Abstract
This paper presents a method for breast density classification using local quinary patterns (LQP) in mammograms. LQP operators are used to capture the texture characteristics of the fibroglandular disk region (\(FGD_{roi}\)) instead of the whole breast region as the majority of current studies have done. To maximise the local information, a multiresolution approach is employed followed by dimensionality reduction by selecting dominant patterns only. Subsequently, the Support Vector Machine classifier is used to perform the classification and a stratified ten-fold cross-validation scheme is employed to evaluate the performance of the method. The proposed method produced competitive results up to \(85.6\%\) accuracy which is comparable with the state-of-the-art in the literature. Our contributions are two fold: firstly, we show the role of the fibroglandular disk area in representing the whole breast region as an important region for more accurate density classification and secondly we show that the LQP operators can extract discriminative features comparable with the other popular techniques such as local binary patterns, textons and local ternary patterns (LTP).
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
1 Introduction
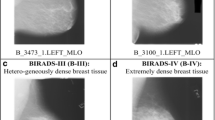
In 2014 there were more than 55,000 malignant breast cancer cases diagnosed in the United Kingdom (UK) with more than 11,000 mortality [1]. In the United States (US), an estimate of more than 246,000 malignant breast cancer were expected to be diagnosed in 2016 with approximately 16% women expected to die [2]. Many studies have indicated that breast density is a strong risk factor for developing breast cancer [3,4,5, 7,8,9,10,11,12,13] because dense tissues are very similar in appearance to breast cancer this making it more difficult to detect in mammograms. Therefore, women with dense breasts can be six times more likely to develop breast cancer, which means an accurate breast density estimation is an important step during the screening procedure. Although most experienced radiologists can do this task, manual classification is impractical, tiring, time consuming and often suffers from results variability among radiologists. Computer Aided Diagnosis (CAD) systems can reduce these problems providing robust, reliable, fast and consistent diagnosis results. Based on the Breast Imaging Reporting and Data System (BI-RADS), there are four major categories used for classifying breast density: (a) predominantly fat, (b) fat with some fibroglandular tissue, (c) heterogeneously dense and (d) extremely dense. These representations are illustrated in Fig. 1.
One of the earliest approaches to breast density assessment was a study by Boyd et al. using interactive thresholding known as Cumulus, where regions with dense tissue were segmented by manually tuning the greylevel threshold value. The most popular approaches are based on the first and second-order (e.g. Grey Level Co-occurrence Matrix) statistical features as used by Oliver et al. [3], Bovis and Singh [4], Muštra et al. [6] and Parthaláin et al. [7]. Texture descriptors such as local binary patterns (LBP) were employed in the study of Chen et al. [8] and Bosch et al. [9] and textons were used by Chen et al. [8], Bosch et al. [9] and Petroudi et al. [13]. Other texture descriptors also have been evaluated such as fractal-based [5, 11], topography-based [10], morphology-based [3] and transform-based features (e.g. Fourier, Discrete Wavelet and Scale-invariant feature) [4, 9].
Many breast density classification methods in mammograms have been proposed in the literature but only very small number of studies have achieved accuracies above 80%. The methods of Oliver et al. [3] and Parthaláin et al. [7] extract a set of features from dense and fatty tissue regions segmented using a fuzzy c-means clustering technique followed by feature selection before feeding them into the classifier. Oliver et al. [3] achieved 86% accuracy and Parthaláin et al. [7] who used a sophisticated feature selection framework achieved 91.4% accuracy. Bovis and Singh [4] achieved 71.4% accuracy based on a combined classifier paradigm in conjunction with a combination of features extracted using the Fourier and Discrete Wavelet transforms, and first and second-order statistical features. Chen et al. [8] made a comparative study on the performance of local binary patterns (LBP), local greylevel appearance (LGA), textons and basic image features (BIF) and reported accuracies of 59%, 72%, 75% and 70%, respectively. Later, they proposed a method by modelling the distribution of the dense region in topographic representations and reported a slightly higher accuracy of 76%. Petroudi et al. [13] implemented the textons approach based on the Maximum Response 8 (MR8) filter bank. The \(\chi ^2\) distribution was used to compare each of the resulting histograms from the training set to all the learned histogram models from the training set and reported 75.5% accuracy. He et al. [12] achieved an accuracy of 70% using the relative proportions of the four Tabár’s building blocks. Muštra et al. [6] captured the characteristics of the breast region using multi-resolution of first and second-order statistical features and reported 79.3% accuracy.
Based on the results reported in the literature, the majority of the proposed methods achieved below 80% which indicate that breast density classification is a difficult task due to the complexity of tissue appearance in the mammograms such as a wide variation and obscure texture patterns within the breast region. In this paper, texture features were extracted from the \(FGD_{roi}\) only (see Fig. 4 later) to obtain more descriptive information instead of from the whole breast region. The motivation behind this approach is that because in most cases the non \(FGD_{roi}\) contains mostly fatty tissues regardless its BI-RADS class and most dense tissues are located and start to develop within the \(FGD_{roi}\). Therefore, extracting features from the whole breast region means extracting overlapping texture information which makes the extracted features less discriminant in corresponding to the BIRADS classes. The reminder of the paper is organised as follows: We present the technical aspects of our proposed method in Sect. 2 and discuss experimental results in Sect. 3 which covers the quantitative evaluation and comparisons, Sect. 4 presents conclusions and future work.
2 Methodology
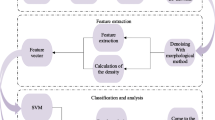
Figure 2 shows an overview of the proposed methodology.
Firstly, we segment the breast area and estimate the \(FGD_{roi}\) from this breast region. Subsequently, we use a simple median filter using a \(3 \times 3\) window size for noise reduction and employ the multiresolution LQP operators to capture the micro-structure information within the \(FGD_{roi}\). Finally, we train the SVM classifier to build a predictive model and use it to test each unseen case.
2.1 Pre-processing
To segment the breast and pectoral muscle region, we used the method in [18] which is based on Active Contours without edges for the breast boundary estimation and contour growing with edge information for the pectoral muscle boundary estimation. The left most image in Fig. 4 shows the estimated \(FGD_{roi}\) area. To extract \(FGD_{roi}\), we find \(B_w\) which is the longest perpendicular distance between the y-axis and the breast boundary (magenta line). The width and the height of the square area of the \(FGD_{roi}\) (amber line Fig. 4) can be computed as \(B_w \times B_w\) with the center located at the intersection point between \(B_h\) and \(B_w\) lines. \(B_h\) is the height of the breast which is the longest perpendicular distance between the x-axis and the breast boundary. \(B_h\) is then relocated to the middle of \(B_w\) to get the intersection point. The size of the \(FGD_{roi}\) varies depending on the width of the breast.
2.2 Feature Extraction
The Local Binary Pattern (LBP) operators were first proposed by Ojala et al. [14] to encode pixel-wise information. Tan and Triggs [16] modified it by introducing LTP operators which thresholds the neighbouring pixels using a three-value encoding system based on a threshold constant set the user. Later, Nanni et al. [15] introduced a five-value encoding system called LQP. The LBP, LTP and LQP are similar in terms of architecture as each are defined using a circle centered on each pixel and a number of neighbours but the main difference is that the LBP, LTP and LQP threshold the neighbouring pixels into two (1 and 0), three (−1, 0 and 1) and five (2, 1, 0, −1 and −2) values, respectively. This means the difference between the grey level value of the center pixel (\(g_c\)) and a neighbour’s grey level (\(g_p\)) can assume five values. The value of LQP code of the pixel (i, j) is given by:
where R is the circle radius, P is the number of pixels in the neighbourhood, \(g_c\) is the grey level value of the center pixel, \(g_p\) is the grey level value of the \(p^{th}\) neighbour, and \(pattern \in \{1, 2, 3, 4 \}\). Once the LQP code is generated, it is split into four binary patterns by considering its positive, zero and negative components, as illustrated in Fig. 3 using the following conditions
In this section, feature extraction is the process of computing the frequencies of all binary patterns and present the occurrences in a histogram which represents the number of appearances of edges, corners, spots, lines, etc. within the \(FGD_{roi}\). This means, the size of histogram is depending on the value of P. To enrich texture information, we extract feature histograms at different resolutions which can be achieved by concatenating histograms using different values of R and P. In this paper resolution means the use of different radii of circle (e.g. different window sizes). Figure 3 shows an example of converting neighbouring pixels to a LQP code and binary code, resulting to four binary patterns.
Figure 4 shows an example of the feature extraction process using multiresolution LQP operators. Note that each resolution produces four binary patterns (see Fig. 3) resulting to four histograms. Subsequently these four histograms are concatenated into a single histogram which represents the feature occurrences of a single resolution. The illustration in Fig. 4 uses three resolutions producing three histograms and finally concatenated to be a single histogram as the final representation of the feature occurrences in the multiresolution approach.
Figure 5 shows four images of binary patterns generated based on the conditions in Eqs. (2), (3), (4) and (5). The LQP code image is generated using the condition in Eq. (6). All histograms from binary patterns are concatenated to produce a single histogram. Similar to LBP and LTP approaches, the LQP approach achieves rotation invariant by rotating the orientation (\(\theta \)) of the circle’s neighbourhood in a clockwise direction as shown in Fig. 6.
2.3 Dominant Patterns
Selecting dominant patterns is an important process for dimensionality reduction purposes due to the large number of features (e.g. concatenation of several histograms). According to Guo et al. [19] a dominant patterns set of an image is the minimum set of pattern types which can cover \(n (0<n<100)\) of all patterns of an image. In other words, dominant patterns are patterns that frequently occurred (or have high occurrence) in training images. Therefore, to find the dominant patterns we apply the following procedure. Let \(I_{1}, I_{2} ... I_{j}\) be images in the training set. Firstly, we compute the multiresolution histogram feature (\(H^{LQP}_{I_{j}}\)) for each training image. Secondly, we perform a bin-wise summation for all the histograms to find the pattern’s distribution from the training set. Subsequently, the resulting histogram (\(H^{LQP}\)) is sorted in descending order and the patterns corresponding to the first D bins are selected. Where D can be calculated using the following equation:
where N is the total number of patterns and n is the threshold value set by the user. For example \(n=97\) means removing patterns which have less than 3% occurrence in \(H^{LQP}\). This means only the most frequently occurring patterns will be retained for training and testing.
2.4 Classification
Once the feature extraction was completed, the Support Vector Machine (SVM) was employed as our classification approach using a polynomial kernel. The GridSearch technique was used to explore the best two parameters (complexity (C) and exponent (e)) by testing all possible values of C and e (\(C=1,2,3...10\) and \(e=1,2,3...5\) with 1.0 interval) and selecting the best combination based on the highest accuracy in the training phase. The SVM classifier was trained and in the testing phase, each unseen \(FGD_{roi}\) from the testing set is classified as BIRADS I, II, III or IV.
3 Experimental Results
To test the performance of the method, we used the Mammographic Image Analysis Society (MIAS) database [17] which consists of 322 mammograms of 161 women. Each image contains BIRADS information (e.g. BIRADS class I, II, III or IV) provided by an expert radiologist. A stratified ten runs 10-fold cross validation scheme was employed, where the patients are randomly split into 90% for training and 10% for testing and repeated 100 times. The metric accuracy (Acc) is used to measure the performance of the method which represents the total number of correctly classified images compared to the total number of images. We evaluate the method using three different parameters combinations (\(LQP_{(R,P)}\)): (a) Small multiresolution (\(LQP^{small}_{((1,8)+(2,12)+(3,16))}\)) (b) Medium multiresolution (\(LQP^{medium}_{((5,10)+(7,14)+(9,18))}\)) and (c) Large multiresolution (\(LQP^{large}_{((11,16)+(13,20)+(15,24))}\)). For the threshold values we have investigated several combinations of \(\tau _{\in \{1, 2\}}\) and found \(\tau _{1}=5\) and \(\tau _{2}=12\) produced the best accuracy for all experiments. However, for the sake of comparison we will report the performance of the method using \([\tau _{1}=4, \tau _{2}=9]\), \([\tau _{1}=3, \tau _{2}=13]\) and \([\tau _{1}=5, \tau _{2}=15]\). We will also show the effect on performance when varying n from 90 to 99.9 with 0.1 interval.
We present the quantitative results of the proposed method in Fig. 7 using small, medium and large multiresolution approaches which show that \(LQP^{medium}\) outperformed \(LQP^{small}\) and \(LQP^{large}\) regardless of the number of dominant patterns selected with the best accuracy of \(84.91\%\) \((n=99.2)\) followed by the small multiresolution approach of 81.81% (n = 98.9).
Figure 8, shows quantitative results of \(LQP_{((5,10)+(7,14)+(9,18))}\) using the following thresholds (\([\tau _{1}, \tau _{2}]\)): [5, 12], [4, 9], [3, 13] and [5, 15] (note that these parameters are determined empirically). It can be observed that threshold values of \([\tau _{1}=5, \tau _{2}=12]\) produced the best accuracy of 84.91% with \(n=99.2\) followed by \([\tau _{1}=5, \tau _{2}=15]\) with \(n=96.8\). The other threshold values still produced good results in comparison to most of the proposed methods in the literature. For performance comparison when extracting features from the whole breast (wb), as majority of the current studies have done, compared to extracting features from \(FGD_{roi}\) only we conducted two experiments both using \(LQP_{((5,10)+(7,14)+(9,18))}\) with \([\tau _{1}=5, \tau _{2}=12]\). Results can be seen in Fig. 9 which suggests that textures from the fibroglandular disk region are sufficient to differentiate breast density. In fact, it produced classification results of between 5–8% better depending on the value of n. Extracting features from the whole breast produced up to 77.88% accuracy with \(n=97.2\) which is \(7\%\) less than when feature extracted from the \(FGD_{roi}\) only.
To investigate the effect on the performance at different orientations (\(\theta \)), we conducted eight experiments by varying \(\theta \) clockwise rotation with the following values: \(0^{\circ }\), \(45^{\circ }\), \(90^{\circ }\), \(135^{\circ }\), \(180^{\circ }\), \(225^{\circ }\), \(270^{\circ }\) and \(315^{\circ }\). The multiresolution approach for \(LQP_{((5,10)+(7,14)+(9,18))}\) was applied with [\(\tau _{1}=5, \tau _{2}=12\)]. Figure 10 shows experimental results on eight different orientations chosen in this study which revealed that \(LQP_{((5,10)+(7,14)+(9,18))}\) with \(\theta =270^{\circ }\) produced the best accuracy of \(85.6\%\) which indicates density patterns are more visible at this orientation followed by \(\theta =180^{\circ }\) with accuracy \(85\%\). Overall results suggest that multiresolution LQP can produce consistent results (>83%) regardless ofthe parameter \(\theta \) with \(97\le n \le 99.5\).
For quantitative comparison with the other methods in the literature we selected those studies that have used the MIAS database [17], four-class classification, and using the same evaluation technique (10-fold cross validation) as in this study to minimise bias. The proposed method achieved up to 85.60% accuracy which is better than the methods proposed by Muštra et al. [6] (79.3%), Chen et al. [8, 10] (59%, 70%, 72%, 75% and 76%), Bovis and Singh [4] (71.4%), and He et al. [12] (70%). However, the methods of Parthaláin et al. [7] and Oliver et al. [3] achieved 91.4% and 86%, respectively. In comparison to the other popular local statistical features such as LBP and textons, Chen et al. [8] reported the best accuracy achieved by these methods as 59% and 75%, respectively using the same evaluation approach and dataset. Recently, Rampun et al. [20] obtained over 82% accuracy using LTP operators, whereas the proposed method achieved up to 85.6%. In this study features were extracted only from one orientation (e.g. \(270^{\circ }\)) resulting in a smaller number of features whereas the method in [20] extracted features from eight different orientations and concatenated them, resulting in a large number of features. Furthermore, this study conducted feature selection by taking of account dominant patterns only which reduced the number of features significantly compared to the study in [20].
4 Conclusion
In conclusion, we have presented and developed a breast density classification method using multireslution LQP operators applied only within the fibroglandular disk area which is the most prominent region of the breast instead of the whole breast region as suggested in current studies [3,4,5, 7,8,9,10,11,12,13]. The multiresolution LQP features are robust in comparison to the other methods such as LBP, texton based approaches and LTP due to the five encoding system which generates more texture patterns. Moreover the multiresolution approach provides complementary information from different parameters which cannot be captured in a single resolution. The proposed method produced competitive results compared to some of the best accuracies reported in the litearture. For future work, we plan to develop a method that can automatically estimate \(\tau _{1}\) and \(\tau _{2}\) as well as combining multiresolution LQP features with features from the texton approach.
References
Cancer Research UK: Breast cancer statistics (2014). http://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/breast-cancer. Accessed 6 Jan 2017
Breast Cancer: U.S. Breast Cancer Statistics (2016). http://www.breastcancer.org/symptoms/understand_bc/statistics. Accessed 6 Jan 2017
Oliver, A., Freixenet, J., Martí, R., Pont, J., Perez, E., Denton, E.R.E., Zwiggelaar, R.: A novel breast tissue density classification methodology. IEEE Trans. Inf Technol. Biomed. 12(1), 55–65 (2008)
Bovis, K., Singh, S.: Classification of mammographic breast density using a combined classifier paradigm. In: 4th International Workshop on Digital Mammography, pp. 177–180 (2002)
Oliver, A., Tortajada, M., Lladó, X., Freixenet, J., Ganau, S., Tortajada, L., Vilagran, M., Sentś, M., Martí, R.: Breast density analysis using an automatic density segmentation algorithm. J. Digit. Imaging 28(5), 604–612 (2015)
Muštra, M., Grgić, M., Delać, K.: A Novel breast tissue density classification methodology. Breast density classification using multiple feature selection. Automatika 53(4), 362–372 (2012)
Parthaláin, N.M., Jensen, R., Shen, Q., Zwiggelaar, R.: Fuzzy-rough approaches for mammographic risk analysis. Intell. Data Anal. 14(2), 225–244 (2010)
Chen, Z., Denton, E., Zwiggelaar, R.: Local feature based mamographic tissue pattern modelling and breast density classification. In: The 4th International Conference on Biomedical Engineering and Informatics, pp. 351–355 (2011)
Bosch, A., Munoz, X., Oliver, A., Martí, J.: Modeling and classifying breast tissue density in mammograms. In: Computer Vision and Pattern Recognition (CVPR 2006), pp. 1552–1558 (2006)
Chen, Z., Oliver, A., Denton, E., Zwiggelaar, R.: Automated mammographic risk classification based on breast density estimation. In: Sanches, J.M., Micó, L., Cardoso, J.S. (eds.) IbPRIA 2013. LNCS, vol. 7887, pp. 237–244. Springer, Heidelberg (2013). doi:10.1007/978-3-642-38628-2_28
Byng, J.W., Boyd, N.F., Fishell, E., Jong, R.A., Yaffe, M.J.: Automated analysis of mammographic densities. Phys. Med. Biol. 41(5), 909–923 (1996)
He, W., Denton, E., Stafford, K., Zwiggelaar, R.: Mammographic image segmentation and risk classification based on mammographic parenchymal patterns and geometric moments. Biomed. Sig. Process. Control 6(3), 321–329 (2011)
Petroudi, S., Kadir, T., Brady, M.: Automatic classification of mammographic parenchymal patterns: a statistical approach. In: Proceedings IEEE Conference Engineering in Medicine Biology Society, vol. 1, pp. 798–801 (2003)
Ojala, T., Pietikainen, M., Maenpaa, T.: Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 24(7), 971–987 (2002)
Nanni, L., Luminia, A., Brahnam, S.: Local binary patterns variants as texture descriptors for medical image analysis. Artif. Intell. Med. 49(2), 117–125 (2010)
Tan, X., Triggs, B.: Enhanced local texture feature sets for face recognition under difficult lighting conditions. In: Analysis and Modelling of Faces and Gestures, pp. 168–182 (2007)
Suckling, J., et al.: The mammographic image analysis society digital mammogram database. In: Proceedings of Exerpta Medica. International Congress Series, pp. 375–378 (1994)
Rampun, A., Morrow, P.J., Scotney, B.W., Winder, R.J.: Fully automated breast boundary and pectoral muscle segmentation in mammograms. Artif. Intell. Med. (2017, under review)
Gio, Y., Zhao, G., Pietikäinen, M.: Discriminative features for feature description. Pattern Recogn. 45, 3834–3843 (2012)
Rampun, A., Winder, R.J., Morrow, P.J., Scotney, B.W.: Breast density classification in mammograms using local ternary patterns. In: International Conference on Image Analysis and Recognition (2017)
Acknowledgments
This research was undertaken as part of the Decision Support and Information Management System for Breast Cancer (DESIREE) project. The project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 690238.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Rampun, A., Morrow, P., Scotney, B., Winder, J. (2017). Breast Density Classification Using Multiresolution Local Quinary Patterns in Mammograms. In: Valdés Hernández, M., González-Castro, V. (eds) Medical Image Understanding and Analysis. MIUA 2017. Communications in Computer and Information Science, vol 723. Springer, Cham. https://doi.org/10.1007/978-3-319-60964-5_32
Download citation
DOI: https://doi.org/10.1007/978-3-319-60964-5_32
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-60963-8
Online ISBN: 978-3-319-60964-5
eBook Packages: Computer ScienceComputer Science (R0)