Abstract
There are an increasing number of human disorders linked to defects in ribosome synthesis collectively known as ribosomopathies. Here we use the prototypical ribosomopathy, Diamond-Blackfan anemia, to explore relationships between the structure of the ribosome, its biogenesis, and the molecular mechanisms that contribute to disease pathology. Other ribosomopathies are discussed as they relate to the genes affected and pathophysiological mechanisms involved in Diamond-Blackfan anemia. The recent finding that several genes affecting ribosome biogenesis are somatically mutated in human tumors implies that understanding the molecular mechanisms underlying this rare group of disorders will likely have much broader implications.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
Ribosomopathies are generally thought of as diseases arising from defects in the process of ribosome biogenesis. Initially, the ribosomopathies showed an intriguing bias toward the inherited bone marrow failure syndromes, suggesting an unusual sensitivity of hematopoiesis to failures in the process of making ribosomes (Liu and Ellis 2006). As the field has continued to mature, a growing number of human diseases have been linked to apparent defects in manufacturing ribosomes. Many of these diseases are idiosyncratic in nature involving a single gene with distinct clinical sequelae. Recent reviews have outlined the growing list of ribosomopathies; and we would direct readers to such a review for an updated list (Yelick and Trainor 2015). Our focus here will be on the contributions of defects in ribosome synthesis to a subset of inherited bone marrow failure syndromes, with particular emphasis on Diamond-Blackfan anemia (DBA), the prototypical ribosomopathy. Other ribosomopathies will be discussed only in the context of how they may contribute to our understanding of the molecular underpinnings of the inherited bone marrow failure syndromes.
Ribosomes are large ribonucleoprotein assemblies engaged in protein synthesis. Many of the fundamental steps involved in peptide bond formation and the process by which the ribosome translocates along an mRNA in synthesizing a nascent polypeptide are thought to reside within the RNA components of the ribosome with the bulk of the ribosomal proteins involved making the process of protein synthesis more accurate and efficient through their interactions with ribosomal RNAs and various factors involved in protein synthesis. The human ribosome is composed of 4 rRNAs and 80 ribosomal proteins distributed between 2 subunits differing in size and shape. The smaller 40S ribosomal subunit is composed of 18S rRNA and 33 ribosomal proteins, whereas the 60S subunit consists of 3 rRNAs, 5S, 5.8S, and 28S, and 47 ribosomal proteins.
5.1.1 Ribosome Biogenesis
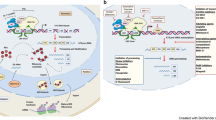
The process by which the 84 structural components of the ribosome assemble with one another to form the individual ribosomal subunits is complex and involves several hundred additional proteins and RNAs working together in a temporal fashion from primary rRNA transcripts in the nucleolus to mature functional subunits in the cytoplasm (de la Cruz et al. 2015). Three of the four rRNAs are transcribed together as a polycistronic RNA by RNA polymerase I. In addition to 18S, 5.8S, and 28S rRNAs, this primary transcript includes flanking sequences 5′ and 3′ to the mature rRNAs and two internal transcribed sequences. The mature rRNAs are liberated from this primary transcript through a series of endo and exonucleolytic cleavage reactions. These cleavages occur as substrates are made available as ribosomal proteins bind and fold pre-rRNAs in concert with a multitude of transiently associated factors that participate in subunit maturation. The fourth rRNA, 5S rRNA is transcribed by RNA polymerase III and forms a ribonucleoprotein (RNP) subcomplex with ribosomal proteins Rpl5(uL18) and Rpl11(uL5) facilitated by additional assembly factors. A number of additional factors contribute to the incorporations of the 5S subcomplex into pre-60S particles.
5.1.2 Ribosome Structure
The ribosome itself took shape before the divergence of the three major domains of life, eubacteria, archaebacteria, and eukaryotes. Somewhat surprisingly, in terms of phylogenetic sequence comparisons, the archaebacteria (which tend to live in extreme environments) and eukarya are more closely related to one another than either are to the more familiar eubacteria. The recent proposal for a universal nomenclature for ribosomal proteins is based upon phylogenetic sequence comparisons, classifying ribosomal proteins as universal if family members are found in all three kingdoms and either B or E if family members are restricted to either the eubacterial or archaeal/eukaryal lineages, respectively (Ban et al. 2014).
In terms of structure, Melnikov et al. have described ribosomes as having a conserved core with bacterial- or eukaryotic-specific shells (Melnikov et al. 2012). The universal ribosomal proteins belong to a common structural core of ribosomes necessary for carrying out the basic steps of protein synthesis shared by ribosomes in the three domains of life. Outside of this common core, new ribosomal proteins unique to eubacteria, archaebacteria/eukarya, and eukarya alone have been recruited to work in conjunction with the common core to perform functions more specialized to a particular domain of life. Eukaryotic ribosomes are much larger than their eubacterial and archaebacterial counterparts: including expansion sequences in rRNAs, additional structural elements added to core proteins, new proteins common to archaeal and eukaryal lineages, and several additional proteins unique to the eukaryal lineage (Gamalinda and Woolford Jr. 2014).
The most dramatic differences between bacterial and eukaryotic ribosomes occur on the solvent exposed surfaces of the ribosomal subunits. These distinctive features are presumably necessary for interactions with the myriad of factors involved in assembling ribosomal subunits, transporting nascent subunits from the nucleolus to the cytoplasm, and for aspects of the translational process differing between the different domains of life. Superimposed on these generalized differences between bacterial and eukaryotic ribosomes are novel ribosomal proteins in eukaryotes that have evolved to give ribosomes unique characteristics in a particular tissue or cell type, creating what are now referred to as specialized ribosomes (Kondrashov et al. 2011).
5.2 Diamond-Blackfan Anemia: The Prototypical Ribosomopathy
To date, 18 genes have been linked to Diamond-Blackfan anemia (Danilova and Gazda 2015). The vast majority of the genes encode ribosomal proteins. The ribosomal proteins affected in DBA include proteins of both the 40S and 60S ribosomal subunit. The 40S subunit proteins known to be affected in DBA include Rps19(eS19), Rps26(eS26), Rps10(eS10), Rps24(eS24), Rps17(eS17), Rps7(eS7), Rps27(eS27), Rps28(e28), and Rps29 (uS14). In parenthesis we include the names for the ribosomal proteins using the new universal system of nomenclature (Ban et al. 2014). Looking at the names for these proteins using the new nomenclature reveals that only one of the nine Rps proteins affected in DBA belongs to the common core of the 40S subunit, Rps29(uS14), with the remaining proteins having evolved after the divergence of archaebacteria and eukarya from the eubacteria. As 15 of the 33 proteins of the eukaryotic 40S subunit belong to the common core, this bias is not simply a reflection of the percentage of 40S subunit proteins belonging to common core relative to those recruited later. Whether there is any significance to this bias against proteins of the common core being affected in DBA remains unknown.
The situation is more complicated for proteins of the 60S subunit. The 60S ribosomal proteins known to be affected in DBA include Rpl5(uL18), Rpl11(uL5), Rpl35A(eL33), Rpl15(eL15), Rpl26(uL24), Rpl27(eL27), and Rpl31(eL31). Here the bias toward protein components of the common core seems to fall off with DBA, though it should be noted that Rpl5(uL18) and Rpl11(uL5) are distinct in terms of forming a subcomplex with 5S rRNA prior to being incorporated into the 60S subunit. Moreover, the 5S rRNA subcomplex has been co-opted as a signaling molecule regulating p53 levels in response to ribosome stress (see pathophysiological mechanisms). Intriguingly, the other 60S subunit protein from the common core so far implicated in DBA is Rpl26(uL24), another protein known to play a role in regulating p53 expression (Chen et al. 2012), although through a mechanism distinct from Rpl5(uL18) and Rpl11(uL5).
5.2.1 Pre-rRNA Processing as a Means of Monitoring Ribosomal Protein Function
The ribosomal protein genes affected in DBA harbor heterozygous loss-of-function mutations resulting in haploinsufficiency for the affected ribosomal protein. Ribosomal protein function is routinely monitored through their role in ribosome biogenesis by assessing the effect of mutant alleles on the process by which mature 18S, 5.8S, and 28S rRNAs are generated from the polymerase I primary transcript. Loss-of-function alleles of ribosomal proteins affected in DBA all display characteristic pre-rRNA processing defects, the nature of which are dictated by where these proteins are located in the structural organization of the 40S subunit relative to ends of mature RNAs (Flygare et al. 2007). As noted by O’Donohue et al. (2010), the three most frequently mutated genes in DBA patients that encode proteins of the 40S ribosomal subunit Rps19(25%), Rps26(6.5%), and Rps10(2.5%) are all involved in the maturation of the 3′ end of 18S rRNA. As shown in Fig. 5.1, these proteins are all located in the head region of the 40S subunit, which also contains the 3′ structural domain of 18S rRNA. This led to the hypothesis that processing defects involved in 3′ end maturation may be less severe than those influencing the 5′ end of 18S, thereby resulting in a disease state rather than embryonic lethality (O’Donohue et al. 2010). As can be seen in Fig. 5.1, DBA proteins are also found on base of the 40S structure with the 5′ domain of 18S rRNA, and studies have shown that these proteins influence the 5′ end of 18S rRNA. Therefore, while the frequency of mutations in small subunit proteins is clearly biased toward those in the head domain, being located on the base and influencing 5′end maturation does not preclude a protein from being involved in DBA. In this regard, it is also noteworthy that components of the small subunit processome carrying out many of the earliest steps in the biogenesis of the 40S subunit have been implicated in other ribosomopathies that do not present as bone marrow failure syndromes but nevertheless are not embryonic lethal (Sondalle and Baserga 2014).
Ribosomal proteins affected in Diamond-Blackfan anemia. The atomic coordinates for the 40S and 60S ribosomal subunits were derived from the work of Khatter et al. (2015) deposited in the protein data bank with the accession code 4UGO. The figure was created using protein workshop software developed by Moreland et al. (2005). Proteins affected in DBA are shown in red with the exception of Rps10, Rps19, and Rps26 of the 40S subunit, which are shown in magenta. Other ribosomal proteins are shown in green. 18S and 28S rRNAs of the 40S and 60S ribosomal subunits, respectively, are shown in gray. 5S and 5.8S rRNAs of the 60S ribosomal subunit are shown in blue
Like the proteins of the 40S ribosomal subunit, proteins of the 60S ribosomal subunit also influence pre-rRNA processing. Some of the more dramatic pre-rRNA processing defects observed with 60S subunit ribosomal proteins are evident by capillary electrophoresis without the need for radio-isotopic detection (Farrar et al. 2014). Quarello et al. have suggested that this technique may be amenable to use as a diagnostic aide for DBA (Quarello et al. 2016).
5.2.2 Non-ribosomal Proteins Causing DBA
To date, only two proteins other than ribosomal proteins have been linked to DBA. Both are located on the X chromosome. The first is GATA1, a transcription factor involved in lineage decisions during hematopoiesis, including erythropoiesis. GATA1 thus represents the first gene identified in DBA that provides a clear molecular basis for erythroid phenotype observed in DBA patients (Sankaran et al. 2012). The other protein is Tsr2, which is involved, at least in yeast, in the maturation of 40S ribosomal subunits (Gripp et al. 2014). This is the first factor involved in ribosome biogenesis other than a ribosomal protein that has been implicated in DBA. Factors involved in ribosome biogenesis are only transiently associated with ribosomes and so in contrast to ribosomal proteins may behave catalytically being continuously regenerated after performing their functions in subunit maturation. Consequently such genes may require two loss-of-function alleles to manifest clinically, which may be an exceptionally rare event, unless such a gene is found on the X chromosome.
5.3 Pathophysiologic Mechanisms
Except for GATA1, it is difficult to reconcile the genes affected in DBA with red cell hypoplasia, the sine qua non of Diamond-Blackfan anemia. How a defect in a process as ubiquitous as ribosome biogenesis can cause highly selective disorders affecting a limited number of tissues remains a mystery for DBA and other ribosomopathies. There are two general models for how defects in ribosome synthesis could be involved in the molecular underpinnings of DBA. The first of these is a ribosome stress model where rogue ribosomal proteins failing to assemble into their respective ribosomal subunits have alternative fates including promoting p53 activation and apoptosis (Ellis and Gleizes 2011). The second model evokes changes in translational output arising from reduced amounts of 40S or 60S subunits caused by ribosomal protein haploinsufficiency. Both models have strong experimental support with some caveats, and it seems highly likely that disease phenotypes may involve a combination of the two mechanisms, which are not mutually exclusive. Each will be discussed in turn below.
5.3.1 Ribosome Stress
Pestov et al. were the first to draw attention to the relationship between defects in ribosome synthesis and the activation of p53 via a process they referred to as nucleolar stress (Pestov et al. 2001). We will use the term ribosome stress to reflect the fact that not all stressors that affect ribosome biogenesis may affect nucleolar structure (Nicolas et al. 2016). The importance of p53 activation to DBA pathophysiology became evident with the finding that loss of p53 function rescued phenotypes in a zebra fish and mouse models of DBA (Danilova et al. 2008; Jaako et al. 2015) and cell death in human cellular models of DBA (Dutt et al. 2011). The mechanisms underlying p53 activation in cells with abortive ribosome synthesis began to take shape with studies showing ribosomal proteins were capable of binding to and inhibiting MDM2, a ubiquitin ligase targeting p53 for proteasomal degradation responsible for maintaining low levels of p53 expression in unstressed cells (Marechal et al. 1994). Inhibition of MDM2 via interaction with ribosomal proteins diverted from their normal fate of being assembled into ribosomal subunits to now interacting with MDM2, leads to p53 stabilization and activation. A potentially straightforward model by which a limited number of ribosomal proteins play a role in signaling p53 activation in response to ribosome stress has given way to an embarrassment of riches with an ever-growing list of ribosomal proteins shown to interact with MDM2 (Kim et al. 2014; Xu et al. 2016). Ribosomal proteins known to interact with MDM2 directly leading to p53 activation include Rps3(uS3), Rps7(eS7), Rps14(uS11), Rps15(u19), Rps20(uS10), Rps25(eS25), Rps26(eS26), Rps27(eS27), Rps27a(eS31), Rps27L, Rpl5(uL18), Rpl11(uL5), Rpl23(uL14), Rpl26(uL24), and Rpl37(eL37).
Among these proteins, Rpl5(uL18) and Rpl11(uL5) stand out as being critical among the direct effectors in mediating p53 activation in response to ribosome stress (Donati et al. 2013). With all the other ribosomal proteins potentially involved in ribosome stress signaling, it seems worthwhile to consider whether the signaling mechanism centered on Rpl5/Rpl11 could be rheostatically controlled. In a normal electronic rheostat, resistance is variable controlled through a moving knob or slider, whereas for a ribosome stress model, the moving knob or slider would be the nature, number, and amount of free ribosomal proteins available for interacting with MDM2 and other growth control molecules. Adaptation of a rheostat model to ribosome stress signaling could potentially explain both the surplus of ribosomal protein modulators and the bias toward noncore ribosomal proteins involved in Diamond-Blackfan anemia.
If we assume that Rpl5 and Rpl11 are critical regulators of ribosome stress signaling as part of the 5S RNP subcomplex, one can envision how a defect in 60S subunit biogenesis could lead to the failure of incorporating this subcomplex into nascent 60S particles making it now available for interaction with MDM2 and p53 induction. Less clear was how a defect in 40S subunit biogenesis could signal ribosomal stress through the 5S RNP subcomplex. Fumagalli et al. resolved this issue by showing that the translation of TOP-containing ribosomal protein mRNAs, including Rpl5 and Rpl11, was increased in cells with impaired 40S subunit biogenesis (Fumagalli et al. 2009).
What remains unclear, however, is our knowledge of threshold of ribosome stress signaling required for p53-induced apoptosis, particularly in vivo in different cell types. If the level by which components of the 5S RNP subcomplex were induced by impaired 40S biogenesis were below a particular threshold for p53-induced apoptosis, there could be a role for small subunit ribosomal proteins acting either on their own or in concert with the 5S RNP in inhibiting MDM2 and activating p53.
The nature, number, and amount of small subunit ribosomal proteins available for this rheostatic control of the MDM2-p53 axis could be influenced by which ribosomal protein was limiting for 40S subunit biogenesis under conditions of haploinsufficiency. The elegant studies by Nomura and colleagues in the 1960s revealed that the assembly of ribosomal subunits was a hierarchical process with some primary assembly proteins binding directly to rRNA and other secondary and tertiary binding proteins being more reliant on RNA binding sites created by folding events mediated by the primary binding proteins and protein/protein interactions (Held et al. 1974). The loss of a tertiary binding protein would still allow some degree of assembly of primary and secondary binding proteins and only affect the binding of a limited number of other tertiary binding proteins into the abortive complex. In contrast, the loss of a critical primary binding protein affects the incorporation of a far greater number of proteins into the abortive complex, thus releasing a far greater number of proteins to potentially signal ribosome stress.
While we do not have an assembly map of human 40S subunits comparable to that worked out for the bacterial 30S subunit, viewing human 40S subunit as having a common core with a eukaryotic shell could give us an analogous, if somewhat cruder, perspective. Very little is known regarding the degradation of abortive intermediates that accumulate when ribosomal subunit assembly is compromised, although it does appear that there are cellular quality control mechanisms involved in the degradation of misassembled or nonfunctional ribonucleoprotein complexes (Fujii et al. 2012). Consequently, proteins that get assembled into abortive complexes may be degraded along with rRNA as part of this quality control mechanism. Thus, we would have two general classes of ribosomal proteins during abortive assembly, those that get assembled into the abortive complex and are presumably degraded and those failing to assemble, which could then be free for ribosome stress signaling. Fewer proteins may be available for ribosome stress signaling if assembly were aborted by haploinsufficiency for a protein of the outer shell, whereas the number of proteins released if a protein of the common core were affected may be incompatible with life.
Rps29(uS14) is the current exception to the finding that ribosomal proteins of the 40S subunit affected in DBA belong to the eukaryotic shell. Intriguingly, bacterial S14 is a tertiary binder in the Nomura assembly map (Held et al. 1974) and in eukaryotes Rps29 is incorporated in a late cytoplasmic step in the maturation of 40S subunits (de la Cruz et al. 2015). Thus, the relationship between ribosomal subunit assembly dependencies and the release of ribosomal proteins for various roles in controlling cell cycle progression and apoptosis is clearly more nuanced, allowing certain proteins within the conserved core of the ribosome to be affected in DBA.
As appealing as the ribosome stress model is as an underlying mechanism for the pathogenesis of DBA, there are significant caveats. Foremost among these caveats is that Rpl5(uL18) and Rpl11(uL5), central mediators of ribosome stress signaling, are themselves DBA proteins (Gazda et al. 2008). Thus, there would have to be alternative means of signaling to p53 activation when either of these two proteins is absent, or that p53-independent mechanisms may also contribute to phenotypes in DBA. In a recent tour de force, the Lafontaine laboratory has monitored the effect of reducing the expression of each of the human ribosomal proteins on nucleolar structure and p53 induction (Nicolas et al. 2016). Based on their cutoff criteria for significant induction of p53 (fivefold increase over basal levels), the loss of function of many ribosomal proteins, including virtually all DBA proteins, did not result in what was considered a significant increase in steady-state levels of p53. These rather shocking results, which in some cases differ from previous published reports, may relate to their rather stringent cutoff of significance at fivefold induction, which again points to our lack of knowledge of critical thresholds for p53 activation in physiologically relevant settings. The ever-mounting complexity of factors feeding into models of ribosome stress continues to confound any consensus regarding the role of misassembled ribosomal proteins and p53 activation in DBA pathophysiology.
Before going on to discuss the role of translational alterations in DBA pathophysiology, it is worthwhile discussing other ribosomopathies in the context of ribosome stress. The first of these is the bone marrow failure syndrome Shwachman-Diamond syndrome. Shwachman-Diamond syndrome and Diamond-Blackfan anemia differ in clinical presentation, which is covered elsewhere in this volume. To date, there is a single gene known to be affected in Shwachman-Diamond syndrome given the acronym SBDS, for Shwachman-Diamond-Bodian syndrome. SBDS is a conserved protein found in eukaryal and archaeal lineages, which plays a role in the release of eukaryotic initiation factor 6 (eIF6) from nascent 60S subunits exiting the nucleus to the cytoplasm. The presence of bound eIF6 serves as an anti-association factor preventing the joining of 40S and 60S ribosomal subunits until the latter are fully matured (Weis et al. 2015). Thus, the function of SBDS in the release of eIF6 is one of the final steps in 60S subunit maturation before the subunit becomes part of the 60S subunit pool active in translation.
The failure to release eIF6 from nascent 60S subunits in cytoplasm interferes with the recycling of eIF6 to the nucleus where it performs a function in transporting nascent subunits from the nucleus to the cytoplasm. In this manner, the loss of SBDS has a secondary effect on 60S subunit maturation by impeding the transport of 60S subunits from the nucleus to the cytoplasm (Menne et al. 2007). In contrast to DBA where the loss of a ribosomal protein can interfere with the assembly of other ribosomal proteins into nascent subunits leading to the degradation of partially assembled subunits, the 60S subunits that accumulate in the nucleus of cells deficient in SBDS and its yeast ortholog Sdo1 are relatively stable and likely have a relatively full complement of ribosomal proteins, except for those assembling in the cytoplasm (Moore et al. 2010). Importantly, the failure to transport 60S subunits from the nucleus to the cytoplasm does not appear to interfere with the incorporation of the 5S RNP subcomplex into nascent subunits. These fundamental differences in the manner in which ribosomal proteins and SBDS interfere with the maturation of ribosomal subunits likely play a role in the differences in clinical phenotypes between Shwachman-Diamond syndrome and Diamond-Blackfan anemia patients.
Any model for ribosome stress in the pathophysiology of Diamond-Blackfan anemia must deal with the Treacher Collins syndrome conundrum. Treacher Collins is a syndrome characterized by craniofacial anomalies. Importantly for our discussion, patients with Treacher Collins syndrome do not manifest bone marrow failure. This is surprising because the genes affected in TCS interfere with the transcription of rRNAs, and like DBA activation of p53 appears to play a central role in the pathophysiology of TCS. Ribosomal RNA forms the base of any assembly pathway for ribosomal subunits, and reductions in rRNA should interfere with subsequent incorporation of all ribosomal proteins into nascent subunits. Therefore, one might expect that ribosomal proteins failing to assemble as a consequence of reduced transcription of rRNA would play a role in activating p53 similar to that seen in DBA and as such affect erythropoiesis. And yet, there is no evidence for a defect in erythropoiesis in TCS patients.
Transcription of rDNA by RNA polymerase I plays a critical role in defining ribosome levels in cells, and although all three RNA polymerases are thought to be coordinated by various levels of crosstalk during ribosome production, there are likely transient windows when expression of ribosomal RNAs and proteins is uncoupled, leading to imbalances in the ribosome substituents before new steady-state conditions are achieved. Consequently there may be additional levels of control on ribosome stress-mediated signaling when rRNA transcription is affected in a physiological setting, as opposed to the signaling mechanisms involved when there is haploinsufficiency for a ribosomal protein.
5.3.2 Translational Alterations
The Ruggero group has shown that loss of DKC1, the gene affected in X-linked dyskeratosis congenita (DC), can influence the translation of IRES-containing mRNAs and contribute to the pathogenesis of the X-linked form of this bone marrow failure syndrome (Bellodi et al. 2010). While the loss of telomere function is the primary cause of most forms of DC, DKC1 mutations affect a number of additional processes including ribosome synthesis and function (Angrisani et al. 2014). These additional functions of DKC1 presumably contribute to the severity of X-linked forms of the disease (Townsley et al. 2014), and so at least the X-linked form of DC could also be classified as a ribosomopathy.
It seems reasonable to infer that changes in translational output in cells affected by haploinsufficiency for ribosomal proteins could also contribute to the pathogenesis of DBA. Each of the ribosomal proteins affected in DBA has been shown to influence pre-rRNA processing and in many cases has been shown to influence the levels of their respective ribosomal subunit (Flygare et al. 2007; Nicolas et al. 2016; Choesmel et al. 2007). Such losses of ribosomal subunits have been shown to affect global translation in various cellular models of DBA and in some cases have been shown to affect the translation of specific mRNAs encoding proteins involved in hematopoiesis (Horos et al. 2012). One of the more telling examples of an effect on translation that could give rise to the clinical features of DBA were the studies of Ludwig et al. showing that haploinsufficiency for ribosomal proteins affected in DBA could adversely affect the translation of GATA1 mRNA (Ludwig et al. 2014). These studies linked DBA caused by ribosomal protein haploinsufficiency with DBA caused by mutations in GATA1. The mutations in GATA1 found in DBA result in an amino-terminal truncated protein retaining residual activity but lacking a critical domain required for erythropoiesis (Sankaran et al. 2012). Importantly, in cells with normal GATA1 but haploinsufficient for different ribosomal proteins affected in DBA, Ludwig et al. showed that the translation of full-length GATA1 was decreased providing a mechanism whereby reduced levels of ribosomal proteins could selectively affect erythropoiesis.
The exact nature by which ribosomal protein haploinsufficiency affects the translation of GATA1 remains to be identified, but it is worthwhile noting that the effect on GATA1 translation is independent of which subunit is affected. It is difficult to envision a specific translational control mechanism that would give similar effects regardless of which subunit was affected. Thus, it seems possible that a general reduction in protein synthetic capacity could somehow be involved in the pathogenesis of DBA.
Support for a role of general reduction of protein synthesis in the pathogenesis of DBA has recently come from studies from the Abkowitz group. Yang et al. have recently published an elegant study on the timing of heme and globin synthesis in developing erythrocytes (Yang et al. 2016). These studies reveal that cells haploinsufficient for ribosomal proteins affected in DBA exhibit a delay in globin synthesis while heme synthesis continues unabated, leading to heme excess. Heme excess, in turn, leads to increased reactive oxygen species and promotes ferroptosis/apoptosis of erythroid progenitors. Further support for the role of heme toxicity in the pathogenesis of DBA comes from the observation that reducing heme synthesis to parallel globin synthesis or an increase in heme export from cells can rescue phenotypes caused by haploinsufficiency for ribosomal proteins in CD34+ cultures induced to differentiate along the erythroid lineage.
Yang et al. also provide a model for how mechanisms of cell death induced by heme toxicity could synergize with ribosome stress signaling to preferentially affect erythropoiesis. In this model, p53 activation in response to ribosome stress could potentially interfere with the synthesis of certain antioxidant defense mechanism sensitizing cells to reactive oxygen species induced by heme excess. Thus, it seems increasingly likely that the two mechanisms implicated in the pathophysiology of DBA interact to preferentially effect erythropoiesis and therefore begin to explain how disrupting a ubiquitous process like ribosome synthesis could engender tissue selectivity in terms of clinical phenotypes.
5.4 Ribosomopathies: A Gateway to Tumorigenesis
The two main models proposed to explain defective erythropoiesis in DBA have also been advocated to account for high incidence of cancer in DBA patients. An increased risk to develop myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), and solid tumors has been observed by studying a large cohort of DBA patients (Vlachos et al. 2012). Shwachman-Diamond syndrome, dyskeratosis congenita, and 5q− syndrome, an acquired condition due to heterozygous loss of the Rps14(uS11) gene (Ebert et al. 2008), located on the long arm of chromosome 5, are all known to be associated with MDS and progression to leukemia. Most recently, next-generation sequencing technologies led to the discovery that somatic mutations in RP genes are rather common in different cancer types. For example, Rpl5(uL18), Rpl10(uL16), and Rpl22(eL22) were found mutated in T-acute lymphoblastic leukemia (De Keersmaecker et al. 2013; Rao et al. 2012), whereas other RPs were mutated in endometrial cancer, colorectal carcinoma, and glioma (Goudarzi and Lindstrom 2016). Heterozygous mutations in several RPs also cause tumors in zebra fish (Amsterdam et al. 2004) and in other animal models including Drosophila and mouse (Kazerounian et al. 2016; Stewart and Denell 1993). Therefore, there is rising evidence that ribosome dysfunction can drive malignant transformation.
Tumorigenesis is typically accompanied by stimulation of protein synthesis, which is critical to support unrestrained proliferation in cancer cells. It seems paradoxical that pathologies characterized by hypoproliferative phenotypes may be associated with elevated cancer risk. The causal link between ribosomopathies and cancer might be, once again, the dysregulation of p53-dependent pathways. Persistent activation of p53 in RP depleted cells may provide selective pressure for the expansion of clones with spontaneous mutations that confer proliferative advantage. The acquisition of any compensatory mutation that counterbalances the effects of ribosome stress and prevents apoptosis may contribute to tumorigenesis. Interestingly, a similar mechanism where a clone with growth advantage prevails over the population of cells carrying only the original RP mutation may also be responsible for the spontaneous remission observed in some cases of DBA.
A high cancer risk in cells subjected to ribosome stress may also be the result of alterations in the translation of specific messenger RNAs, particularly oncogenes and tumor suppressors. Other mechanisms have been suggested to explain why RP depletion would increase cancer predisposition. A reduction in the number of ribosomes or qualitative alterations in their composition could impact translational fidelity and lead to cellular transformation (Sulima et al. 2014); moreover, some RPs may have extraribosomal functions relevant for cancer development.
5.5 Concluding Thoughts
There are an increasing number of human diseases caused by mutations in genes encoding either structural components of the ribosome or factors involved in ribosome biogenesis. These diseases collectively known as ribosomopathies continue to provide challenges in terms of underlying pathogenic mechanisms. In some cases it still remains to be seen whether defective ribosome biogenesis plays a major role in disease phenotype, or instead in cases where the factors involved have multiple functions within cells whether defects in ribosome synthesis play a role in modifying disease phenotype. In other diseases, where defects in ribosome synthesis are more clearly defined as having a role in disease pathogenesis, the link between defects in a process as ubiquitous as ribosome biogenesis and seemingly tissue-selective disease phenotypes remains elusive. Many studies have supported a role for p53 activation via ribosome stress in disease pathogenesis, but many questions remain because of the remarkable myriad of factors that have been implicated in ribosome stress signaling. Recent studies on the timing of heme and globin synthesis in erythroid progenitors have also provided fascinating new insights as to how general defects in protein synthesis linked to haploinsufficiency for ribosomal proteins could potentially account for the erythroid selectivity of Diamond-Blackfan anemia. Finally, adding fuel to growing interest in ribosomopathies is the link between many of these diseases and tumorigenesis as evidenced by the cancer predisposition in certain ribosomopathies and the finding of somatic mutations in ribosomal protein genes in tumor genotyping.
References
Amsterdam A et al (2004) Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol 2(5):E139
Angrisani A et al (2014) Human dyskerin: beyond telomeres. Biol Chem 395(6):593–610
Ban N et al (2014) A new system for naming ribosomal proteins. Curr Opin Struct Biol 24:165–169
Bellodi C et al (2010) Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res 70(14):6026–6035
Chen J, Guo K, Kastan MB (2012) Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J Biol Chem 287(20):16467–16476
Choesmel V et al (2007) Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood 109(3):1275–1283
Danilova N, Gazda HT (2015) Ribosomopathies: how a common root can cause a tree of pathologies. Dis Model Mech 8(9):1013–1026
Danilova N, Sakamoto KM, Lin S (2008) Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood 112(13):5228–5237
De Keersmaecker K et al (2013) Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 45(2):186–190
Donati G et al (2013) 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep 4(1):87–98
Dutt S et al (2011) Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117(9):2567–2576
Ebert BL et al (2008) Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451(7176):335–339
Ellis SR, Gleizes PE (2011) Diamond Blackfan anemia: ribosomal proteins going rogue. Semin Hematol 48(2):89–96
Farrar JE et al (2014) Exploiting pre-rRNA processing in Diamond Blackfan anemia gene discovery and diagnosis. Am J Hematol 89(10):985–991
Flygare J et al (2007) Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood 109(3):980–986
Fujii K et al (2012) 40S subunit dissociation and proteasome-dependent RNA degradation in nonfunctional 25S rRNA decay. EMBO J 31(11):2579–2589
Fumagalli S et al (2009) Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol 11(4):501–508
Gamalinda M, Woolford JL Jr (2014) Deletion of L4 domains reveals insights into the importance of ribosomal protein extensions in eukaryotic ribosome assembly. RNA 20(11):1725–1731
Gazda HT et al (2008) Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet 83(6):769–780
Goudarzi KM, Lindstrom MS (2016) Role of ribosomal protein mutations in tumor development (Review). Int J Oncol 48(4):1313–1324
Gripp KW et al (2014) Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A 164A(9):2240–2249
Held WA et al (1974) Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem 249(10):3103–3111
Horos R et al (2012) Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood 119(1):262–272
Jaako P et al (2015) Disruption of the 5S RNP-Mdm2 interaction significantly improves the erythroid defect in a mouse model for Diamond-Blackfan anemia. Leukemia 29(11):2221–2229
Kazerounian S et al (2016) Development of Soft Tissue Sarcomas in Ribosomal Proteins L5 and S24 Heterozygous Mice. J Cancer 7(1):32–36
Khatter H et al (2015) Structure of the human 80S ribosome. Nature 520(7549):640–645
Kim TH, Leslie P, Zhang Y (2014) Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget 5(4):860–871
Kondrashov N et al (2011) Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145(3):383–397
de la Cruz J, Karbstein K, Woolford JL Jr (2015) Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem 84:93–129
Liu JM, Ellis SR (2006) Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood 107(12):4583–4588
Ludwig LS et al (2014) Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med 20(7):748–753
Marechal V et al (1994) The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol 14(11):7414–7420
Melnikov S et al (2012) One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 19(6):560–567
Menne TF et al (2007) The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet 39(4):486–495
Moore JBt et al (2010) Distinct ribosome maturation defects in yeast models of Diamond-Blackfan anemia and Shwachman-Diamond syndrome. Haematologica 95(1):57–64
Moreland JL et al (2005) The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics 6:21
Nicolas E et al (2016) Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat Commun 7:11390
O’Donohue MF et al (2010) Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol 190(5):853–866
Pestov DG, Strezoska Z, Lau LF (2001) Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol 21(13):4246–4255
Quarello P et al (2016) Ribosomal RNA analysis in the diagnosis of Diamond-Blackfan Anaemia. Br J Haematol 172(5):782–785
Rao S et al (2012) Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 120(18):3764–3773
Sankaran VG et al (2012) Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest 122(7):2439–2443
Sondalle SB, Baserga SJ (2014) Human diseases of the SSU processome. Biochim Biophys Acta 1842(6):758–764
Stewart MJ, Denell R (1993) Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol Cell Biol 13(4):2524–2535
Sulima SO et al (2014) Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc Natl Acad Sci U S A 111(15):5640–5645
Townsley DM, Dumitriu B, Young NS (2014) Bone marrow failure and the telomeropathies. Blood 124(18):2775–2783
Vlachos A et al (2012) Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood 119(16):3815–3819
Weis F et al (2015) Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol 22(11):914–919
Xu X, Xiong X, Sun Y (2016) The role of ribosomal proteins in the regulation of cell proliferation, tumorigenesis, and genomic integrity. Sci China Life Sci 59(7):656–672
Yang Z et al (2016) Delayed globin synthesis leads to excess heme and the macrocytic anemia of Diamond Blackfan anemia and del(5q) myelodysplastic syndrome. Sci Transl Med 8(338):338ra67
Yelick PC, Trainor PA (2015) Ribosomopathies: global process, tissue specific defects. Rare Dis 3(1):e1025185
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Aspesi, A., Ellis, S.R. (2018). Ribosomopathies Through a Diamond Lens. In: Kupfer, G., Reaman, G., Smith, F. (eds) Bone Marrow Failure. Pediatric Oncology. Springer, Cham. https://doi.org/10.1007/978-3-319-61421-2_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-61421-2_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-61420-5
Online ISBN: 978-3-319-61421-2
eBook Packages: MedicineMedicine (R0)