NPC and chromatin, it will be possible to shed light on the cellular pathways underlying AIDS pathology. Overall, new studies on this topic will undoubtedly gain insights into HIV-1 replication mechanisms and could serve in the development of new antiviral strategies.
References
Ahmed S, Brickner DG, Light WH et al (2010) DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 12(2):111–8
Aiken C (1997) Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol 71(8):5871–7
Aiken C (2006) Viral and cellular factors that regulate HIV-1 uncoating. Curr Opin HIV AIDS 1(3):194–9
Ambrose Z, Aiken C (2014) HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology 454-455:371–9
Arhel NJ, Souquere-Besse S, Munier S et al (2007) HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J 26(12):3025–37
Bichel K, Price AJ, Schaller T et al (2013) HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358. Retrovirology 10:81
Blobel G (1985) Gene gating: a hypothesis. Proc Natl Acad Sci U S A 82(24):8527–9
Brass AL, Dykxhoorn DM, Benita Y et al (2008) Identification of host proteins required for HIV infection through a functional genomic screen. Science 319(5865):921–6
Brickner DG, Cajigas I, Fondufe-Mittendorf Y et al (2007) H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5(4):e81
Burdick RC, Delviks-Frankenberry KA, Chen J, Janaka SK, Sastri J, Hu WS, Pathak VK. (2017) PLoS Pathog 13(8):e1006570.
Bushman FD (2003) Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115(2):135–8
Butler SL, Hansen MS, Bushman FD (2001) A quantitative assay for HIV DNA integration in vivo. Nat Med 7(5):631–4
Cabal GG, Genovesio A, Rodriguez-Navarro S et al (2006) SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441(7094):770–3
Campbell EM, Hope TJ (2015) HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13(8):471–83
Cardarelli F, Lanzano L, Gratton E (2012) Capturing directed molecular motion in the nuclear pore complex of live cells. Proceedings of the National Academy of Sciences 109(25):9863–9868
Chen NY, Zhou L, Gane PJ et al (2016) HIV-1 capsid is involved in post-nuclear entry steps. Retrovirology 13:28
Cherepanov P, Maertens G, Proost P et al (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278(1):372–81
Chin CR, Perreira JM, Savidis G et al (2015) Direct visualization of HIV-1 replication intermediates shows that capsid and CPSF6 modulate HIV-1 intra-nuclear invasion and integration. Cell Rep 13(8):1717–31
Chubb JR, Boyle S, Perry P et al (2002) Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12(6):439–45
Ciuffi A, Llano M, Poeschla E et al (2005) A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11(12):1287–9
Cohen S, Au S, Pante N (2011) How viruses access the nucleus. Biochim Biophys Acta 1813(9):1634–45
Copeland AM, Newcomb WW, Brown JC (2009) Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J Virol 83(4):1660–8
Craig EA, Marszalek J (2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell Mol Life Sci 59(10):1658–65
Cremer T, Cremer M (2010) Chromosome territories. Cold Spring Harb Perspect Biol 2(3):a003889
Dharan A, Talley S, Tripathi A et al (2016) KIF5B and Nup358 cooperatively mediate the nuclear import of HIV-1 during infection. PLoS Pathog 12(6):e1005700
Di Nunzio F (2013) New insights in the role of nucleoporins: a bridge leading to concerted steps from HIV-1 nuclear entry until integration. Virus Res 178(2):187–96
Di Nunzio F, Danckaert A, Fricke T et al (2012) Human nucleoporins promote HIV-1 docking at the nuclear pore, nuclear import and integration. PLoS One 7(9):e46037
Di Nunzio F, Fricke T, Miccio A et al (2013) Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology 440(1):8–18
Dieppois G, Iglesias N, Stutz F (2006) Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 26(21):7858–70
Elis E, Ehrlich M, Prizan-Ravid A et al (2012) p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes. PLoS Pathog 8(12):e1003103
Farnet CM, Haseltine WA (1991) Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol 65(4):1910–5
Fassati A (2012) Multiple roles of the capsid protein in the early steps of HIV-1 infection. Virus Res 170(1-2):15–24
Fassati A, Goff SP (2001) Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75(8):3626–35
Feldherr CM, Akin D (1994) Role of nuclear trafficking in regulating cellular activity. Int Rev Cytol 151:183–228
Forshey BM, von Schwedler U, Sundquist WI et al (2002) Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76(11):5667–77
Francis AC, Marin M, Shi J et al (2016) Time-Resolved Imaging of Single HIV-1 Uncoating In Vitro and in Living Cells. PLoS Pathog 12(6):e1005709
Fricke T, Valle-Casuso JC, White TE et al (2013) The ability of TNPO3-depleted cells to inhibit HIV-1 infection requires CPSF6. Retrovirology 10:46
Garcia-Oliver E, Garcia-Molinero V, Rodriguez-Navarro S (2012) mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim Biophys Acta 1819(6):555–65
Hilditch L, Towers GJ (2014) A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Curr Opin Virol 4:32–6
Hsieh MJ, White PJ, Pouton CW (2010) Interaction of viruses with host cell molecular motors. Curr Opin Biotechnol 21(5):633–9
Hulme AE, Kelley Z, Okocha EA et al (2015) Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. J Virol 89(1):643–51
Hulme AE, Perez O, Hope TJ (2011) Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A 108(24):9975–80
Ibarra A, Hetzer MW (2015) Nuclear pore proteins and the control of genome functions. Genes Dev 29(4):337–49
Iglesias N, Tutucci E, Gwizdek C et al (2010) Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev 24(17):1927–38
Jacques DA, McEwan WA, Hilditch L et al (2016) HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 536(7616):349–53
João I. Mamede, Gianguido C. Cianci, Meegan R. Anderson, Thomas J. Hope, (2017) Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proceedings of the National Academy of Sciences 114(34):E7169–E7178
Jordan A, Defechereux P, Verdin E (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 20(7):1726–38
Kalverda B, Pickersgill H, Shloma VV et al (2010) Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140(3):360–71
Katz RA, Greger JG, Boimel P et al (2003) Human immunodeficiency virus type 1 DNA nuclear import and integration are mitosis independent in cycling cells. J Virol 77(24):13412–7
Koh Y, Wu X, Ferris AL et al (2013) Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol 87(1):648–58
Konig R, Zhou Y, Elleder D et al (2008) Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135(1):49–60
Krishnan L, Matreyek KA, Oztop I et al (2010) The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J Virol 84(1):397–406
Krull S, Dorries J, Boysen B et al (2010) Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 29(10):1659–73
Lee K, Ambrose Z, Martin TD et al (2010) Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7(3):221–33
Lelek M, Casartelli N, Pellin D et al (2015) Chromatin organization at the nuclear pore favours HIV replication. Nat Commun 6:6483
Lelek M, Di Nunzio F, Henriques R et al (2012) Superresolution imaging of HIV in infected cells with FlAsH-PALM. Proc Natl Acad Sci U S A 109(22):8564–9
Lesbats P, Botbol Y, Chevereau G et al (2011) Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog 7(2):e1001280
Lewinski MK, Bisgrove D, Shinn P et al (2005) Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol 79(11):6610–9
Liang Y, Franks TM, Marchetto MC et al (2013) Dynamic association of NUP98 with the human genome. PLoS Genet 9(2):e1003308
Light WH, Brickner DG, Brand VR et al (2010) Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell 40(1):112–25
Light WH, Freaney J, Sood V et al (2013) A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol 11(3):e1001524
Lim RYH, Fahrenkrog B, Koser J, Schwarz-Herion K, Deng J, Aebi U (2007) Nanomechanical Basis of Selective Gating by the Nuclear Pore Complex. Science 318(5850):640–643
Lin DH, Zimmermann S, Stuwe T et al (2013) Structural and functional analysis of the C-terminal domain of Nup358/RanBP2. J Mol Biol 425(8):1318–29
Llano M, Saenz DT, Meehan A et al (2006) An essential role for LEDGF/p75 in HIV integration. Science 314(5798):461–4
Lucic B, Lusic M (2016) Connecting HIV-1 integration and transcription: a step toward new treatments. FEBS Lett 590(13):1927–39
Luo T, Douglas JL, Livingston RL et al (1998) Infectivity enhancement by HIV-1 Nef is dependent on the pathway of virus entry: implications for HIV-based gene transfer systems. Virology 241(2):224–33
Ma Y, Kanakousaki K, Buttitta L (2015) How the cell cycle impacts chromatin architecture and influences cell fate. Front Genet 6:19
Maeshima K, Yahata K, Sasaki Y et al (2006) Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci 119(Pt 21):4442–51
Marini B, Kertesz-Farkas A, Ali H et al (2015) Nuclear architecture dictates HIV-1 integration site selection. Nature 521(7551):227-31
Matreyek KA, Yucel SS, Li X et al (2013) Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog 9(10):e1003693
McDonald D, Vodicka MA, Lucero G et al (2002) Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 159(3):441–52
Mercer J, Schelhaas M, Helenius A (2010) Virus entry by endocytosis. Annu Rev Biochem 79:803–33
Miller MD, Farnet CM, Bushman FD (1997) Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol 71(7):5382–90
Ocwieja KE, Brady TL, Ronen K et al (2011) HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog 7(3):e1001313
Pante N, Kann M (2002) Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell 13(2):425–34
Peng K, Muranyi W, Glass B et al (2014) Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. Elife 3:e04114
Perelson AS, Neumann AU, Markowitz M et al (1996) HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271(5255):1582–6
Price AJ, Fletcher AJ, Schaller T et al (2012) CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog 8(8):e1002896
Price AJ, Jacques DA, McEwan WA et al (2014) Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS Pathog 10(10):e1004459
Raices M, D’Angelo MA (2012) Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol 13(11):687–99
Rasaiyaah J, Tan CP, Fletcher AJ et al (2013) HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503(7476):402–5
Razooky BS, Pai A, Aull K et al (2015) A hardwired HIV latency program. Cell 160(5):990–1001
Rodriguez-Navarro S, Fischer T, Luo MJ et al (2004) Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116(1):75–86
Saito A, Ferhadian D, Sowd GA et al (2016) Roles of capsid-interacting host factors in multimodal inhibition of HIV-1 by PF74. J Virol 90(12):5808–23
Schaller T, Ocwieja KE, Rasaiyaah J et al (2011) HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7(12):e1002439
Schmitz A, Schwarz A, Foss M et al (2010) Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog 6(1):e1000741
Schneider WM, Brzezinski JD, Aiyer S et al (2013) Viral DNA tethering domains complement replication-defective mutations in the p12 protein of MuLV Gag. Proc Natl Acad Sci U S A 110(23):9487–92
Shun MC, Raghavendra NK, Vandegraaff N et al (2007) LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21(14):1767–78
Sowd GA, Serrao E, Wang H et al (2016) A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc Natl Acad Sci U S A 113(8):E1054–63
Stanley GJ, Fassati A, Hoogenboom BW (2017) Biomechanics of the transport barrier in the nuclear pore complex. Semin Cell Dev Biol. 68:42-51
Taddei A, Gasser SM (2004) Multiple pathways for telomere tethering: functional implications of subnuclear position for heterochromatin formation. Biochim Biophys Acta 1677(1-3):120–8
Trotman LC, Mosberger N, Fornerod M et al (2001) Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat Cell Biol 3(12):1092–100
Valle-Casuso JC, Di Nunzio F, Yang Y, Reszka N, Lienlaf M, Arhel N, Perez P, Brass AL, Diaz-Griffero F (2012) J Virol 86(10):5931–6
Van Lint C, Bouchat S, Marcello A (2013) HIV-1 transcription and latency: an update. Retrovirology 10:67
Vaquerizas JM, Suyama R, Kind J et al (2010) Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 6(2):e1000846
Varadarajan P, Mahalingam S, Liu P et al (2005) The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr. Mol Biol Cell 16(4):1823–38
Wang GP, Ciuffi A, Leipzig J et al (2007) HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 17(8):1186–94
Wight DJ, Boucherit VC, Wanaguru M et al (2014) The N-terminus of murine leukaemia virus p12 protein is required for mature core stability. PLoS Pathog 10(10):e1004474
Wu X, Li Y, Crise B et al (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300(5626):1749–51
Yamashita M, Perez O, Hope TJ et al (2007) Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog 3(10):1502–10
Yang Y, Fricke T, Diaz-Griffero F (2013) Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J Virol 87(1):683–7
Acknowledgements
This work was supported by ANRS (grant N. ECTZ4469), Sidaction and the Pasteur Institute in Paris. A special thanks to Julie and Yann Ravel for all support and to inspire me to write the current chapter.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this chapter
Cite this chapter
Di Nunzio, F. (2018). Nuclear Pore Complexes, Genome Organization and HIV-1 Infection. In: D’Angelo, M. (eds) Nuclear Pore Complexes in Genome Organization, Function and Maintenance. Springer, Cham. https://doi.org/10.1007/978-3-319-71614-5_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-71614-5_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-71612-1
Online ISBN: 978-3-319-71614-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)