Abstract
With the greater importance of parallel architectures such as GPUs or Xeon Phi accelerators, the scientific community has developed efficient solutions in the bioinformatics field. In this context, FPGAs begin to stand out as high performance devices with moderate power consumption. This paper presents and evaluates a parallel strategy of the well-known Smith-Waterman algorithm using OpenCL on Intel/Altera’s FPGA for long DNA sequences. We efficiently exploit data and pipeline parallelism on a Intel/Altera Stratix V FPGA reaching upto 114 GCUPS in less than 25 watt power requirements.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Field Programmable Gate Array
- Graphic Processor Unit
- Field Programmable Gate Array Implementation
- Latency Memory Access
- Dedicated Memory
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
In recent years, genome research projects have produced a vast amount of biological data. In fact, biologists are working in conjunction with computer scientists to extract relevant biological information from these experiments. The comparison of millions of sequences [13] is one of the most useful mechanisms known in Bioinformatics, commonly solved by heuristic methods.
Smith-Waterman (SW) algorithm compares two sequence in an exact way and corresponds to the so-called local methods because it focuses on small similar regions only. Besides, this method has been used as the basis for many subsequent algorithms and is often used as basic pattern to compare different alignment techniques. However, one of the main drawbacks is the cost of this approach in computing time and memory requirements which makes it unsuitable in some cases.
Regarding on the performance aspect, many approaches, such as BLAST [1] and FASTA [14] considerably reduce execution time at the expense of not guaranteeing the optimal result. Nevertheless, accelerating the SW algorithm is still a great challenge for the scientific community. SW is usually used to align two DNA sequences or a protein sequence to a genomic database. A single matrix must be built for each alignment and the matrix size depends on sequence lengths. In DNA alignment, the matrix can be huge since these sequences can have upto hundreds of million nucleotides. As protein sequences are shorter, multiple small matrices are usually computed simultaneously since they are independent among them [16].
In the last years, we have witnessed attempts to parallelize for both schemes. These efforts reduce SW execution time through the exploitation of High-Performance Computing (HPC) architectures. However, most implementations focus on short sequences, particularly protein sequences [15]. For very long sequences, as the DNA case, few implementations are nowadays available. In the hardware accelerators scenario, we highlight SW# [9] and CUDAlign [18] (and its newer versions [4, 5]) that focus on the alignment of huge DNA sequences with multi CUDA-enabled Graphics Processor Units (GPUs). Meanwhile, Liu et al. have presented SWAPHI-LS [12] a highly optimized hand-tuned implementation for Intel Xeon Phi accelerators. In addition, several proposals based on Field Programmable Gate Array (FPGA) speedup sequence alignments [2, 22] in the context of DNA comparison. Moreover, Wienbrandt presents a study of multi-FPGAs version in [21].
Nowadays, HPC capabilities are changing in data-centers scenario. FPGAs are being integrated with CPUs due to accelerators consolidation in HPC community as a way of improving performance while keeping power efficiency. Recently, Microsoft announced that their data-centers equipped with FPGAs increase dramatically Bing’s engine capacities, and they have also incorporated them to Azure ecosystem [6]. In the same way, Amazon has included FPGAs in its Amazon Web Services [10]. With the acquisition of Altera in 2015, Intel plans to incorporate FPGA capabilities in the next Xeon’s server processors because they expect to be used by at least 30% of data-center in next years [11].
Our paper proposes and evaluates a SW implementation, capable of aligning DNA sequences of unrestricted size, for Altera’s FPGA. Unlike previous works on literature, we employed the novel OpenCL paradigm on FPGAs. Although Altera’s developers promote a similar implementation in [19], no-real sequence data of fixed, limited size (n = 256 residues) are used. As this issue can radically differ from real bioinformatic contexts, the observed behavior becomes unpredictable for these scenarios, especially in alignment of long DNA sequences. We would like to point out that, unlike GPU or Xeon Phi accelerators which need to be purchased separately, the hardware used in this study will be available soon in the next processor generation at null cost. In that sense, this study can be taken as starting point for future hybrid CPU-FPGA solutions.
The rest of the paper is organized as follows. Section 2 describes SW algorithm. Section 3 introduces Altera’s OpenCL programming extension while Sect. 4 addresses our parallelization of the SW algorithm using OpenCL on FPGAs. Section 5 presents experimental results and finally Sect. 6 outlines conclusions and future lines of work for this novel viability study.
2 Smith-Waterman Algorithm
The SW algorithm is used to obtain the optimal local alignment between two sequences and was proposed by Smith and Waterman [20]. This method employs a dynamic programming approach and its high sensitivity comes from exploring all the possible alignments between two sequences.
Given two sequences \(S_1\) and \(S_2\), with sizes \(|S_1|=m\) and \(|S_2|=n\), the recurrence relations for the SW algorithm with affine gap penalties [7] are defined below.
\(H_{i,j}\) contains the score for aligning the prefixes \(S_1[1..i]\) and \(S_2[1..j]\). \(E_{i,j}\) and \(F_{i,j}\) are the scores of prefix \(S_1[1..i]\) aligned to a gap and prefix \(S_2[1..j]\) aligned to a gap, respectively. SM is the scoring matrix which defines match/mismatch scores between nucleotides. \(G_{oe}\) is the sum of gap open and gap extension penalties while \(G_{e}\) is the gap extension penalty. The recurrences should be calculated with \(1 \le i \le m\) and \(1 \le j \le n\), after initialising H, E and F with 0 when \(i = 0\) or \(j = 0\). The maximum value in the alignment matrix H is the optimal local alignment score.
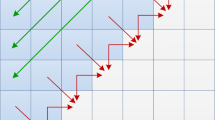
It is important to note that any cell of the matrix H can be computed only after the values of the upper, left and upper-left cells are known, as shown in Fig. 1. These dependences restrict the ways in that H can be computed.
3 OpenCL Extension on Altera’s FPGA
OpenCL is a framework for developing parallel programs across heterogeneous platforms. It is currently supported by several hardware devices such as CPUs, GPUs, DSPs and FPGAs. The OpenCL is based on host-device model, where host is in charge of managing device memory, transferring data from/to device and launching the kernel code.
Kernel corresponds to a piece of code which expresses the parallelism of a program. OpenCL programming model divides a program workload into work-groups and work-items. Usually, in the denoted data parallel programming model, work-items are grouped into work-groups, which are executed independently on a processing element. Data-level parallelism is ordinarily exploited in SIMD way, where each work-item is mapped to a lane width of the target device.
OpenCL memory model uses a particular memory hierarchy which is also characterized by the access type, performance and scope. Global memory is read-write accessible by all work-items which implies a high latency memory access. Local memory is a shared read-write memory that can be accessed from all work-items of a single work-group. Besides, it usually involves a low latency memory access. Constant memory is a read-only memory visible by all work-items across all work-groups, and private memory as the name suggests it is only accessible by a single work-item.
OpenCL allows a programmer to express parallelism abstracting the target platform details. We can highlight portability and reduction in development time as the main advantages. FPGAs permit programming networks composed of logic elements, memory blocks and specific DSP blocks. In order to verify and create digital designs, Hardware Description Languages (HDLs) are generally used, which are complex, error prone and affected by an extra abstraction layer as they contain the additional concept of time.
Regarding the execution model, Altera’s OpenCL SDK [3] recommends the use of task parallel programming model, where the kernel consists of a single work-group with a unique work-item. The Altera OpenCL Compiler (AOC) is capable of extracting parallelism from each loop iteration in a loop-pipelined way allowing to process the loop in a high-throughput fashion.
Altera’s OpenCL extension also take advantage of I/O channels and kernel channels as in OpenCL 2.0 by means of pipes [8]. Altera’s channel extension allows to transfer data between work-item’s in the same kernel or between different kernels without host interaction.
4 SW Implementation
In this section we will address the programming aspects and optimizations applied to our implementations on FPGA accelerated platforms. Algorithm 1 shows the pseudo-code for the host implementation. Memory management is performed in OpenCL by means of clCreateBuffer (memory allocation and initialization) and clEnqueueReadBuffer (memory transfer to host). Kernels are invoked through the clEnqueueTask function.


The kernel is implemented following the task parallel programming model mentioned in Sect. 3. Algorithm 2 shows the pseudo-code for our kernel. The alignment matrix is divided into vertical blocks (BW means Block Width) and each block is computed in row-by-row manner: from top to bottom, left to right direction (see Fig. 2). Besides improving the data locality, this blocking technique reduces the memory requirements for block execution, which favors the exploitation of the private low-latency memory. In that sense, we employed two buffers to store one row for matrices H and F. Additionally, both sequences are partially copied to private memory.
Fully unrolling of the inner loop represents an essential aspect of this kernel from performance point of view. This technique allows the AOC to exploit loop instruction pipelining and, in consequence, more operations per clock cycle are performed. As the compiler needs to know the number of iterations at compile phase, \(S_2\) sequence must be extended with dummy symbols to make its length a multiple of fixed BW value.
Due to the data dependencies mentioned in Sect. 2, each block needs the last column H and E values of the previous block. Global memory buffers are employed to communicate these data. To avoid read-write dependences in global memory, separate buffers are used: one for reading the values from the previous block and one for writing the values for the next block, so after each kernel invocation buffers are swapped in the host. It is important to mention that, although in OSWALD implementation [17] Altera OpenCL channels are used to exchange these information, the use of this technique is not affordable in DNA context with million of nucleotide bases involved.
Moreover, host-side buffers are allocated to be 64-byte aligned. This fact improves data transfer efficiency by means of Direct Memory Access. Both sequence are transferred when creating the device buffers and optimal score is retrieved after all kernels finished.
5 Experimental Results
5.1 Experimental Platforms and Tests Carried Out
Tests have been performed on different platforms running CentOS (release 6.6):
-
A server with two Intel Xeon CPU E5-2670 8-core 2.60 GHz, 32 GB main memory and an Altera Stratix V GSD5 Half-Length PCIe Board with Dual DDR3 (two banks of 4 GByte DDR3).
-
A server with two Intel Xeon CPU E5-2695 v3 16-core 2.30 GHz, 64 GB main memory and two NVIDIA GPU cards: one Tesla K20c (Kepler architecture, 2496 CUDA cores, 5 GB dedicated memory and Compute Capability 3.5) and one GTX 980 (Maxwell architecture, 2048 CUDA cores, 4 GB dedicated memory and Compute Capability 5.0).
-
A server with two Intel Xeon CPU E5-2695 v3 16-core 2.30 GHz, 128 GB main memory and a single Xeon Phi 3120P coprocessor card (57 cores with 4 hw thread per core, 6 GB dedicated memory).
We have used the Intel’s ICC compiler (version 16.0.3) with the -O3 optimization level by default. The synthesis tool used is Quartus II DKE V12.0 2 with OpenCL SDK v14.0 and the CUDA SDK version is 7.5.
To provide the most relevant study, tests were made with the real DNA sequences retrieved from the National Center for Biotechnology Information (NCBI)Footnote 1, ranging from thousands to millions of nucleotide bases. The accession numbers and sizes of the sequences are presented in Table 1. Also, for the sake of validation, optimal alignment scores are included in Table 1. The scoring scheme used was: +1 for match; \(-3\) for mismatch; \(-5\) for gap open; and \(-2\) for gap extension. Each particular test was run ten times and performance was calculated with the average of ten execution times to avoid variability.
5.2 Performance and Resource Usage Evaluation
The metric GCUPS is used to performance assessment in the Smith-Waterman scenario [18]. In order to evaluate FPGA performance rates, we have considered different kernel implementations according to integer data type and BW value. We detail below the main differences:
-
The name prefix denotes the integer data type used; i.e. int, short and char represent 32, 16 and 8 bit integer data types, respectively.
-
The name suffix denotes the BW value used; e.g. bw32 means that BW value was set to 32.
Table 2 presents FPGA resource utilization and the performance achieved for our OpenCL kernels implementations. Larger BW means better performance but also higher resource consumption. Adaptive logic modules (ALMs) are the most influenced resources; registers (Regs) and RAM blocks (RAMs) are slightly increased while DSP blocks (DSPs) stay intact. In spite of this fact, there are still available resources that allow increasing BW parameter. Nevertheless, larger values could not be used because AOC reports non-real read-write dependences in private memory associated to H and F matrices. In fact, these false dependences reduce the number of operations per clock cycle decreasing considerably performance rates achieved.
Regarding integer data type, as can be observed, smaller data type not only improves performance but also decreases resource consumption. This behavior is clearly exposed when comparing int_bw256 and short_bw256 kernels: using the same BW configuration, short_bw256 reports an increase from up to 1.22\({\times }\) in performance with a reduction from up to 0−0.28\({\times }\) in resource usage against to int_bw256 counterpart. A similar behavior is observed with short_bw512 and char_bw512 kernels: char_bw512 presents an increase from up to 1.28\({\times }\) in performance with a reduction from up to 0−0.36\({\times }\) in resource usage respect to short_bw512. However, it is also important to take into account that the use of narrower integer data types also involves an important reduction in representation range. Due to this fact, there are three alignment scores out of ten that can not be represented when using 16 bit integer data. It is also observed for the experiments with 8-bits data type where only three experiments can be carried outFootnote 2.
From sequence length point of view, all kernels benefit from larger workloads regardless of sequences similarity. The best performances achieved are 37.68, 74 and 113.78 GCUPS for int, short and char kernels, respectively.
5.3 Performance and Power Efficiency Comparison with Other SW Implementations
In this subsection our developed version is compared with other optimized SW implementations in the HPC scenario: the Xeon Phi-based SWAPHI-LS program (v1.0.12) [12] and the GPU-based SW# [9] and CUDAlign (v3.9.1.1024) [4] programs. We would like to note that both GPU implementations were configured to perform only their score version.
Table 3 presents performances for the SWAPHI-LS, SW# and CUDAlign implementations. Comparing our FPGA implementation against Xeon-Phi accelerator, SWAPHI-LS yields an average performance of 25.89 GCUPS and a peak of 34.38 GCUPS being outperformed by int_bw256 version in all cases. In particular, the most impressive performance difference occurs for smaller matrix sizes where int_bw256 runs on average 21.8\({\times }\) faster. For the rest of the tests, the performance gain decreases but still improves on average 1.45\({\times }\).
Unlike our FPGA kernels, both GPU implementations are sensitive to sequences similarity; better results are obtained on alignments with higher scores. On Tesla K20c, SW# achieves an average performance of 44.38 GCUPS and a maximum performance of 90.15 GCUPS, outperforming CUDAlign by a factor of 1.78\({\times }\) on average. int_bw256 improves SW# on the three shortest alignments while the latter runs 1.62\({\times }\) faster on average than the former on the largest ones. In contrast, SW# only beats CUDAlign on the four shortest alignments on GTX 980. CUDAlign reports 71.23 GCUPS on average and a peak of 163.77 GCUPS, achieving an average speedup of 1.24\({\times }\) on the six largest matrix sizes. Just as the previous case, int_bw256 improves both GPU implementations on the three shortest alignments while is outperformed in the rest. In that sense, CUDAlign runs 2.76\({\times }\) faster on average than int_bw256.
In FPGA context, a theoretical comparison with other OpenCL implementations is the only possibility since the absence of available source codes. We would like to note that the Altera staff implementation [19] uses 32-bit integer data type and imposes a fixed, limited size (n = 256 residues) to \(S_2\) sequence, which substantially differs from the common ones used in DNA analysis. In spite of this fact, Altera’s staff implementation reported a peak of 24.7 GCUPS on Stratix V meanwhile our approach achieves an average of 33.8 GCUPs.
In order to complete this study, we have also analyzed the power efficiency. Table 4 presents power efficiency ratios considering the GCPUS peak performance and Thermal Design Power (TDP) in each accelerator. It can be seen that the worst ratio observed is for SWAPHI-LS due to low performance rates and high TDP on the Xeon Phi. GPU implementations place at an intermediate position because they obtain the highest performance peak but at the expense also of higher power requirements. As expected, both GPU implementations obtain better GCUPS/Watts ratios on GTX 980 in comparison with Tesla K20c since its better performance and power capacities. We would like to remark that FPGA implementation reaches the best GCUPS/Watts ratios. Despite not achieving the highest performance peak, its low power consumption leads to the best choice taking this aspect into account. Further, the lowest performance FPGA kernel considering integer data type (int_bw256) outperforms SWAPHI-LS implementation by a factor of 11.62\({\times }\) and to the GPU implementations by a range of 1.56–3.78\({\times }\). It is important to mention that if this analysis is carried out considering average GCUPS instead of GCUPS peak, larger differences in favor of FPGA implementations will be found.
6 Conclusions
In this paper, we addressed the benefits of a parallel SW implementation using OpenCL on Intel/Altera’s FPGA, not only from performance perspective but also from power efficiency point of view. To the best of the author’s knowledge, this is the first time that a paper examines an implementation of this kind with real, long DNA sequences. In addition, as Intel will incorporate FPGA capabilities in its next Xeon processors in a free manner, this study can be useful as an starting point for future hybrid CPU-FPGA implementations.
Among main contributions of this research we can summarize:
-
Data type exploitation is crucial to achieve successful performance rates. Narrower data-types reports better performance rates with less resource impact, but at expense of decreasing representation width. In fact, a peak of 114 GCUPS is reached when using 8 bit integers.
-
From performance perspective, our most successful 32-bit implementation reaches 37.7 GCUPS peak, running 1.53\({\times }\) faster than Altera’s staff implementation [19]. Despite being competitive in performance terms respect to other solutions on accelerators (such as GPUs or Xeon Phi), our implementation is significantly better from power efficiency perspective. In particular, the fastest 32-bit FPGA kernel outperforms SWAPHI-LS by a factor of 11.62\({\times }\) and the GPU implementations by a range of 1.56–3.78\({\times }\) in efficiency.
Taking into account these encouraging results, as future lines we will consider three aspects:
-
As all kernels developed still have free available resources, we will try to exploit them to improve performance rates. On one hand, we expect to solve performance limitations imposed by AOC with larger BW values. On the other hand, we plan to combine distinct integer data width kernels to explore different configurations in order to find the best performance-representation width trade-off.
-
As OpenCL allows multiple devices exploitation, we would like to extend this work to a multi-FPGA implementation and explore most successful workload distribution.
-
As real power consumption on accelerators can differ from TDP values due to a variety of reasons, we plan to measure the instant power consumption in all devices in order to make a more fairly performance vs. power analysis.
Notes
- 1.
Sequences are available in http://www.ncbi.nlm.nih.gov.
- 2.
The symbol ‘-’ indicates an alignment that can not be computed because the optimal score exceeds the corresponding maximum value.
References
Altschul, S.F., Madden, T.L., Schffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J.: Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acid Res. 25(17), 3389–3402 (1997)
Caffarena, G., Pedreira, C.E., Carreras, C., Bojanic, S., Nieto-Taladriz, O.: FPGA acceleration for DNA sequence alignment. J. Circuits Syst. Comput. 16(2), 245–266 (2007)
Altera Corporation: Altera SDK for OpenCL Programming Guide, v14.0 (2014)
de Oliveira Sandes, E.F., Miranda, G., Alves de Melo, A.C.M., Martorell, X., Ayguadé, E.: CUDAlign 3.0: parallel biological sequence comparison in large GPU clusters. In: CCGRID, pp. 160–169. IEEE Computer Society (2014)
de Oliveira Sandes, E.F., Miranda, G., Martorell, X., Ayguad, E., Teodoro, G., Alves de Melo, A.C.M.: CUDAlign 4.0: incremental speculative traceback for exact chromosome-wide alignment in GPU clusters. IEEE Trans. Parallel Distrib. Syst. 27(10), 2838–2850 (2016)
Feldman, M.: Microsoft goes all in for FPGAs to build out AI cloud (2016). https://www.top500.org/news/microsoft-goes-all-in-for-fpgas-to-build-out-cloud-based-ai/
Gotoh, O.: An improved algorithm for matching biological sequences. J. Mol. Biol. 162, 705–708 (1981)
Khronos Group: The OpenCL Specification, version 2 (2014)
Korpar, M., Sikic, M.: SW# - GPU-enabled exact alignments on genome scale. Bioinformatics 29(19), 2494–2495 (2013)
Leopold, G.: AWS Embraces FPGAs, Elastic GPUs (2016). https://www.hpcwire.com/2016/12/02/aws-embraces-fpgas-elastic-gpus/
Leopold, G.: Intels FPGAs target datacenters, networking (2016). https://www.hpcwire.com/2016/10/06/intels-fpgas-target-datacenters-networking/
Liu, Y., Tran, T.T., Lauenroth, F., Schmidt, B.: SWAPHI-LS: Smith-Waterman Algorithm on Xeon Phi coprocessors for long DNA sequences. In: IEEE International Conference on Cluster Computing (CLUSTER), pp. 257–265 (2014)
Mount, D.W.: Bioinformatics: Sequence and Genome Analysis. Cold Spring Harbor Laboratory Press, Mount (2004)
Pearson, W.R., Lipman, D.J.: Improved tools for biological sequence comparison. Proc. Nat. Acad. Sci. U.S.A. 85(8), 2444–2448 (1988)
Rucci, E., García, C., Botella, G., Giusti, A., Naiouf, M., Prieto-Matías, M.: State-of-the-Art in Smith–Waterman protein database search on HPC platforms. In: Wong, K.-C. (ed.) Big Data Analytics in Genomics, pp. 197–223. Springer, Cham (2016). doi:10.1007/978-3-319-41279-5_6
Rucci, E., Garca, C., Botella, G., De Giusti, A., Naiouf, M., Prieto-Matas, M.: An energy-aware performance analysis of SWIMM: SmithWaterman implementation on Intel’s multicore and manycore architectures. Concurrency Comput. Pract. Exp. 27(18), 5517–5537 (2015)
Rucci, E., Garca, C., Botella, G., De Giusti, A., Naiouf, M., Prieto-Matas, M.: OSWALD: OpenCL Smith-Waterman Algorithm on Altera FPGA for large protein databases. Int. J. High Perform. Comput. Appl. (2016). doi:10.1177/1094342016654215
de Oliveira Sandes, E.F., Alves de Melo, A.C.M: CUDAlign: using GPU to accelerate the comparison of megabase genomic sequences. In: Proceedings of the 15th ACM SIGPLAN Symposium on Principles and Practice of Parallel Computing, PPoPP 2010, pp. 137–146. ACM, New York (2010)
Settle, S.O.: High-performance dynamic programming on FPGAs with OpenCL. In: IEEE High Performance Extreme Computing Conference (HPEC 2013), pp. 1–6 (2013)
Smith, T.F., Waterman, M.S.: Identification of common molecular subsequences. J. Mol. Biol. 147(1), 195–197 (1981)
Wienbrandt, L.: Bioinformatics applications on the FPGA-based high-performance computer RIVYERA. In: Vanderbauwhede, W., Benkrid, K. (eds.) High-Performance Computing Using FPGAs, pp. 81–103. Springer, New York (2013). doi:10.1007/978-1-4614-1791-0_3
Yamaguchi, Y., Tsoi, H.K., Luk, W.: FPGA-based Smith-Waterman Algorithm: analysis and novel design. In: Koch, A., Krishnamurthy, R., McAllister, J., Woods, R., El-Ghazawi, T. (eds.) ARC 2011. LNCS, vol. 6578, pp. 181–192. Springer, Heidelberg (2011). doi:10.1007/978-3-642-19475-7_20
Acknowledgments
This work has been partially supported by Spanish government through research contract TIN2015-65277-R and CAPAP-H5 network (TIN2014-53522).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Rucci, E., Garcia, C., Botella, G., De Giusti, A., Naiouf, M., Prieto-Matias, M. (2017). Accelerating Smith-Waterman Alignment of Long DNA Sequences with OpenCL on FPGA. In: Rojas, I., Ortuño, F. (eds) Bioinformatics and Biomedical Engineering. IWBBIO 2017. Lecture Notes in Computer Science(), vol 10209. Springer, Cham. https://doi.org/10.1007/978-3-319-56154-7_45
Download citation
DOI: https://doi.org/10.1007/978-3-319-56154-7_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-56153-0
Online ISBN: 978-3-319-56154-7
eBook Packages: Computer ScienceComputer Science (R0)