Abstract
We design an evolutionary heuristic for the combinatorial problem of de-novo DNA assembly with short, overlapping, accurately sequenced single DNA reads of uniform length, from both strands of a genome without long repeated sequences. The representation of a candidate solution is a novel segmented permutation: an ordering of DNA reads into contigs, and of contigs into a DNA scaffold. Mutation and crossover operators work at the contig level. The fitness function minimizes the total length of scaffold (i.e., the sum of the length of the overlapped contigs) and the number of contigs on the scaffold. We evaluate the algorithm with read libraries uniformly sampled from genomes 3835 to 48502 base pairs long, with genome coverage between 5 and 7, and verify the biological accuracy of the scaffolds obtained by comparing them against reference genomes. We find the correct genome as a contig string on the DNA scaffold in over 95% of all assembly runs. For the smaller read sets, the scaffold obtained consists of only the correct contig; for the larger read libraries, the fitness of the solution is suboptimal, with chaff contigs present; however, a simple post-processing step can realign the chaff onto the correct genome. The results support the idea that this heuristic can be used for consensus building in de-novo assembly.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Sequencing technologies have increasingly low cost and feasible runtimes; they can generate deep coverage of mammal genomes within days, such that many new projects attempt to sequence previously unsequenced organisms. The raw sequence data generated in such a project is a set of overlapping, either single or paired-end DNA reads, from either of the two reverse-complementary strands of a DNA molecule. Single reads are either short (\(10^2\) bases for most available second-generation sequencers, with high single-read accuracy: 99.9% for Illumina dye sequencing [1]) or long (\(10^4\) bases with lower single-read accuracy: 80–90% for third-generation SMRT [2] and MinION sequencing [3], with predominantly insertion and deletion errors). Short-read second-generation sequencers are in wide use; a sufficient number of overlapping short reads need to be obtained in order to cover the underlying genome multiple times over. This coverage c is the average coverage of each base on the DNA strand, \(c=n\cdot r/G\), where n is the number of reads, r the read length, and G the expected length of the genome.
We look at the problem of assembling accurate, single, short DNA reads with uniform length and a medium depth of coverage of the original genome. The overlapping DNA reads (essentially, strings over a four-letter alphabet) must be totally linearly ordered such that each pair of adjacent reads overlaps. Continuously overlapping sequences of short reads form a DNA contig, and the contigs obtained are ordered on a DNA scaffold which equates the genome. This is a combinatorial problem, yielding a computationally hard assembler; the larger the genome sequenced and the deeper its coverage, the larger the read set in input and the more difficult the computation. Some genomes and read sets are comparatively more difficult to assemble than others; current DNA assembling algorithms are themselves not optimal, and may obtain different solutions for the same input [4,5,6].
De-novo assembly refers to sequencing DNA reads in the absence of a reference genome, i.e., a closely related genome whose internal structure is essentially the best accuracy metric with which to verify the assembly obtained: the newly assembled genome must align back onto the reference. In absence of a reference genome, only an estimate of the length of the reference genome can replace it as an imperfect accuracy metric. In practice, consensus is used: a newly sequenced genome is considered correct if a number of different assembly algorithms (or parameter settings to the algorithms) computed it. Obtaining consensus is effectively the current accuracy metric in absence of a reference genome.
We employ all three accuracy metrics above to verify our method: the length of the genome as a fitness function, aligning back to the reference genome as a post-factum evaluator for the correctness of a single assembly, and consensus as a post-factum evaluator for the method as a whole.
The novel points in this study are:
-
Algorithmic. We build on prior work using Evolutionary Computation in DNA assembly. We introduce a new representation for a candidate solution which models the scaffold as a segmented permutation, with DNA contigs as building blocks. The corresponding genetic operators mutate and crossover DNA contigs natively, i.e., operate at macro level rather than over individual reads. This representation closely models a real genome, and allows assembly metrics to be written as computationally simple fitness functions.
-
Fitness functions. A new fitness function is used: the length of the DNA scaffold, together with the number of contigs on the scaffold (to be minimized). This fitness function is the closest option to an accuracy metric for a de-novo problem, and also makes possible to write a stopping condition for the computation when the length of the scaffold matches the estimate length of the reference genome.
-
Correct assemblies. We evaluate this method with read sets uniformly sampled from existing consensus genomes 3835 to 48502 base pairs long and genome coverage between 5 and 7. In over 95% of all assemblies, even with suboptimal fitness, the correct genome is obtained as a contig on a multi-contig scaffold; a simple realignment of the “chaff” contigs in the solution then yields a correct genome.
-
Consensus among assemblies. Since the algorithm often obtains the correct genome across repeated assemblies with different random seeds and parameter settings, the algorithm can serve effectively as a consensus builder for a new genome.
2 Related Work
On a theoretical note, assuming that the shortest assembled genome is desired, the assembly problem has been recast into the shortest common superstring (SCS) problem, i.e., the problem of finding, for n given finite strings \(s_1, s_2, \ldots , s_n\) over an alphabet of size greater than 2, a shortest superstring S such that every string \(s_i\) can be obtained by deleting zero or more elements from S. The SCS problem is known to be NP-complete [7], as is the simpler version of SCS in which the n strings are totally ordered in a superstring [8].
In this section, we summarize results from two research areas: the performance of existing, commercial-grade assemblers, and the closely related prior work on using artificial genetic algorithms to sequence natural genetic material.
2.1 Lessons Learned from Existing Assemblers
Current short-read DNA sequencers implement heuristics which order short DNA reads onto contigs based on maximizing the overlap between adjacent reads, on maximizing the sum of overlaps across the scaffold, and on incorporating the most reads in the final assembly. As summaries of these techniques, we point to recent comparative studies such as [4,5,6], and give an overview of knowledge they gained from these comparative campaigns, which can act as a baseline of expectations for our own method. [4] experimented with 8 assemblers for paired-end short reads, and compared the results against reference genomes. [5] compared 43 assemblies from 21 teams over both short- and long-read, single- and paired-end read sets, without reference genomes. In [6] long-read MinION sequence data was assembled with 4 assemblers from all algorithmic classes.
No Optimal Assembly Strategy Exists. In [4], endemic problems were found across the board: the contiguity of an assembled genome varied wildly among both assemblers and genomes under sequence. When comparing the assemblies obtained to a reference genome, the following issues we found: many “chaff” contigs (of length under twice the read length), unnecessarily duplicated contigs, compressions of true repeat sequences (a widespread problem for short-read assemblers), contig “misjoins” (i.e., the assembler joined two contigs which are in fact distant in the reference genome), many indels (insertions and deletions in the assembly compared to the reference). In [5], a high degree of variability was found among assemblies, to the extent that the result of a single assembler and set of assembler parameters is not to be trusted. This supports the idea of consensus: an assembly obtained by different methods is likelier to be accurate.
Assembly Metrics are Error-prone; Use Accuracy Metrics. The metrics used to evaluate the assembled genome in absence of a reference genome are error-prone [4]; these assembly metrics (e.g., the number of contigs obtained, or various basic statistics over the sizes of the contigs) were found not to correlate very well with accuracy metrics, which compare the assembly against a true reference genome (when available), by computing the degree to which the reference genome is covered by the assembly, and the percentage of contigs alignable onto the reference genome. It remains unclear how to assess the quality of the assembled genomes, because no assembler performed well by all assembly metrics and all genomes [5]. In the evaluation of an assembly, one should not trust a single assembly metric. In [6], assemblies of poor accuracy were seen: for one species, all assemblers obtained very low genome coverage (between 0% and 12%); a greedy assembler had both the genome coverage and the percentage of alignment under 5% for both species.
Some Genomes and Read Sets are More Difficult to Assemble. In [4], the accuracy of the reads had larger negative effect upon the quality of the genome assembled than the assembler itself. A uniform read length r and reads uniformly distributed across the genome both improve the efficiency and accuracy of the assembly [9]. Also, a widespread problem is that genomes with repeated subsequences are more difficult to assemble: repeat sequences longer than the read length may create a gap, a misjoin, or may be compressed [4].
In consequence, we take the following decisions for this study:
-
We simplify the problem to an extent by sequencing genomes or genome segments which are small and have no repeats longer than the read length.
-
We control the shape of the read set by artificially sampling it using a random uniform sampler, from a reference genome.
-
We use accuracy metrics (the reference genome, and consensus among assemblies) to evaluate our method both via the runtime fitness and post-factum.
2.2 Previous Genetic Representations and Operators
A major issue with prior work sequencing DNA using an evolutionary approach is that their fitness functions only model assembly metrics which state nothing about the accuracy of the result (also a widespread issue across other assemblers, per Sect. 2.1). Table 1 summarizes prior work using an evolutionary approach.
[8] builds on genetic algorithms for the TSP problem. If a candidate were to be represented as a permutation of n read identifiers, then classic two-point crossover leads to illegal offspring, so a complicated individual representation is used, not directly linked to the order in which the reads are placed in the assembly. In the evaluation, it was found that this candidate representation doesn’t construct increasingly improved solutions, and is thus not useful.
Two fitness functions model the total overlap between adjacent fragments in a layout: \(F_1 = \sum _{i=1}^{n-1} w(i, i+1)\), where w(i, j) is the degree of overlap between fragments i and j (linear, and to be maximized), and \(F_2\) (square) a variant minimizing the total degree of overlap w(i, j) among all fragment pairs i and j at distant locations in the assembly. Neither of these functions, when evaluated over a candidate solution, can directly tell how closely the assembly relates to the true genome. A greedy algorithm outdid this decisively in contig counts and marginally in terms of \(F_1\).
In [10, 11] (with further parameter tuning in [12]) the fitness functions from [8] are kept, but a simpler and more effective permutation representation is used, i.e., an ordered list of read identifiers as appearing in the assembly. This requires specialized genetic operators listed below (in some cases renamed, to clarify that we preserve the semantics of 3 out of 5 in this work). Each operator is either micro, if it operates over reads, or macro, when at contig level:
-
Order crossover (micro): random read indices l and r are selected, the subsequence between l and r on the first parent is copied into the offspring preserving absolute position, and the remaining slots in the offspring are filled from left to right with the reads not yet in the offspring, in the relative order in which they appear in the second parent.
-
Edge recombination (micro): greedily attempts to preserve read adjacencies from the parents into the offspring. Selects the first read r from the first parent, and follows it with that read s which (i) is adjacent to r in both parents, or, failing that, (ii) has the most adjacencies left.
-
Swap (micro): two random reads are swapped; in [12], late swaps (to avoid local optima) become greedy by overlap rather than randomly.
-
Reverse-complement (macro): reverts a random contig (with the contig bounds being probabilistic in [12]).
-
Inshift (macro): moves a random contig between a random two previously adjacent contigs.
The number of contigs is not used as a fitness function. From the functions used, \(F_1\) is found to be adequate (and better than \(F_2\)), but not ideal; the authors leave the design of a more appropriate function for future research. Neither function can be seen as an accuracy metric.
PALS [13] adds as fitness to \(F_1\) the number of contigs and obtains one contig for all but the largest read set. In this candidate representation, the calculation of the number of contigs has high time complexity, and their method to estimate this is done via delta-increments from the application of operators at each iteration.
In our evaluation, we experiment with the genomes used in this prior work (under \(10^5\) base-pairs long, with low coverage between 5 and 7, a cutoff value for overlap of 30 base pairs, and read sets generated artificially from a reference genome without long repeats). More importantly, while all prior work makes an effort to compare against other assemblers in terms of the number of contigs obtained (and occasionally the abstract assembly metric \(F_1\)), it makes no effort to measure how well the reference genome is actually covered by the best assemblies they obtained. Here, we bridge this gap between the abstract evaluation of the algorithm, and the practical, domain-specific evaluation of the solutions.
3 Methodology
In Fig. 1 and Table 2, we summarize our design choices in terms of the genetic representation, operators, and fitness functions. Previous work borrowed from evolutionary-inspired solutions to the TSP problem, and represented a candidate solution as a simple permutation of DNA reads (a comparative overview was given in Table 1 in the related-work Sect. 2.2). This representation made it natural to then design primarily micro genetic operators, i.e., operations changing the location of individual DNA reads in the permutation. Macro operators, modifying a DNA contig, required more effort in recovering the contig structure from this representation. We build on these early designs, and lift the candidate representation and all operators to macro level, to then be able to write computationally simple fitness functions and stopping conditions, both of which reflect the metrics measuring the accuracy of a DNA assembly, learnt from recent studies of commercial DNA sequencers (Sect. 2.1). These design choices are detailed in the remaining of this section.
3.1 Representation of Candidate Solution
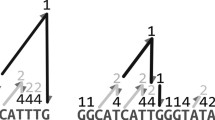
A segmented permutation arranges the DNA reads in the input read set in an order, to model a DNA scaffold, as a simple permutation would also do. However, it also logically segments this scaffold into DNA contigs, where each is a subarray (of length at least one) of DNA read identifiers. This essentially memorizes the boundaries of contigs at the level of the candidate solution, and also allows to write a fitness function and a stopping condition for the evolutionary process which compare a candidate solution’s scaffold length against its expected length, as given by a reference genome (i.e., an accuracy metric from Sect. 2.1). A candidate (with a sketch in Fig. 1) is an object with the following attributes:
-
DNA scaffold: a segmented permutation of the unmodified DNA reads in the input set, of which the reads in any segment (modelling a DNA contig) will have adjacent overlaps above a certain minimum overlap threshold.
-
Reverse-complement bits: for each DNA read, a Boolean variable which is True if the corresponding read is used it its reverse-complemented form in this candidate scaffold with respect to the input read set.
-
DNA contig strings: for each DNA contig on the scaffold, the overlapped contig (i.e., a relatively short string). Initially, all contigs contain a single read. The sum of the lengths of these contig strings will thus start at value \(n\cdot r\), where n is the number of reads in the input set, and r is the read length, and will decrease in time, as contigs merge.
3.2 Operators Over Candidate Solutions
All genetic operators work natively over DNA contigs. This is a generalization rather than a limitation in comparison to the micro operators from the related work, since a contig here may well consist of a single read, and any DNA scaffold may be obtained via either set of operators. However, macro operators may be more efficient in that they apply a mutation (such as a transposition of a portion of DNA from one place in the scaffold to another) for an entire existing contig, in one step, rather than read by read.
Mutation (top) and crossover (bottom) operators. From the candidate representation, only the DNA scaffold and the reverse-complement bits are shown (a backward arrow models a True value). Indices are 1-indexed. This crossover example uses index 2, i.e., preserves the first two contigs from the first parent into the first offspring.
Our set of genetic operators are listed below, and shown in Fig. 2.
-
Reverse-complement: as the corresponding operator in prior work [10,11,12]. The corresponding DNA contig string in the representation becomes the reverse-complement of the original.
-
Inshift: also as in prior work. It preserves the internal structure of all contigs.
-
Split: a randomly selected contig is split (if it consists of at least two reads) at a random point, forming two new adjacent contigs. Two new DNA contig strings are computed for the scaffold.
-
Random Merge: a random pair of adjacent contigs is merged into a new contig, with a new DNA contig string computed for the scaffold. The merge only occurs for pairs of contigs which overlap by a number of base pairs above a configurable threshold (the higher this threshold, the fewer the merge options are). The two contigs may overlap by a number of base pairs longer than the read length: in this case, the newly formed contig is an interleaving of the two original contigs. We also experimented with a variant of this operator:
Greedy Merge: among the list of all possible merges on a given candidate scaffold, the highest 50% in terms of overlap are selected, and a random choice is made out of these.
-
Order crossover: a random index is selected between two adjacent contigs on the first parent. All the contigs to the left of this point are preserved (in both position and internal structure) in the first offspring. The remaining reads for this offspring will come from the second parent, in the order in which that parent has them on its scaffold; two of these reads will be adjacent in the same contig only if they were arranged as such on the second parent (otherwise, they will form separate contigs).
We abbreviate these operators by their initials, R, S, I, M, O. They are applied in the evolutionary process to a candidate solution in a configurable order (e.g., RSIMO), each operator at a configurable rate of application.
With one exception, each operator performs a modification of a candidate solution which cannot be achieved by any combination of other operators—in other words, this operator set is minimum. The exception is the crossover operation, which may be modelled by a long sequence of mutation operators; the innate advantage of crossover is speed, i.e., it drastically raises the rate of change in the population.
The time complexity of applying each operator to a candidate with n total reads and r read length, and copying in the offspring, is up to O(nr) in all cases except Greedy Merge, where an extra \(O(n \,\text {log}\,n)\) worst-case factor is needed (NB: this factor may be dominated by O(nr)).
3.3 Candidate Fitness
Good options for fitness functions are accuracy metrics from the literature. Since it has been observed that the assembly metrics regularly used in comparative studies do not correlate well with accuracy metrics [4], we use fitness functions which borrow from both categories.
The length of the candidate scaffold, i.e., the sum of lengths of DNA contig strings, is the main fitness, to be minimized. This is essentially an assembly metric, except if an estimate length of the reference genome is known: then it also acts as an accuracy metric.
The number of DNA contigs is the second, less crucial, component of the fitness, to be minimized. The two functions are summed up. This fitness reflects purely an assembly metric; unlike the first fitness component, it states nothing about the accuracy of the solution, since many assemblies may exist, of various scaffold lengths, which will have exactly one contig. For this reason, most of the early related work [8, 10,11,12] does not use this fitness function at all, while the more recent work [13,14,15,16] uses it in conjunction with another assembly metric.
The time complexity of computing the scaffold length over a candidate with c contigs on the scaffold, in our candidate representation, is \(\varTheta (c)\): the length of the scaffold is the sum of the lengths of the DNA contig strings included in the representation (Sect. 3.1). Computing the number of contigs is constant-time if the length of the data structure modelling the segmented permutation is stored explicitly in the implementation.
4 Evaluation
4.1 Datasets
We obtain read libraries from existing consensus genomes. The reads are generated artificially, as in some of the prior work, rather than being selected from sequenced read libraries. This brings several advantages (argued as desirable in Sect. 2.1):
-
This gives a reference genome, so we can compute accuracy metrics to evaluate any assembly obtained against the original genome itself.
-
We know the raw reads to be accurate.
-
We can verify that adjacent reads have an overlap above 30 base pairs.
-
The genome lacks repeat sequences longer than the read length.
The genomes used in the evaluation (Table 3) were also used in the related work. X60189 and M15421 are fragments of human DNA (in the case of M15421, sequenced from messenger RNA); J02459 is the whole genome of a phage virus, the Enterobacteria phage lambda. The difference to [10, 11, 13, 17] is that we take reads of uniform length, as uniform-length reads of length on the order of \(10^2\) are most common in current commercial sequencers, and we use the entirety of the J02459 (rather than the first 40% of the sequence, as done previously in [10, 11, 13, 17]); we thus have a larger read library for J02459. We generated 10 distinct read libraries for X60189 and M15421, and one read library for J02459. Each read library was assembled 20 times, using different random seeds for the genetic algorithm.
4.2 Genetic Parameters and Implementation
Candidate solutions are selected via tournament (of tournament size 5 across all experiments), and 5 elites are kept in the population. Two stopping conditions are placed upon the evolutionary process: the fitness reaching the expected, optimal fitness for that genome (i.e., the length of the scaffold: an accuracy metric), and a stagnation condition upon the number of generations without improvement in fitness; the latter is set equal to the population size. The population size and the number of stagnating generations are always 100.
The genetic operators are applied sequentially in the basic order RSIMOM, i.e., with a Merge mutation after each chain of other operators which is likely to break down contigs (i.e., Split and Order crossover); the default is a Random Merge. The rates of application for the operators are 0.5 for all mutation operators, and 0.1 for the crossover.
The software implementation uses the inspyred [21] Python framework for bio-inspired algorithms, which is easily extensible with new candidate representations and genetic operators.
4.3 Results and Evaluation via Fitness Values
For the smallest read sets and genomes, the algorithm, when run repeatedly with different random seeds, always obtains the optimal solution. The optimal fitness value is the length of the reference genome plus one (the latter being the contig count). For the second genome, the optimal fitness is obtained in a majority of the runs; for the third, no run was optimal. We discuss this outcome later.
Assemblies obtained for 10 different read sets sampled from the X60189 genome (top) and the M15421 genome (bottom). For each of the 10 read sets, 20 assemblies were run with different random seeds. Parameters as in Sect. 4.2. Optimal assembly length shown in red (grey in print). (Color figure online)
We first gain an insight as to the frequency and the cost of reaching the optimal fitness: in Fig. 3, we show assemblies for 10 read sets obtained from the X60189 genome, and for 10 read sets from the M15421 genome. For each of these read sets, 20 assemblies are run, with different random seeds, for a total of 200 per plot.
All assemblies used the parameter settings in Sect. 4.2. Each data point shows the best solution obtained by an assembly, in terms of fitness; the fitness is shown with its two components explicitly separate: the scaffold length (in base pairs) on the vertical axis, and the number of contigs as an annotation to each point. The optimal number of contigs is naturally one, and the optimal scaffold length is shown as a threshold line. On the horizontal axis, the number of generations required to reach the best assembly per run gives a measure of the method’s runtime. For all the read sets, some or all of the assemblies found an optimal solution, i.e., the reference genome in either forward or reverse-complemented form. Not surprisingly, the smaller the read set, the likelier optimality is. For the largest genome (J02459), no assembly was optimal in fitness.
4.4 Results and Evaluation via Accuracy Metrics
Here, we evaluate the best assemblies obtained against the ground truth provided by the reference genome. Figure 4 shows the internal breakdown into contigs of the 20 assemblies obtained from a read set of the two largest genomes. The scaffold is shown with the longest contig first (without loss of information: as they are disjunct, contigs may be rearranged). Almost all assemblies in Fig. 4 computed a contig of length equal to the reference genome, which is a strong hint that this contig also aligns fully with the genome, and is thus an accurate assembly by itself.
Scaffolds obtained as best assemblies for the largest genomes, for one read set per genome. For each scaffold, the lengths of the internal contigs are shown, with the longest contig first. A check mark is placed on all contigs which are identical to the reference genome. Optimal assembly length shown in red (grey in print). (Color figure online)
We verify this in a separate step. All assemblies for X60189 obtained a single contig identical to the reference genome. Similarly, 131 out of 200 assemblies for M15421 obtained a single optimal contig, with the remaining assemblies consisting of a multi-contig scaffold, where the longest contig is identical to the reference; these results, together with a comparison with results from the related work and an account of the runtime of our method in the current implementation, are summarized in Table 4. The related work in [17], which also proved the optimality of the solution, is based on particle swarm optimization. We executed the greedy heuristic SSAKE [22] ourselves on these read sets; the parameters were -w 10 -m 20.
The results support the idea that the evolutionary algorithm can be used for consensus building.
There remains the problem of chaff contigs obtained in the assembly alongside the correct genome; the chaff can be seen in Fig. 4, and is loosely understood here as contigs of lengths shorter than 3–4 times the read length. This is a weakness of the evolutionary algorithm, also encountered in the prior work [12, 13]: the best solution obtained by the GA may be suboptimal in fitness. However, suboptimal solutions are easily refinable into optimal ones: it is provable that, when a long contig is the accurate solution, a simple, deterministic, post-processing step can align each individual read left in chaff contigs back onto the long contig. This remains for future work.
4.5 Random vs. Greedy Merge
We found some variability in the results of assembly runs due to the type of the Merge operator. Figure 5 (left) shows the fitness values for the 20 assemblies of a M15421 read set, executed separately with either of the Merge operators. While a Random Merge contributed to an optimal fitness in slightly over half the runs (and did construct a correct contig in all the runs), the Greedy Merge under the same GA parameters never led to optimal fitness, but still did construct a correct contig in half the runs — Fig. 5 (right) shows the scaffolds obtained with the Greedy Merge. A greedy behaviour in this method may thus lead to local minima, depending on the read library assembled; the type of Merge made no difference in assembling the smaller X60189 genome.
The effect of the type of Merge upon assemblies over one read set for the M15421 genome. (left) 20 assembly runs with either Random Merge or Greedy Merge; (right) the scaffolds obtained with Greedy Merge. Parameters as in Sect. 4.2. Optimal assembly length shown in red (grey in print). (Color figure online)
5 Conclusions
We summarize our results as follows:
-
An evolutionary algorithm will often find an accurate solution to the assembly problem, for genomes and read sets of type and size used here.
-
For large problems, chaff contigs is unavoidable, and an optimal fitness unattainable. However, even in these cases, the accurate genome is obtained after a simple post-processing refinement step.
-
Accuracy metrics must be used to evaluate an assembly algorithm against reference genomes. De novo assemblies (without the advantage of reference genomes) can be validated by consensus.
-
Evolutionary algorithms are eminently suitable as consensus builders: repeated executions will yield a number of large DNA contigs, among which the correct genome is found in majority.
Many practical difficulties remain for future work, in order that this method applies in commercial-grade assembly: (a) a uniform distribution is unlikely to be obtained in practice due to bias in some DNA fragmentation methods [9]; (b) the read sets to assemble may have far larger sizes and smaller reads, leading to a more complex problem; finally, (c) largest genomes will have long DNA repeats, which pose difficulties to all short-read assembly algorithms.
References
Illumina: Illumina sequencing technology, October 2016. http://www.illumina.com/documents/products/techspotlights/techspotlight_sequencing.pdf
Chin, C.S., Alexander, D.H., Marks, P., Klammer, A.A., Drake, J., Heiner, C., Clum, A., Copeland, A., Huddleston, J., Eichler, E.E., Turner, S.W., Korlach, J.: Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10(6), 563–569 (2013)
Ip, C., Loose, M., Tyson, J., de Cesare, M., Brown, B., Jain, M., Leggett, R., Eccles, D., Zalunin, V., Urban, J., Piazza, P., Bowden, R., Paten, B., Mwaigwisya, S., Batty, E., Simpson, J., Snutch, T., Birney, E., Buck, D., Goodwin, S., Jansen, H., O’Grady, J., Olsen, H.: MinION analysis and reference consortium: phase 1 data release and analysis. F1000Research 4, 1075 (2015)
Salzberg, S.L., Phillippy, A.M., Zimin, A., Puiu, D., Magoc, T., Koren, S., Treangen, T.J., Schatz, M.C., Delcher, A.L., Roberts, M., Marçais, G., Pop, M., Yorke, J.A.: GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22(3), 557–567 (2012)
Bradnam, K., Fass, J., Alexandrov, A., Baranay, P., Bechner, M., Birol, I., Boisvert, S., Chapman, J., Chapuis, G., Chikhi, R., et al.: Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2, 10 (2013)
Cherukuri, Y., Janga, S.C.: Benchmarking of de novo assembly algorithms for Nanopore data reveals optimal performance of OLC approaches. BMC Genom. 17(Suppl. 7), 507 (2016)
Räihä, K.J., Ukkonen, E.: The shortest common supersequence problem over binary alphabet is NP-complete. Theoret. Comput. Sci. 16(2), 187–198 (1981)
Parsons, R., Burks, C., Forrest, S.: Genetic algorithms for DNA sequence assembly. In: International Conference in Intelligent Systems for Molecular Biology (1993)
Poptsova, M.S., Il’icheva, I.A., Nechipurenko, D.Y., Panchenko, L.A., Khodikov, M.V., Oparina, N.Y., Polozov, R.V., Nechipurenko, Y.D., Grokhovsky, S.L.: Non-random DNA fragmentation in next-generation sequencing. Sci. Rep. 4(4532), 1697–1712 (2014)
Parsons, R.J., Forrest, S., Burks, C.: Genetic algorithms, operators, and DNA fragment assembly. Mach. Learn. 21(1–2), 11–33 (1995)
Parsons, R., Johnson, M.E.: DNA sequence assembly and genetic algorithms - new results and puzzling insights. In: Proceedings of the Third International Conference on Intelligent Systems for Molecular Biology, Cambridge, United Kingdom, 16–19 July 1995, pp. 277–284 (1995)
Parsons, R., Johnson, M.E.: A case study in experimental design applied to genetic algorithms with applications to DNA sequence assembly. Am. J. Math. Manag. Sci. 17(3–4), 369–396 (1997)
Alba, E., Luque, G.: A new local search algorithm for the DNA fragment assembly problem. In: Cotta, C., Hemert, J. (eds.) EvoCOP 2007. LNCS, vol. 4446, pp. 1–12. Springer, Heidelberg (2007). doi:10.1007/978-3-540-71615-0_1
Nebro, A., Luque, G., Luna, F., Alba, E.: DNA fragment assembly using a grid-based genetic algorithm. Comput. Oper. Res. 35(9), 2776–2790 (2008). Part Special Issue: Bio-inspired Methods in Combinatorial Optimization
Dorronsoro, B., Alba, E., Luque, G., Bouvry, P.: A self-adaptive cellular memetic algorithm for the DNA fragment assembly problem. In: 2008 IEEE Congress on Evolutionary Computation (IEEE World Congress on Computational Intelligence), pp. 2651–2658, June 2008
Firoz, J.S., Rahman, M.S., Saha, T.K.: Bee algorithms for solving DNA fragment assembly problem with noisy and noiseless data. In: Proceedings of the 14th Annual Conference on Genetic and Evolutionary Computation, GECCO 2012, pp. 201–208. ACM, New York (2012)
Mallén-Fullerton, G.M., Fernández-Anaya, G.: DNA fragment assembly using optimization. In: 2013 IEEE Congress on Evolutionary Computation, pp. 1570–1577, June 2013
NCBI: Human MHC class III region DNA with fibronectin type-III repeats (2016). https://www.ncbi.nlm.nih.gov/nuccore/X60189
NCBI: Human apolipoprotein B-100 mRNA, complete cds (2016). https://www.ncbi.nlm.nih.gov/nuccore/M15421
NCBI: Enterobacteria phage lambda, complete genome (2016). https://www.ncbi.nlm.nih.gov/nuccore/J02459
Garret, A.L.: Inspyred: a framework for creating bio-inspired computational intelligence algorithms in Python (2017). https://pypi.python.org/pypi/inspyred
Warren, R.L., Sutton, G.G., Jones, S.J.M., Holt, R.A.: Assembling millions of short DNA sequences using SSAKE. Bioinformatics 23(4), 500–501 (2007)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Bucur, D. (2017). De Novo DNA Assembly with a Genetic Algorithm Finds Accurate Genomes Even with Suboptimal Fitness. In: Squillero, G., Sim, K. (eds) Applications of Evolutionary Computation. EvoApplications 2017. Lecture Notes in Computer Science(), vol 10199. Springer, Cham. https://doi.org/10.1007/978-3-319-55849-3_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-55849-3_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-55848-6
Online ISBN: 978-3-319-55849-3
eBook Packages: Computer ScienceComputer Science (R0)