Abstract
The human brain is intrinsically a high-dimensional and multi-variant system. Simultaneous EEG and fMRI recoding offers a powerful tool to examine the electrical activity of neuron populations at high temporal and frequency resolution, and concurrent blood oxygen level dependent (BOLD) responses at high spatial resolution. Joint analysis of EEG and fMRI data could thus help comprehensively understand the brain function at multiple scales. Such analysis, however, is challenging due to the limited knowledge on the coupling principle of neuronal-electrical and hemodynamic responses. A rich body of works have been done to model EEG-fMRI data during event related design and resting state, while few have explored concurrent data during natural stimulus due to the complexity of both stimulus and response. In this paper, we propose a novel method based on N-way decomposition to jointly analyze simultaneous EEG and fMRI data during natural movie viewing. Briefly, a 4-way tensor from the EEG data, constructed in four dimensions of time, frequency, channel and subject, is decomposed into group-wise rank one components by canonical polyadic decomposition (CPD). We then used the decomposed temporal features to constrain a 2-way sparse decomposition of fMRI data for network detection. Our results showed that the proposed method could effectively decode meaningful brain activity from both modalities and link EEG multi-way features with fMRI functional networks.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Simultaneous
- EEG
- fMRI
- Natural stimulus
- Joint analysis
- Canonical polyadic decomposition
- Sparse decomposition
1 Introduction
The human brain is a complex system that involves massive electrical and metabolic processes, which is high-dimensional and multi-variant [1]. We now could record brain signals using EEG and fMRI simultaneously, which opened a new window to model the brain processes across multiple dimensions [2]. EEG records the massive neuronal-electrical activities that are reflected on the scalp, the high sampling rate of which enables the multi-way time-frequency-space feature extraction [2,3,4]. In contrast, the fMRI imaging captures the hemodynamic response of brain with low temporal resolution but high spatial resolution, which could be modeled as functional networks on brain structures [1,2,3,4]. The neuronal-electrical activities are believed to couple with blood oxygen level changes [2], so joint analysis of both modalities could link the two processes, provide comprehensive understanding of brain function, and benefit decoding the pathology of some neurology diseases, like epilepsy.
However, joint modeling of EEG and fMRI data is challenging because even though the two modalities are derived from the same brain activity, the coupling principle of them is poorly understood [2,3,4]. While many studies have developed joint analytical strategies for event-related design and resting state paradigm [4], few methods have been reported for analyzing simultaneous EEG and fMRI data during natural stimulus [5]. In recent years, the use of natural stimuli becomes increasingly popular in neuroscience research [4, 5], therefore, methodological innovation is urgently needed in this field to fuse EEG and fMRI activity during such complexity of stimulus and response.
In this paper, we proposed a novel approach based on N-way decomposition towards linking concurrent EEG and fMRI data analysis. Our method includes 4-way canonical polyadic decomposition (CPD) [6] on EEG data and 2-way sparse decomposition [7] on fMRI data. The EEG data was first transformed into time-frequency domain [8]. Considering electrode channel and subject as two additional dimensions, we organize the group-wise EEG data as a 4D tensor. Using the 4-way CPD, the tensor is decomposed into numbers of rank one components [6] which have features of time, frequency, channel topology and subject contribution. Then the temporal features convolved by hemodynamic function (HRF) serve as explanatory variables for the 2-way sparse decomposition of fMRI data, which reconstructs functional networks. Since the two decomposition share the same temporal feature, the EEG frequency and spatial topology features are associated with fMRI networks. Our method was applied to a continuous movie watching dataset, and meaningful features are linked between EEG and fMRI.
2 Materials and Method
2.1 Overview
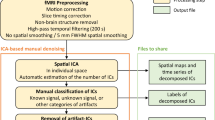
The framework of the proposed method is summarized in Fig. 1. The whole method includes 4-way CPD decomposition on EEG data and 2-way sparse decomposition on the fMRI data. First, the EEG data from each subject was transformed into the time-frequency domain (t and f), and the spatial topology of the channels are preserved in the dimension “c” as shown in Fig. 1a. Since the data records the responses of multiple subjects to the same movie, we assume the fourth dimension “s” to account for the similarity and variability of subjects. The 4D tensor was decomposed into R rank one components (Fig. 1b), or four factor matrices (Fig. 1c). We call them R components from the EEG data. Each component includes four factors – \( t_{i} \), \( c_{i} \), \( f_{i} \) and \( s_{i} \), which show features in different domains and are associated with each other. For example, in Fig. 1h and i, each topological distribution is associated with one frequency power spectrum density (PSD) histogram. The most important factor is the temporal one, as it serves as a bridge to link the fMRI data. The signals in the T matrix of Fig. 1c was convolved with HRF and become E matrix, which constructs the explanatory variables to model fMRI networks. In Fig. 1d, the whole brain signals of each subject are organized into a 2D matrix \( Y_{j} \), which was further decomposed by the E matrix with sparse constraint and a corresponding coefficient matrix \( A_{j} \) is optimized. By repeating the decomposition for each subject and performing T-test on the coefficient matrices across subjects, we reconstruct functional networks at the group level (Fig. 1e and g). As these networks are modeled based on the T factor from EEG data, they inherit the association with the EEG topological distribution and frequency PSD, therefore, our method effectively links EEG features and fMRI networks.
2.2 Data Acquisition and Preprocessing
Twenty right-handed volunteers (mean age 27 ± 2.7) were recruited from University of Queensland. The study was approved by human ethics research committee of the University, in which simultaneous EEG and fMRI data was acquired during resting state (8 min) and passive viewing of a movie named “The Butterfly Circus” (20 min).
Functional MRI images were acquired from a 3T Siemens Trio scanner equipped with a 12-channel head coil. The acquisition parameters are: TR = 2200 ms, TE = 30 ms, FA = 79°, FOV of 134 × 134 mm, 64 × 64 acquisition matrix, 44 axial slices, and 3 × 3 × 3 mm3 voxel resolution. fMRI data was preprocessed using SPM12 (http://www.fil.ion.ucl.ac.uk/spm/), which include slice-timing and motion correction, temporal filtering (0.0085–0.15 Hz), spatial smoothing and registration to MNI space.
The EEG data were acquired using an MR-compatible EEG system (BrainProducts, Germany) with 64 scalp channels positioned according to the international 10–20 system. The signals were recorded at a sampling rate of 5000 Hz, and filtered online via a low-pass hardware filter at 250 Hz. The BrainProducts SyncBox ensured the EEG system were synchronized precisely to the MRI system. The EEG data was pre-processed using the EEGLAB toolbox [8] and the preprocessing include gradient-switching artifacts correction, head motion removal, band-pass filtering (0.5–45 Hz), down sampling to 500 Hz, ballistocardiogram artifacts and eye movement artifacts removal [8].
2.3 EEG Time-Frequency Transformation
Dynamic power spectrum density from the EEG data was calculated during the whole movie watching in the following way. First, the preprocessed EEG signals were divided into time windows with the length of TR (2.2 s) in fMRI. The EEG signals was normalized to zero mean and standard deviation of 1 in each window on each electrode. The power spectrum density (PSD) was estimated in each time window by Welch’s method [9] with the frequency resolution of 1 Hz. Inter-subject consistency analysis was performed on the PSD time series for each frequency interval and each electrode channel. The inter-subject consistency of the time-frequency decomposition provides a basis for the tensor decomposition, which will be discussed in Sect. 3.1.
2.4 Four-way Canonical Polyadic Decomposition on EEG
In Fig. 1, the group-wise EEG time-frequency data is organized into a 4-way tensor, \( X \in {\mathbb{R}}^{{d_{1} \times d_{2} \times d_{3} \times d_{4} }} \). The CPD aims to decomposed X into R rank one components [6]:
The operator \( \otimes \) denotes outer product of vectors. In each component, there are four factors, i.e., \( t_{i} \), \( f_{i} \), \( c_{i} \), and \( s_{i} \) correspondingly in time, frequency, channel and subject domain. Equation (1) could be rewritten into a tensor-matrix product form:
Where I is an identical tensor, \( W = [\lambda_{1} ,\lambda_{2} , \ldots \lambda_{R} ] \in {\mathbb{R}}^{R \times 1} \) is a weight vector to balance the contribution of normalized rank one tensors [6]. \( T \in {\mathbb{R}}^{{R \times d_{1} }} \), \( F \in {\mathbb{R}}^{{R \times d_{2} }} \), \( C \in {\mathbb{R}}^{{R \times d_{3} }} \), and \( S \in {\mathbb{R}}^{{R \times d_{4} }} \) are four factor matrices. The cost function is summarized as:
The advantage of CPD comparing with other type of tensor decomposition is that the solution is unique [6]. In this work, we employ the Tensorlab toolbox (www.tensorlab.net/) to estimate the CPD of EEG data. The toolbox provides various structure constraint for the CPD based on parametric transformations [6], and here regarding the characteristics of our data, we apply nonnegative constraint to matrix of \( W,T,F,C,S \). In addition, to make the decomposed features more distinct, and to decompose fMRI data effectively, we apply the orthogonal constraint on the \( T \) matrix.
2.5 Two-way FMRI Sparse Decomposition and Statistics
The T matrix from Sect. 2.4 is composed of group-wise temporal features from the EEG data, which share the same time resolution with the fMRI data as illustrated in Sect. 2.3. To deal with the lag in the fMRI signal comparing with EEG electrical activity, we convolve the HRF (h(t)) to the signals in T to construct the explanatory variables E, which is used to decompose the fMRI signal matrices. Here, for the i-th signal: \( E_{i} (t) = T_{i} (t) \star h(\tau ) \), where “\( \star \)” denotes convolution.
The 2D fMRI signal matrix \( Y_{j} \) of each subject (j = 1,2…s) is then decomposed by E. A L1 sparse penalty is introduced in the optimization to avoid overfitting and improve the feature selection. So, the final function to optimize is:
This sparse coding problem [7] can be solved by using the SPAMS toolbox [7]. Coefficient matrix \( A_{j} \) is then optimized for each subject, j = 1,2…s (j: subject ID). Two tailed t-test was applied on \( A_{j} \) across subjects, which derive R functional networks with group significance. Null hypothesis was rejected with p value threshold of 0.05.
2.6 Feature Selection
So far, we have linked multi-way features from EEG data and fMRI data, i.e., association is built up among components in the domains of time, frequency, scalp topology and fMRI networks. However, there are various sources of noises in both EEG and fMRI data, therefore, not all the components from our analysis are meaningful. Thus, feature selection is essential. We used the following steps to eliminate artifact or noise components: (1) We performed non-zero hypothesis test for the subject factor matrix S from EEG, and the components without significant non-zero mean were removed; (2) The components whose time course in the T matrix with abnormal fluctuation, like with one single peak in ten times intensity were removed; (3) Components with no significant fMRI networks were removed; (4) Components were removed if topological distribution was not smooth. These four steps combined thus selected meaningful results.
3 Results
3.1 Inter-subject Consistency Analysis for EEG Time-Frequency Data
The CPD decomposition is usually based on low rank assumption, i.e., there are a lot of repeated information in the data [6]. To test this low-rank assumption across subjects, we examined inter-subject consistency in the EEG data. Here we employed Cronbach’s α [10] as a measurement of consistency. In our experiment, the raw EEG signals during natural stimulus show surprisingly low inter-subject consistency. Only after transforming the data into time-frequency domain, we found significant consistency amongst many electrodes and frequency bands (Alpah: 7–14 Hz, Beta: 16–28 Hz and Gamma: 29–40 Hz) during natural stimulus as shown in Fig. 2b. As a comparison, inter-subject consistency from resting state activity is very low (Fig. 2a). Therefore, it’s feasible to apply 4-way CPD to the time-frequency EEG data during natural stimulus.
3.2 The Linked EEG Components and FMRI Networks
Our proposed method and feature selection steps in Sect. 2 were able to decode and associate meaningful EEG components and fMRI networks, as shown in Figs. 3, 4, 5 and 6. According to the pattern of the fMRI networks, we categorized the components into visual components (Fig. 3), auditory components (Fig. 4), attention components (Fig. 5) and others with interaction of multiple networks (Fig. 6). Interestingly, the EEG components within category show similar topology of electrode spatial patterns (Figs. 3a, 4a, 5a and 6a). As shown in Figs. 3b, 4b, 5b and 6b, these components are primarily related to Alpha band activity, and a few of them involve Beta band activity (C7–C9). The predominant presence of Alpha band reflects its roles in the attention and sensory processing [11], and evidently multiple sensory systems (visual& auditory) could possibly invoke Alpha oscillation [11].
It’s interesting that in C1–C4 (Fig. 3) different parts of visual cortex are associated with low, middle and high Alpha frequencies. Meanwhile, auditory networks in C5 and C6 (Fig. 2) are more associated with low Alpha frequencies. The attention networks in Fig. 3 focused in Alpha band but also involve certain level of Beta oscillation, which could be related to the role of Beta band in emotional and cognitive processes [12]. The networks from fMRI in Fig. 6 involve multiple networks, e.g. C10 with visual and auditory networks; C11 with auditory and attention networks; C12 with visual, auditory and attention networks. These components show evidence of interaction among visual, auditory and attention networks (Fig. 6) during natural stimulus. We didn’t find Gamma components with our method even though we see high inter-subject consistency in Fig. 2, possibly because of huge variability in Gamma activities. Note that, based on two tailed t-test activations in the fMRI networks were not further categorized as positive or negative, but treated as equally important in this work.
4 Discussion
In this paper, we proposed a joint analyzing method for concurrent EEG and fMRI data based on N-way decomposition. Experiment results have shown that our method could detect meaningful EEG components associated with fMRI networks. This work could open a new window to link two data modalities and provide a comprehensive way to understand brain function during natural stimulus. Future efforts will be devoted to further interpret the associated EEG components and fMRI networks.
References
Bullmore, E., Sporns, O.: Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10(3), 186–198 (2009)
Ullsperger, M., Debener, S.: Simultaneous EEG and fMRI: recording, analysis, and application. Oxford University Press, New York (2010)
Moosmann, M., et al.: Joint independent component analysis for simultaneous EEG–fMRI: principle and simulation. Int. J. Psychophysiol. 67(3), 212–221 (2008)
Huster, R.J., et al.: Methods for simultaneous EEG-fMRI: an introductory review. J. Neurosci. 32(18), 6053–6060 (2012)
Whittingstall, K., et al.: Integration of EEG source imaging and fMRI during continuous viewing of natural movies. Magn. Reson. Imaging 28(8), 1135–1142 (2010)
Sorber, L., et al.: Structured data fusion. IEEE J. Sel. Top. Sign. Process. 9(4), 586–600 (2015)
Mairal, J., et al.: Online learning for matrix factorization and sparse coding. J. Mach. Learn. Res. 11, 19–60 (2010)
Delorme, A., Makeig, S.: EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 134(1), 9–21 (2004)
Dumermuth, G., Molinari, L.: Spectral analysis of the EEG. Neuropsychobiology 17(1–2), 85–99 (1987)
Bland, J.M., Altman, D.G.: Statistics notes: Cronbach’s alpha. BMJ 314(7080), 572 (1997)
Foxe, J.J., Snyder, A.C.: The role of alpha-band brain oscillations as a sensory suppression mechanism during selective attention. Frontiers Psychol. 2, 154 (2011)
Rowland, N., et al.: EEG alpha activity reflects attentional demands, and beta activity reflects emotional and cognitive processes. Science 228(4700), 750–752 (1985)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Lv, J., Nguyen, V.T., van der Meer, J., Breakspear, M., Guo, C.C. (2017). N-way Decomposition: Towards Linking Concurrent EEG and fMRI Analysis During Natural Stimulus. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10433. Springer, Cham. https://doi.org/10.1007/978-3-319-66182-7_44
Download citation
DOI: https://doi.org/10.1007/978-3-319-66182-7_44
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66181-0
Online ISBN: 978-3-319-66182-7
eBook Packages: Computer ScienceComputer Science (R0)