Abstract
The ability to combine heterogeneous data distributed across the globe is critically important to boost research on rare diseases, but it presents a number of methodological, representational and automation challenges. In this scenario, biomedical ontologies are of critical importance for enabling computers to aid in information retrieval and analysis across data collections.
This chapter presents an approach to preparing rare disease data for integration through the application of a global standard for computer-readable data and knowledge. This includes the use of common data elements, ontological codes and computer-readable data. This approach was developed under a number of domain-relevant requirements, such as controlled access to data, independence of the original sources, and the desire to combining the data sources with other computational workflows and data platforms.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Rare diseases present a driving use-case for the development of methods that help to efficiently combine data from disparate and dispersed resources (clinical and physiological data such as blood pressure and phenotype; molecular data such as gene expression and genotype; biobank data; and model organism/disease model information). The ability to do this efficiently with data distributed across the globe is critically important to boost research on rare diseases (RD).
Correctly combining data from disparate, heterogeneous sources presents a number of challenges that broadly split into three types: methodological challenges, representational challenges, and automation challenges.
With respect to methodological challenges, these generally relate to the act of gathering the original data. For example, what measurements were performed and how? Were the same methods or instruments used in all locations? Do instruments share identical calibrations? Were survey results collected using the same questions? If measurements were not exactly the same, at what level may they still be compared? For instance, if smoking habits were measured differently, is there a unifying measure of smoking that the datasets can be mapped-to for comparison?.
Representational challenges relate mainly to the data’s “transparency” and encoding. For example, is it clear what data from each source is, in fact, comparable? Which spreadsheet columns contain which type of data? If a clinical coding system is used, is that same coding system used by both datasets? For example, can a data analyst be absolutely sure that a ‘2’ under the column header ‘smoking habit score’ in one data file is equivalent to the ‘2’ in another data file under the header ‘smoking score’? This may seem trivial, but is a source of many errors. Data analysts lose a lot of time correcting mistakes and redoing analyses because they misinterpreted the meaning of the data in disparate datasets. It is important to see that if the encoding between data sets is ambiguous, the harmonization of data gathering methods is rendered futile.
The methodological challenge and the representational challenge are both aspects that relate to data quality, and high-quality data will meet both of these challenges. We might use the Orphanet database as an example. Orphanet is curated according to a set of Standard Operating Procedures (SOPs) to ensure optimal and consistent quality of its data about rare diseases [13, 15]. These SOPs address both the methodological and the representational challenge. However, if Orphanet had not focused on the representational challenge, and its curators had chosen to use French disease names to represent diseases in their database, then the data would be nearly unusable for many data scientists. Orphanet addressed this representational challenge by providing orphacodes linked to the Orphanet Rare Disease Ontology (ORDO) to uniquely identify diseases for applications across the globe. Thus, this database is both methodologically rigorous, as well as representationally transparent, and as such, is highly reusable by other researchers.
The third challenge relates to the need to combine numerous data sets. To achieve the scale of data integration required by the rare disease case, the number of datasets that must be interpreted and parsed quickly scales beyond the ability for manual manipulation. In that case we need computers to ‘know’ what the structures and values in the data represent, in order to combine them correctly. For example, a row in a table with motor score, phenotype, and gene expression, does not explicitly state how motor score, phenotype, and gene expression are related to a person and to each other. This may be obvious when an expert inspects a table, and a data analyst can ask the expert who drafted a table, but that is too time consuming and error-prone for more than two or three data sets. Achieving clarity on what data means for both humans and computers is therefore a critical challenge in speed and quality-control in rare disease research. Lack of such clarity can even entirely block the reuse of the data if the person who created and managed a data set is no longer available for assistance. As such, this third challenge requires that the data be computer readable (structurally) and computer interpretable (semantically). It extends the representational challenge by requiring that all data and their interrelationships are available in a form that conforms to a global framework for data linking.
Fortunately, the technology experts of the World Wide Web have had to address this challenge before and created such a framework: the Resource Description Framework (RDF). This framework enables, for example, linkage of the information in a specialized registry on ring-14 syndrome in Italy to the curated information in Orphanet in Paris, and to relevant biobank information stored in Graz. This occurs when all three sites use the Web address of the code for ring-14 disease, defined by Orphanet. Sharing common codes, based on their Web addresses, also referred to by as Uniform Resource Locators (URLs), enables a study on the symptoms of epileptic attacks across all three data sources without the need to explicitly coordinate between them. In this way, we ‘virtually augment’ the potentially sparse ring-14 data in the specialized registry with the highly curated and detailed information in two remote knowledge bases. We note that in practice, this layer of interoperability is often added as a complement to a more local data representation. It is also important to point out that RDF reuses Web technology, but without any implication that this makes data open or public. Data encoded by URLs is still data, and is as safely stored as it was without URLs.
It is our observation that while the first challenge is well-understood by the rare disease researcher or registry/biobank host, and the second challenge is becoming increasingly recognized as ‘best practice’ by this community, the third challenge poses problems that are unfamiliar to rare disease domain experts. Nevertheless, the interlinking between related Web data and knowledge resources, and the ‘virtual augmentation’ that results, ensures that each participating data host is maximally useful, both for their local users, and for the broader rare disease research community. As such, we have worked with the rare disease community to establish some guidelines and workflows that will simplify this third challenge, hopefully to the point that the data hosts are comfortable undertaking this challenge on their own.
In summary, in this chapter we present an approach to prepare data for integration by enabling rare disease data to be exchanged on the basis of a global standard for computer-readable knowledge and data. We explain how this enables cross-resource research and creation of a robust infrastructure of rare disease data resources that are Findable, Accessible, Interoperable, and Reusable for humans and computers – FAIR [24] – at the source.
2 The Bio-ontology Forest
Ontologies play an important role in the scenario described above. ‘Ontology’ is an ancient concept in philosophy that has been adopted by computer scientists to describe a particular approach to making knowledge computer-readable. Real-world concepts are represented by a concept hierarchy where each concept is called a “class” and the subclasses – those further down the hierarchy – become increasingly more specific (e.g. humans are a more specific subclass of mammals). It is a best practice to publish ontologies that cover a specific part of reality. For instance, the Human Phenotype Ontology (HPO) covers only human phenotypes. As such, there are numerous ontologies; effectively, one for every top-level concept in the domain. For example, in the rare disease domain there would be ontologies describing disease symptoms, genetics, hospital staff, diagnostic equipment, etcetera. Things in the real-world – for example, individual researchers, or individual pieces of equipment, are called “instances” of these classes. Properties (also referred to as relations or predicates) describe how instances of these classes relate to each other. For example: one of the authors of this chapter is an instance of the class ‘Researcher’ and has a relation ‘hasSurname’ with ‘Roos’, which is an instance of the class ‘Family Name’. Thus, a machine could, without human intervention, find these two instances in the database, and know that one instance is a ‘Researcher’, and that the researcher has the family name ‘Roos’. A full record of the researcher ‘Roos’ would, therefore, have facets encoded by a wide range of ontologies, spanning multiple kinds of data such as medical history, address information, and various identification numbers. Globally defined and shared properties enable these ontologies to be unambiguously connected, such that a functionally interlinked knowledge network can arise. It is important to realize that the current consensus is that an ontology should cover a facet of reality in depth, and be linkable to other ontologies to cover the breadth of an application. For example, it makes little sense to expect concepts for drugs or genes in HPO, as they are not human phenotypes. Thus, so-called ‘application ontologies’ or ‘semantic archetypes’ select a subset of terms and properties from a number of ontologies to cover the breadth of an application [9].
Numerous ontologies already exist for the biomedical community. Although general search engines such as Google may be used to create a list of existing biomedical ontologies, the easiest way to locate them is the use of public ontology repositories. Ontology repositories are usually more specific than search engines and they offer tools that may be focused on the type of applications the repository was designed for. The leading repository of biomedical ontologies is the BioPortal (http://bioportal.bioontology.org/) [23], developed by the National Center for Biomedical Ontology (NCBO), which is one of the National Centers for Biomedical Computing funded under the NIH Roadmap Initiative. BioPortal provides access to a library of biomedical ontologies and terminologies via the NCBO services. Ontologies from a number of different groups are published in BioPortal, including the Consultative Group on International Agriculture Research, the Open Biomedical Ontologies (OBO) Foundry (http://www.obofoundry.org) [20], the WHO Family of International Classifications, the Cancer Biomedical Informatics Grid, the Proteomics Standards Initiative, the Clinical and Translational Science Awards, the Biodiversity Information Standards and the Unified Medical Language System. The Web services allow multi-layered access to the ontology content, spanning functionality such as getting all terms in a ontology to retrieving the definition of a single term.
Two of the most important domains of ontology for RD clinical medicine and research are those defining phenotypic or clinical features (signs, symptoms, and findings of diseases), and ontologies defining specific disease classifications or groups. Beyond these critical core ontologies, additional ontologies and standards will be required for various RD data repositories depending on their data collection process, potentially including ontologies or standards for mutation nomenclature, biobanking, clinical trials, natural history, as well as for RD medications and treatments [4].
Given the large number of ontologies which currently exist, and given that RD data hosts will generally lack experience in exploring ontologies and selecting terms, it would be useful to highlight a set of reference ontologies to facilitate the selection of ontological codes to use in the registry/biobank. The OBO Foundry is a collaborative experiment involving developers of science-based ontologies who are establishing a set of principles for ontology development, and creating a suite of reference ontologies in the biomedical domain. Ontology developers have agreed to work together on an evolving set of design principles that can foster interoperability between ontologies, and ensure a gradual improvement of quality and formal rigor, in ways designed to meet the increasing needs of data and information integration in the biomedical domain. The OBO Foundry also works to minimize overlap and redundancy between ontologies, encouraging members to share and reuse terminologies within their specialist domains, rather than creating new, but redundant ontological classes. In so doing, there is community convergence on a single reference ontology that already assists in finding and selecting the best ontological term. Nevertheless, it would be useful to undertake an additional filtering step to more precisely define the optimal ontologies for the rare disease domain. This is an area of active investigation in this field. For example, we propose to share ‘semantic archetypes’: small models comprised of terms and properties from different ontologies that are selected by ontology experts for encoding a specific set of related data elements, such as for the data gathered by a case report form [17].
3 Preparing Data at the Source for Analysis Across Resources
Preparing data for integration can be viewed from different perspectives. For instance, health professionals may see this as a matter of harmonizing operating procedures and/or clinical measurement protocols (the methodological challenge), while IT (“Information Technology”) professionals may wish to agree on data elements and their exchange format (the representational challenge). Attention to both of these is critical for accurate integration, but here we will focus on the latter, as the former perspective is best managed by health experts.
From the IT perspective, we divide the problem into three distinct considerations, according to the aforementioned challenges: (i) what is measured or observed and how (methodological challenge), (ii) how measurements (observations) are encoded in data collections (representational challenge), (iii) how we make data computer-readable (automation challenge). We note that these three considerations pertain only to preparing data for integration. Downstream analyses will likely require additional data transformations (e.g. R will require data in the form of R data frames for statistical analysis); however, analyses can often not begin until the data from multiple sites has been accurately located, retrieved, and integrated, so that is our focus in this chapter. We also note that the considerations are, in effect, hierarchical, and we will present them as such.
3.1 Consideration 1: Consensus on Common Data Elements
It is typical for specialist communities to reach consensus on what should be measured and how, but the importance of this step cannot be understated. Deciding on common data elements (CDEs) across resources is mostly a social process, and is common practice in consortia that are formed to perform a large study, for instance a GWAS (Genome-Wide Association Study) consortium. It is the first step towards integrative analyses within the consortium for the duration that it is funded.
Consensus, however, has limitations with respect to reusing the data outside of the consortium and/or beyond its lifespan, which is usually coupled to a grant. For instance, if a consortium of cystic fibrosis researchers reaches consensus on measuring forced expiratory volume in 1 s (and how), this may differ from the consensus of measurements and methodology in a primary ciliary dyskinesia consortium. Nevertheless, comparison of these very similar diseases could lead to significant insights.
Striving for global consensus between all researchers in all domains to accommodate all future uses of data is unrealistic and overly rigid (different domains legitimately have different requirements). While lack of widespread consensus does limit the ease and power of cross-resource data comparisons and analyses, it does not thwart it completely. Applying the solution proposed in Consideration 2, below, mitigates this problem by moving the requirement from consensus to compromise with respect to the way that these common data elements are encoded. This will clearly be more acceptable, and therefore effective, than attempting to enforce a rigid set of common data elements that all resources must have.
3.2 Consideration 2: Ontological Encoding
Health research has a long history of the use of nosologies (classifications of diseases). Similarly, healthcare organizations use coding systems both for patient care as well as for billing and other administrative tasks. Biomedical ontologies are very similar to these familiar approaches to knowledge capture and classification, with the extension that contemporary ontologies utilize formal logics in their code definitions, and are thus able to be processed and interpreted by machines. Consideration 2, therefore, proposes the use of globally unique identifiers [10] and ontologies when exchanging data elements. For instance, when HPO identifiers are used as the codes for phenotypes in disparate disease databases, then phenotypic features in these databases can be unambiguously compared and, when commonalities are found, the data may be selected for integration. Resources in different countries may have used different terms or languages, but the agreement to use HPO codes as the unifying descriptor – the “Rosetta Stone” – can easily reveal that two entries are referring to the same concept, regardless of language. Ontologies, therefore, play a key role in rare disease data collections. They provide standard terms by which the common data element values can be compared. ‘HP:0002072’, the identification number for the concept which is, in English, called “chorea”, is the same in all resources that use the HPO to define phenotypes. One caveat remains: codes for phenotypes such as HPO codes are by themselves not necessarily uniquely identifiable across the globe if the codes do not conform to some globally defined schema. For instance, without the context of knowing that we are discussing diseases, we cannot tell if the string of characters “HP:0002072” refers to the HPO term for ‘chorea’ or perhaps to some Hewlett and Packard component number. This particular requirement is addressed in the next level of the hierarchy, Consideration 3. The technology that we add to ontological encoding enables data to be made unambiguous. The positive consequence of this is that, if a data element is unambiguous, and shared between multiple resources, it becomes unambiguously linkable with those resources, much like the shared keys between database tables. Thus, it eliminates the need/desire to explicitly combine data in one central warehouse separately from the sources, an undertaking that is costly in terms of finances, human effort, and risks to privacy.
3.3 Consideration 3: Machine Readable Data and Knowledge
This consideration pertains to making data, and the meaning of the data, computer-readable using a structured data representation model combined with a more formal approach to representing ontological (and other) concepts. The purpose is to enable computers to aid in combining data from multiple rare disease resources across the globe.
To prepare data for integration at the source, we advise the framework that is recommended by the Semantic Web initiative and the ‘Linked Data’ principles – Resource Description Framework (RDF). Both of these integrative initiatives reuse the core technology that underlies the World Wide Web itself (i.e. the HTTP protocol). The use of RDF together with HTTP allows machines to “surf” the Web in a meaningful way; much like how grammatical rules define how words can be assembled into meaningful sentences, RDF explains how to structure ontological concepts, and other entities such as individual patients and their specific interventions or treatments, into relationships whose meaning can then be interpreted by software. This requires, simply, that all aforementioned codes (for specific phenotypes, diseases, genes, etc.), but also data types such as the general class ‘Human phenotype’ for all human phenotypes, patient identifiers, and relation types such as ‘binds to’, are represented by a Uniform Resource Identifier (URI). Biologically and clinically meaningful statements are then constructed using “Triples” of URIs. For example, in RDF ‘chorea is-a-manifestation-of Huntington’s Disease’, becomes an unambiguous statement – a Triple – understood by both humans and machines, because each element of that Triple is represented as a URI, and all parties, globally, use the same URI to refer to the same concept or relationship. If the ontological concepts and relationships within these “sentences” are further formalized in description logics, they can be even more powerfully processed by computers, where, for example, a computer could automatically define the category for a new data entry, or could infer consequences from certain combinations of data points that were not explicitly entered into the database. Defining relations between data elements in terms of these Triples further mitigates the need for a rigid set of globally common data elements. The encoding by description logics allows any inferable commonality at any level to be exploited, instead of only the values of pre-defined common data elements. Nevertheless, it does not replace the solutions for Considerations 1 and 2. URIs and Triples of URIs only represent what researchers have decided to measure, encode and define relations between, such that computers can help to perform accurate analysis across any number of data sets. The stack of solutions is most powerful when all three levels are addressed.
4 Requirements for Preparing Rare Disease Data for Integration
We constrain our pursuit of an integrative solution by the following requirements and desiderata [17]:
-
1.
When access to data is granted, ‘linkable data’ must be trivial to query and/or analyze across (large numbers of) independent data sources, by both humans and machines.
-
2.
All originating sources must retain their independence; i.e. the solution-space cannot depend on centralized data warehouses or portals.
-
3.
Data sources should be easily combined with existing computational workflows and data platforms such as those developed by the RD-Connect project [21]. The solution should avoid proprietary or de novo interfaces and formats (data silos).
-
4.
The technology that we propose to make rare disease data linkable should complement existing technologies and protocols being used at-source, and not interfere with them.
These desiderata and requirements impose certain challenges. The first requirement –the ability to dynamically integrate large numbers of potentially linkable resources- poses significant demands on the knowledge representation that we apply, confirming the aforementioned representation and automation challenges. Effectively, at larger scales, human assessment of the meaning of the data in each of the resources should not (and cannot) be required. The second desiderata, that all sources should remain independent, does not exclude the use of global services to facilitate data integration scenarios, such as initiatives that make it easier to find and access registries and biobanks through creating centralized indexes [7]. It does, however, exclude the wholesale warehousing and en masse integration of the data, as has been the norm in the biomedical domain for many years, in lieu of retaining the data at its original source.
We point-out, in addition, that these requirements surpass simply finding data. Making data discoverable is often considered lower-hanging fruit, because it requires only the information about the data source in a standard form (‘metadata’). Examples are the disease that a data set pertains to, how many subjects it contains, the type of material that was collected, etcetera. Our driving research questions, however, require more than information about data. For instance, finding tissue samples of patients with ring-formation in chromosome 14 (the defining feature of ring-14 syndrome) requires interrogation of the specific karyotype of a patient, which goes beyond simply knowing that karyotype information was collected. Furthermore, we need to enable researchers to exploit relevant biomedical information. For example, information associated with the ring-14 karyotype may be the link to rich sets of information about model systems that researchers can exploit to find new treatments for the disease.
5 Backbone: Linkable Data and Ontologies
The backbone for our approach to make data linkable and computer readable at the source is, as we noted in Consideration 3, provided by the recommendations of the World Wide Web Consortium (W3C): Linked Data principles [1], Ontologies, and the Resource Description Framework (RDF). RDF is a generic data model that was designed with the objective of creating qualified networks of data, upon which increasingly complex domain models can be overlaid to assist with interpretation of that data. For instance, the Human Phenotype Ontology and the Orphanet Rare Disease Ontology are available in RDF, as are most ontologies in the biomedical domain. We therefore consider this the best way to facilitate integrative biological and translational research across rare disease resources. In addition to tools that exploit the use of ontologies, such as the Exomizer [19], MatchMaker Exchange (MME; [14]), and Monarch [11], we see an increasing amount of life science data resources that use RDF to support data linking, such as the RDF platform of the European Bioinformatics Institute (EBI; [6]) and the Open Phacts [25]. RDF is capable of representing disease specimen identifiers, patient/disease personal and clinical information, and molecular data, thus the choice of this singular technological framework helps reduce the overall cost of data integration for rare disease resources.
6 Building on the Backbone: A Reference Model for Data Integration
The process to prepare data for analysis across resources entails recoding values by ontology codes, adding ontology terms to describe the meaning of values, and adding relation terms (also from ontologies) to define how values are related and what they represent. This is not a trivial process. While many ontologies exist in the biomedical domain, choosing the appropriate ontology terms requires substantial understanding of ontologies, and substantial understanding of what the data represents. We recommend consulting an ontology expert to collaboratively choose the correct terms. However, this in itself does not guarantee that the same ontology terms will be used by all resource providers. There are often multiple ontologies that appear to have appropriate, even identical terms. Moreover, to increase efficiency for the large amount of data resources in the rare disease domain, it is important that we can reuse the ontology choices of one resource for other resources with similar data.
To mitigate these issues, our approach entails the development of reusable reference models for data integration (‘semantic archetypes’) that are composed of terms from recommended ontologies. These models differ from typical ontologies in that their purpose is to provide a common schema for multiple types of data for a particular application, not to conceptualize a particular part of reality. Publishing these semantic archetypes, for instance via FAIRsharing.org, allows reuse of previous effort and thereby stimulate greater commonality between ontology-based data sets.
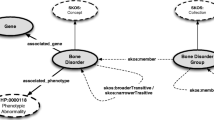
As an example, we have created a first version of a semantic archetype for a subset of identifier types in rare disease databases for the purpose of enabling answering questions across patient registries and biobanks. We constructed the model as a stack of modules to cater for increasingly complex applications of the archetype (Fig. 9.1; [17]). The model and our selection of ontologies can subsequently serve as reference for new cases that involve similar data.
Semantic archetype for rare disease data integration. The model is constructed hierarchically from modules that can be used for increasingly complex cases. From bottom to top: ‘Values’ represent data in multiple resources; ‘rdc-core’ provides simple classes for database identifiers; ‘rdc-meta’ supplies immediately relevant classes and properties to denote the meaning of identifiers and their interlinks; ‘rdc-meta-extended’ provides logical definitions from the reused ontologies as needed for computational reasoning; the top semi-circle represents the ‘foundational ontologies’ that the reused ontologies refer to (they are not directly part of the semantic archetype)
7 Composition of the Prototype Reference Model
The starting point for crafting the semantic reference model was to list the core set of identifiers that will likely exist in RD registries/biobanks (the dark grey semicircle at the center of Fig. 9.1). These are:
-
Biobanks
-
Patients
-
Sample donors
-
Experiments
-
Samples (biological specimens)
The next task (‘rdc-meta’ in Fig. 9.1) was to provide a model that describes the meaning of these identifiers and their interrelationships in computer readable terms. The following ontologies contain classes that could be used to add meaning to the kinds of identifiers above:
Ontology for BIoBanking (OBIB; [2]):
-
Human being
-
Patient/donor role
-
Identifier
-
Object properties
Open Archives Initiative Object Reuse and Exchange (OAI-ORE; [8]):
-
Aggregation
-
Aggregate properties
EMBRACE Data and Models, an ontology of bioinformatics operations, types of data, data identifiers, data formats, and topics (EDAM*; [5]):
-
Specific types of identifiers (e.g. biobank ID, stock accession ID, person ID)
-
Standard terms for genes, proteins, DNA, and other biological entities
-
Standard terms for analytical methodologies
Information Artefact Ontology (IAO*; [3]):
-
Specific types of identifiers (e.g. biobank ID, stock accession ID, person ID)
* EDAM and IAO both provide an identifier class. Including them both in the semantic archetype increases the reusability of the model. While EDAM is widely used, IAO provides the convenient link to the OBO Foundry suite of ontologies.
Dublin Core ontology (DC; [22]):
-
Identifier properties
-
Authorship and other contact information
-
Basic descriptive information
Simple Knowledge Organization System (SKOS; [12]):
-
Mappings (for instance, to SNOMED terms)
From these ontologies, the following semantic modules were created (see the layers in Fig. 9.1):
-
1.
rdc-core: the minimal set of classes and properties to map to the data in the sources. Because of the task at hand the focus is on identifiers. Rdc-core represents little more that the lowest level types of the identifiers.
-
2.
rdc-meta: the minimal semantic model, defined as much as possible using the aforementioned ontologies (Fig. 9.2). Ontology experts will note that this module lacks the complete set of logical definitions (so-called axioms) to be able to use the concepts.
Fig. 9.2 Semantic archetype for enabling questions across registries and biobanks (the class hierarchy). The call-outs indicate the ontologies from which the classes were used. The complete ORE can be found on https://www.openarchives.org/ore/. The complete versions of the other ontologies can be found on http://bioportal.bioontology.org/
-
3.
rdc-meta-extended: this module includes the axioms and the extra subclasses and properties that are required to reason over the semantic archetype if and when required by computational scientists [18].
These modules (and others currently under construction) provide support for the stepwise migration of data in RD registries/biobanks. Each module provides a constrained set of ontological choices, based on the task-at-hand, and on the most prevalent data types encountered in RD data repositories. For example, in Fig. 9.2, “Phenotips patient ID” is one of only six options provided for the data-type “Identifier”; however, the original ontology from which these six options were derived (EDAM) has many dozens of additional options. We believe that constraining the choices to only a few possibilities specific for RD data sources will dramatically ease the burden of making RD data interoperable. We hope that, with proper tools, we can arrive at the point where RD registry/biobank owners can undertake this task without expert assistance.
8 Summary
What we present here is a general approach for preparing data for integration that enables to address the current driving research questions, but also future applications beyond the scope of a single project. Compared to projects where, for instance, data is prepared for integrative analysis in R or SPSS, it adds an intermediate step. This is undeniably extra work, but it makes the harmonization effort of a project reusable. It quickly becomes the more efficient approach when we desire data collections to be used many times, realizing that without preparation at the source, the harmonization step is carried out by each user of the data again and again with high risk of errors.
Ontologies are of critical importance for enabling computers to aid in information retrieval and analysis across data collections. They play a key role in speeding up the overall research process towards better understanding of a disease, new treatments, and diagnostic biomarkers.
Linked Data with strong ontological underpinnings, and a clear model for achieving proper access control, is our first ambition for preparing the relatively small, but numerous and disparate, rare disease data sets for wide-scale data integration. Sharing and reusing semantic archetypes developed by ontology experts mitigates an immediate and major bottleneck: the current sparsity of expertise in the community to make informed decisions about which ontological concepts to use for their data annotations. Searching for a concept, e.g. in NCBO’s bioportal or EBI’s ontology lookup service, typically returns too many “hits” for a non-ontologist to choose-from. Specific ontologies may be advised by experts, but the breadth of data types across data sets is large. For example, in a recent workshop [16] organized for RD patient registries owners and computer experts, we could easily list at least 10 ontologies relevant for just a subset of a registry’s data, and not all of these are included in the BioPortal or EBI search services. Here, we propose finding a middle-ground and providing an early workflow towards that goal. Domain experts first select a subset of the most appropriate and common ontological classes used for each of the data types encountered in a rare disease resource that we need to make FAIR, such as for the data types of a typical rare disease registry. Only these limited (but relevant) options are presented to the data curator, in a stepwise, and contextually-sensitive manner, as they undertake their data transformation.
References
Bizer C, Heath T, Berners-Lee T (2009) Linked data-the story so far. Int J Semant Web Inf Syst 5(3):1–22
Brochhausen M, Zheng J, Birtwell D, Williams H, Masci AM et al (2016) OBIB-a novel ontology for biobanking. J Biomed Semant 7(May):23
Ceusters W (2012) An information artifact ontology perspective on data collections and associated representational artifacts. Stud Health Technol Inform 180:68–72
International Rare Disease Research Consortium (IRDiRC) Policies and Guidelines, Long version (2013). Available from: http://www.irdirc.org/wp-content/uploads/2013/06/IRDiRC_policies_24MayApr2013.pdf. Accessed Dec 2016
Ison J, Kalas M, Jonassen I, Bolser D, Uludag M et al (2013) EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics 29(10):1325–1332
Jupp S, Malone J, Bolleman J, Brandizi M, Davies M et al (2014) The EBI RDF platform: linked open data for the life sciences. Bioinformatics 30(9):1338–1339
López E, Thompson R, Gainotti S, Wang M, Rubinstein Y et al (2016) Overview of existing initiatives to develop and improve access and data sharing in rare disease registries and biobanks worldwide. Expert Opin Orphan Drugs 4(7):729–739
Lynch C, Parastatidis S, Jacobs N, Van de Sompel H, Lagoze C (2007) The OAI-ORE Effort: Progress, Challenges, Synergies. Proceedings of the 7th ACM/IEEE-CS joint conference on digital libraries 80-80
Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J et al (2010) Modeling sample variables with an experimental factor ontology. Bioinformatics 26(8):1112–1118
McMurry J, Blomberg N, Burdett T, Conte N, Dumontier M et al (2015) 10 Simple rules for design, provision, and reuse of identifiers for web-based life science data. Zenodo. Available from: https://doi.org/10.5281/zenodo.31765. Accessed Dec 2016
McMurry J, Köhler S, Washington NL, Balhoff JP, Borromeo C et al (2016) Navigating the phenotype frontier: the monarch initiative. Genetics 203(4):1491–1495
Miles A, Bechhofer S (2009) SKOS Simple Knowledge Organization System Reference. World Wide Web Consortium. Available from: http://www.w3.org/TR/skos-reference/. Accessed Dec 2016
Orphanet Standard Operating Procedures, Version 02.1 (2016) Available from: http://www.orpha.net/orphacom/special/eproc_SOPs_V2.pdf. Accessed Dec 2016
Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA et al (2015) The matchmaker exchange: a platform for rare disease gene discovery. Hum Mutat 36(10):915–921
Rath A, Olry A, Dhombres F, Brandt MM, Urbero B et al (2012) Representation of rare diseases in health information systems: the orphanet approach to serve a wide range of end users. Hum Mutat 33(5):803–808
RD-Connect “Bring Your Own Data (BYOD)” Workshop to Link Rare Disease Registries (September 29–30, 2016) National centre for rare diseases, Istituto Superiore di Sanità, Rome. Available from: http://www.iss.it/binary/cnmr4/cont/RD_Connect_BYOD_2016_preliminary_programme_rev12.07.2016.pdf. Accessed Dec 2016
Roos M, Wilkinson MD, Kaliyaperumal R, Thompson M, Carta C et al (2016) Registries of domain-relevant semantic reference models help bootstrap interoperability in domains with fragmented data resources. Proceedings of the 9th International Semantic Web Applications and Tools for the Life Sciences (SWAT4LS) Conference. Available from: http://www.swat4ls.org/wp-content/uploads/2016/10/paper-16.pdf. Accessed Dec 2016
Samadian S, McManus B, Wilkinson MD (2012) Extending and encoding existing biological terminologies and datasets for use in the reasoned semantic web. J Biomed Semant 3(1):6
Smedley D, Jacobsen JOB, Jäger M, Köhler S, Holtgrewe M et al (2015) Next-generation diagnostics and disease-gene discovery with the exomiser. Nat Protoc 10:2004–2015
Smith B, Ashburner M, Rosse C, Bard J, Bug W et al (2007) The OBO foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25(11):1251–1255
Thompson R, Johnston L, Taruscio D, Monaco L, Béroud C et al (2014) RD-Connect: an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J Gen Intern Med 29(S3):S780–S787
Weibel S, Kunze J, Lagoze C, Wolf M (1998) Dublin core metadata for resource discovery. Available from: http://www.rfc-editor.org/info/rfc2413. Accessed: Dec 2016
Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C et al (2011) BioPortal: enhanced functionality via new web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res 39(Web Server issue):W541–W545
Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M et al (2016) The FAIR guiding principles for scientific data management and stewardship. Sci Data 3(March):1600018
Williams AJ, Harland L, Groth P, Pettifer S, Chichester C et al (2012) Open PHACTS: semantic interoperability for drug discovery. Drug Discov Today 17(21–22):1188–1198
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Roos, M., López Martin, E., Wilkinson, M.D. (2017). Preparing Data at the Source to Foster Interoperability across Rare Disease Resources. In: Posada de la Paz, M., Taruscio, D., Groft, S. (eds) Rare Diseases Epidemiology: Update and Overview. Advances in Experimental Medicine and Biology, vol 1031. Springer, Cham. https://doi.org/10.1007/978-3-319-67144-4_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-67144-4_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-67142-0
Online ISBN: 978-3-319-67144-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)