Abstract
Influenza virus is an infectious agent, belonging to the virus family Orthomyxoviridae, that causes a respiratory tract infection in vertebrates. There are three main species, and of these, influenza A viruses cause the most virulent infections in humans and are also a common cause of zoonotic infections. Although the influenza seasonal strains constitute a substantial public health concern, influenza strains far more lethal than the seasonal strains emerge sporadically. These virulent strains produced three global pandemics in the past century, the worst of which occurred in 1918. Influenza also infects a variety of animal species in addition to humans. Some of these influenza strains are species specific, but new strains may spread from other animals to humans. The 2009 influenza pandemic was a recombinant influenza involving a mix of swine, avian, and human gene segments. Influenza outbreaks usually occur in winter in temperate climates. Influenza type A viruses have a mutation rate two or three times higher than that of type B viruses. Because of this, WHO analyzes the vaccine components each year and makes any necessary changes on the basis of worldwide trends. These vaccine recommendations are based on numerous factors, including global influenza virological and epidemiological surveillance, genetic and antigenic characterization, antiviral susceptibility, and the availability of candidate vaccine viruses for production. Although vaccines are the best means of protection for influenza, antivirals constitute an important tool to control these infections. Neuraminidase inhibitors are approved for chemoprophylaxis and treatment of influenza, and they act by reducing the severity and duration of illness significantly, although potential adverse effects and higher costs should be considered.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Influenza virus
- Influenza surveillance
- Influenza pandemic
- Genetic drift and shift
- Transmission
- Swine influenza
- Avian influenza
- Prophylaxis and prevention
1 Introduction
Influenza viruses are RNA viruses of negative polarity and segmented, belonging to the family Orthomyxoviridae . This family includes six genera: Influenzavirus A; Influenzavirus B; Influenzavirus C; Isavirus; Thogotovirus, which includes the Thogoto virus and Dhori virus including Infectious salmon anemia virus; and Quaranjavirus [17]. Influenza A viruses are classified into subtypes based on the antigenicity of the surface molecules hemagglutinin (H) and neuraminidase (N). Influenza A viruses infect a wide variety of birds and mammals . These viruses continually mutate, exhibiting well-studied patterns, such as antigenic drift and reassortment of genomic segments. In the past century, influenza A viruses have caused three pandemics [14], as they are known to be epidemics that extend over more than one continent. Influenza B viruses infect humans, and there is only one subtype; they have a low potential to cause pandemics , although they can cause severe respiratory illnesses. Influenza C viruses infect humans and pigs and in general cause mild respiratory illness.
Wild aquatic birds, such as ducks and gulls among others, are the most important natural reservoir of these viruses [33]. In this type of bird circulate all H and N subtypes, and it is thought that wild birds are the source of the transmission of the virus to other animal species, including poultry. Many of the subtypes of influenza A viruses infect birds asymptomatically, that is, without causing disease or causing mild symptoms; however, some virus infections, for example, H5 and H7 subtypes , can cause severe illness and death in some species of wild and domestic birds, such as chickens and turkeys [34]. Pigs are also natural reservoirs of influenza A viruses, from which a limited number of subtypes have been isolated.
Every year, around 500 million people worldwide fall ill because of influenza virus A infection, of which between 3 and 5 million become severe cases leading to about 300,000 deaths. These cases of influenza occur regularly during the cold or rainy seasons each year and are known as epidemics or outbreaks of seasonal influenza.
Since 1977, in the human population two influenza A virus subtypes, known as H1N1 and H3N2 , circulate seasonally, taking into account the characteristics of the proteins present on the surface, along with influenza virus B. The frequency of these three groups of viruses varies temporarily and geographically. In 2009, a new subtype H1N1 emerged with great antigenic/genomic changes that allowed them to reach the pandemic state. This new virus emerged from a virus generated by reassortment between a human virus, a porcine virus from North America, a porcine virus from Eurasian, and an avian virus [24].
2 Nomenclature
Influenza viruses belong to the Orthomyxoviridae family and consist of six genera:
-
1.
Influenzavirus A. Type species, Influenza A virus
-
2.
Influenzavirus B. Type species, Influenza B virus
-
3.
Influenzavirus C. Type species, Influenza C virus
-
4.
Isavirus. Type species, Infectious salmon anemia virus
-
5.
Quaranjavirus. Species, Johnston Atoll virus, and type species, Quaranfil virus
-
6.
Thogotovirus. Species, Dhori virus, and type species, Thogoto virus
The Influenzavirus A and Influenzavirus B genera cause epidemics every year. The emergence of a new and different virus in the human population can cause an influenza pandemic . Influenza C virus infections cause mild respiratory disease and are considered not to cause epidemics.
Influenza A viruses are divided into subtypes based on the two surface proteins of the virus: the hemagglutinin (H) and neuraminidase (N). In nature, there are at least 18 different subtypes of H and 11 different subtypes of N [31, 32]. In turn, within each subtype, influenza A viruses can be divided into different strains with different antigenic characteristics . Current influenza A virus subtypes detected in the human population are A(H1N1) and A(H3N2). In the spring of 2009, a new influenza virus A(H1N1) emerged and began to cause illness in people. This virus has major changes in comparison with seasonal influenza A(H1N1) virus and has caused an influenza pandemic after 40 years. The virus, known as A(H1N1)pdm09, has largely replaced the previously circulating H1N1 virus among humans. The influenza B viruses are not divided into subtypes, but different antigenic strain characteristics can be distinguished.
The Centers for Disease Control and Prevention (CDC) , located in Atlanta, GA (USA), follows an internationally accepted nomenclature convention for influenza virus. This convention was recommended by the World Health Organization (WHO) in 1979. This nomenclature establishes the use of the following components:
-
1.
The antigenic type (e.g., A, B, C).
-
2.
The host of origin, for example, pig, horse, chicken, or in the case of environmental samples. It is not specified in the case of human viruses.
-
a.
Geographic origin (e.g., Denver, Taiwan).
-
a.
-
3.
Number assigned by the laboratory of origin to the sample from which the virus was isolated (e.g., 15, 7).
-
4.
Year of isolation (e.g., 1957, 2009).
-
5.
For influenza virus A, the description of H and N antigens appears in brackets [e.g., (H1N1), (H5N1)].
For example :
-
A/duck/Czechoslovakia/1956 (H4N6) for a virus originated in ducks
-
A/Sydney/5/97 (H3N2) for a virus originated in human
3 Structure and Biology
Under the electron microscope, these viruses have a pleomorphic appearance, with an average diameter of 100 nm (one ten-thousandth of a millimeter). The viral particle has an envelope composed of a lipid bilayer in which the H glycoproteins and N and lesser amounts of M2 transmembrane protein are inserted (see Fig. 13.1). Lining the inside of the lipid membrane is a layer formed by the matrix protein M1. Inside the sheath is the viral genome. The genomic segments are covered with nucleoprotein NP and are also associated with viral RNA polymerase, which is a protein complex consisting of two basic subunits (PB1, PB2) and an acidic subunit (PA). The viral genome consists of eight segments of single-stranded RNA of negative polarity, ranging from 890 to 2,350 nucleotides in size, with some variation depending on the virus strain. In total, the genome has approximately 13,600 nucleotides and encodes 11 viral proteins. All segments encode a protein, with the exception of the PB1 gene, which in some strains also encodes the PB1-F2 protein; the gene protein matrix, which encodes two proteins, M1 and M2; and the smallest gene, NS, which codes for proteins NS1 and NS2.
3.1 Viral Tropism
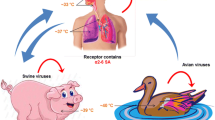
In humans, the virus usually enters the body through the nose or mouth and infects the cells lining the respiratory tract, joining sialic acid (SA) molecules on the surface of the cells to start the infection. The SA is an abundant molecule in all cells, a part of sugar chains bound to proteins or lipids, and defines the influenza virus tropism. This binding occurs because of the specificity of different virus strains for different types of links of SA with sugar, the upstream carbohydrate, which is generally galactose. Thus, human viruses recognize the conformation of SA with galactose in the alpha 2–6 position, whereas avian viruses join SA with union 2.3 [30]. The affinity for SA partly explains the restriction host of influenza viruses. In the swine tracheal epithelial cells, both types of SA links exist, which allow the pig to be naturally infected by swine, avian, or human viruses. This mechanism enables genetic rearrangements among animal species originating influenza viruses with combinations of gene segments from different origins.
3.2 Molecular Determinants of Virulence: Viral Proteins and Pathogenesis
Several viral proteins have an important role in some aspects of pathogenesis and host restriction of influenza viruses, including the ability to modulate the host immune system and the ability to replicate efficiently at different temperatures, among others. The best characterized proteins in order of their pathogenic potential are H, PB1, and PB2. Furthermore, the N and M2 proteins have been widely studied because of their ability to confer resistance to approved antiviral drugs.
3.3 Role of Hemagglutinin in Virulence and Viral Tropism
This glycoprotein, along with N, is one of the major proteins of the viral particle. The most neutralizing antibodies are produced against this main antigen, and these are capable of neutralizing virus infectivity. The H, as its name suggests, is capable of binding erythrocytes, a property that has been used in techniques for the classification of different subtypes . The H has a very important role in virus entry into the host cell, mediating the viral adsorption through its interaction with the SA. H is synthesized as a precursor protein called H0, which is cleaved at a specific site, resulting in the generation of the subunits HA1 and HA2. The cleavage site is characterized by being composed of basic amino acids. Proteolytic cleavage is essential for infectivity because it exposes a hydrophobic peptide at the amino terminus of HA2, which is responsible for mediating the fusion of the viral membrane and endocytic vesicle formed when the particle is internalized into the cell.
With the purpose of describing the spatial dissemination dynamics of influenza A(H3N2) within South America, 316 HA1 sequences of A(H3N2) viruses from Argentina, Brazil, Chile, Paraguay, Uruguay, Bolivia, Colombia, French-Guyana, Peru, and Venezuela collected between 1999 and 2012 were analyzed together with 153 available contemporary sequences from Australia, Hong Kong, the UK, and the U.S. Phylogenetic analyses revealed that influenza A(H3N2) sequences from South America were highly intermixed with sequences from other geographic regions, although a clear geographic virus population structure was detected globally. Fourteen clades mostly (≥80%) composed of influenza sequences from South American countries were identified. Bayesian phylogeographic analyses of those clades support a significant role of both temperate and tropical regions in the introduction and dissemination of new influenza A(H3N2) strains within South America and identify an intensive bidirectional viral exchange between different geographic areas. These findings indicated that seasonal influenza A(H3N2) epidemics in South America are seeded by both the continuous importation of viral variants from other geographic regions and the short-term persistence of local lineages. These results support a complex meta-population model of influenza A(H3N2) dissemination in South America, with no preferential direction in viral movement between temperate and tropical regions [7].
3.4 Viral Replication and Protein PB2
Protein PB1 and PA form the viral replication complex. PB2 has been associated with the transmissibility of the virus through the air and also with the restriction host.
3.5 PB1-F2: A Cell Death Inductor Factor
Protein PB1-F2 is a small protein encoded by the gene that also encodes the subunit viral polymerase PB1. PB1-F2 is not expressed by all influenza strains. It has been found that this protein interacts with the mitochondria membrane, causing its permeation and cytochrome C release, which induces cell death. It has also been reported that PB1-F2 exacerbates inflammatory response during viral infection in mice and increases the frequency and severity of secondary bacterial pneumonias.
3.6 NS1 and Control of the Innate Immune Response
During viral replication, transcriptional activation factors that stimulate the production of interferon are triggered; this is a strategy to prevent cell viral infections. The NS1 protein is a viral protein that antagonizes the interferon response of the cell. It was found that highly virulent strains, such as H5N1 avian, not only confer resistance to antiviral effects of interferon but also induce an exacerbated pro-inflammatory cytokine response . During pregnancy, immunological and hormonal alterations place women at increased risk for influenza-related severe illnesses, including hospitalization and death. Although A(H1N1)pdm09 infection resulted in increased disease severity in pregnant women, the precise mechanisms responsible for this risk have yet to be established. The role of host chemokines and cytokine profiles in A(H1N1)pdm09 infection regarding disease severity in pregnant women with confirmed influenza A(H1N1)pdm09 infection was investigated. Results revealed that pregnancy-related reductions in interferon (IFN)-β and transforming (TGF)-β expression levels and elevated levels of pro-inflammatory cytokines could explain the increased severity of infection and death of pregnant women [26].
3.7 NS2
This protein is also encoded by the viral RNA segment 8, and it has been detected associated with M1 protein in the virion. It is possible to find it in the nucleus and cytoplasm. It possesses a nuclear export signal that would function to export the RNPs from the nucleus.
3.8 M2: A Viral Ion Channel
The M2 protein is the less abundant viral coat. This protein functions as an ion channel that allows the entry of protons into the viral particle, facilitating the release of the viral genome into the cytoplasm. Adamantanes selectively block the channel formed by M2, which inhibits the release and therefore the replication of the viral genome. These compounds have been used against influenza outbreaks for many years; however, it has been found that adamantane resistance appears rapidly and frequently in influenza wild strains. Most human and swine A(H3N2) strains currently circulating are resistant to these antivirals; moreover, the new A(H1N1)pdm09 viruses are also resistant to these drugs.
3.9 Neuraminidase and Viral Dissemination
This glycoprotein, neuraminidase, is a sialidase whose function is to remove not only the SA of the viral glycoproteins of newly synthesized virus but also the SA present on the cell surface, allowing efficient release of virus from the infected cell to infect new cells . Inhibition of this activity produces the de novo synthesized virions that remain attached to the cell surface , inhibiting their spread to other cells. The N is the target of oseltamivir and zanamivir antivirals . These molecules are specific inhibitors of the N-sialidase activity.
4 Genetic Drift and Shift
Influenza viruses are constantly changing, and they can change in two different ways. One way is called “antigenic drift ,” which are small changes in the genes of influenza viruses that happen continually over time as the virus replicates. These small genetic changes usually produce viruses that are quite closely related to one another, which can be illustrated by their close location on a phylogenetic tree. Viruses that are closely related to each other usually share the same antigenic properties, and an immune system exposed to a similar virus will usually recognize it and respond. However, these small genetic changes can accumulate over time and result in viruses that are antigenically different (further away on the phylogenetic tree). When this happens, the body’s immune system may not recognize those viruses. Genetic changes that result in a virus with different antigenic properties are the main reason why people can be infected by influenza virus more than once. This is also why the influenza vaccine composition must be reviewed each year and updated as needed to keep pace with evolving viruses [9]. In Mexico, whole-genome sequencing studies of human A(H3N2) isolates from 2003 to 2012 showed that different viral lineages co-circulate within the same season and can also persist locally in between different influenza seasons, increasing the chance for genetic reassortment events. A novel minor cluster was also identified, named here as Korea, that circulated worldwide during 2003 [11].
The other type of change is called “antigenic shift.” Antigenic shift is an abrupt, major change in the influenza A viruses, resulting in new H or new H and N proteins in influenza viruses that infect humans. Shift results in a new influenza A subtype or a virus with a H or a H and N combination that has emerged from an animal population which is so different from the same subtype in humans that most people do not have immunity to the new (i.e., novel) virus. Such a “shift” occurred in the spring of 2009, when an H1N1 virus with a new combination of genes emerged to infect people and quickly spread, causing a pandemic. When this shift happens, most people have little or no protection against the new virus [9]. A(H1N1)pdm09 was originated from a quadruple viral reassortment: viral genes from North American swine, Eurasian swine, and avian and human populations.
On June 11, 2009, in acknowledgment of sustained global human-to-human transmission, the WHO declared that the novel influenza A(H1N1)pdm09 virus was pandemic. Although this virus has not been reported as more virulent than seasonal influenza A(H1N1), its high transmissibility in an immunologically naïve population implied the potential for substantial morbidity and mortality and mandated close surveillance for evolution toward increased virulence. Although worldwide the pandemic influenza A(H1N1) case fatality rate (CFR) was 0.4%, that rate in Argentina was 4.5%. After performing full-genome sequencing of strains isolated from mild and severe cases, no evidence was observed that the high CFR can be attributed directly to viral changes. Further, no evidence of reassortment with human or animal locally circulating strains, mutations associated with resistance to antiviral drugs, or genetic drift that might contribute to virulence was documented [3].
Analysis of 13 samples of 2009 H1N1 pandemic virus circulating in Paraguay in 2009 showed that these viruses clustered in a single genetic group. Neither the mutation related to exacerbation of disease (D239G in H) nor that related to antiviral resistance (H275Y in neuraminidase), both detected in neighboring countries, was found [12].
To generate information about the disease burden caused by 2009 influenza A(H1N1)pdm09 in children in Argentina, a retrospective case series study was conducted. The results obtained indicated that hospitalization rates were double in comparison with those for seasonal influenza in 2008. The overall rate of death was 1.1 per 100,000 children, as compared with 0.1 per 100,000 children for seasonal influenza in 2007. This investigation concluded that influenza A(H1N1)pdm09 was associated with pediatric death rates that were ten times the rates registered for seasonal influenza in previous years in Argentina [18].
Molecular characterization of circulating influenza A viruses in all regions of the world is essential to detect mutations potentially involved in increased virulence, antiviral resistance, and immune escape. To gain insight into these matters, a phylogenetic analysis of the N gene of 146 pandemic H1N1 (H1N1pdm) influenza A virus strains isolated in Argentina, Brazil, Chile, Paraguay, Peru, and Uruguay from 2009 to 2013 was performed. The comparison of vaccine strain A/California/7/2009 was included in the influenza vaccine recommended for the Southern Hemisphere from 2010 through 2016. Strains differ from vaccine in two predicted B-cell epitope regions present at positions 102–103 and 351–352 of the NA protein. Moreover, vaccine and strains isolated in Paraguay differ also in an epitope present at position 229. The analysis of the N gene of 2009 to 2013 H1N1 South American strains revealed several genetic and antigenic differences in the N of influenza A(H1N1)pdm09 among vaccine and strains circulating in South America [10].
Influenza viruses are changing by antigenic drift all the time, but antigenic shift happens only occasionally. Type A viruses undergo both kinds of changes; influenza type B viruses change only by the more gradual process of antigenic drift.
In the past century, three antigenic shifts occurred in the influenza A virus circulating in humans that were responsible for pandemics : in 1918, with the emergence of an A(H1N1) virus; in 1957, when the A(H1N1) virus was replaced by a virus subtype A(H2N2); and in 1968, when an A(H3N2) virus replaced the A(H2N2) viral subtype. In 1977, the A(H1N1) subtype was reintroduced in humans and did not replace the circulating subtype H3N2. The 1977 (H1N1) strain was replaced by the A(H1N1)pdm09 in 2009 and co-circulates together with A(H3N2) viral subtype up to the present worldwide, and they are responsible for the seasonal outbreaks that occur each year.
5 Viral Entry, Release, and Transmission
The main targets of the influenza virus are the columnar epithelial cells of the respiratory tract. These cells may be susceptible to infection if the viral receptor is present and functional. Thus, viral receptors are determinants of tropism. Viruses replicate in the respiratory epithelium, causing a localized infection. The new virions are overturned in the inner portion of the respiratory tract, transported through secretions, and disseminated out of the body when the individual sneezes, coughs, talks, or laughs.
The disease caused by the influenza virus is highly contagious. Transmission occurs by air in most cases, by coughing, talking, or sneezing, but can also spread through contact with surfaces contaminated with the respiratory secretions of sick individuals. Adults eliminate the virus from 1 day before the onset of symptoms until 5–10 days later; in children, elimination can start several days before and continue for 10 days or more after the onset of symptoms. Immunocompromised people can spread the virus for weeks.
6 Epidemiology and Local and Global Geographic Distribution
Human influenza viruses, including the new A(H1N1)pdm09 virus that has affected the human population since 2009, are distributed worldwide. This virus reaches peak prevalence in winter and rainy seasons. In Argentina particularly, although it is possible to detect viral circulation throughout the year, an increase of viral activity is observed between May and July, depending on the season. The number of positive cases detected and the types and viral subtypes circulating each year also vary by season. In 2012, the transmission pattern of influenza viruses was different in the four countries of the Southern Cone in South America. Surprisingly, peak activities were detected very late in this region. Increased viral circulation in the area was first noted at the end of April to early May in Chile and Paraguay; Argentina and Uruguay begun to report active transmission in early June. Particularly in Argentina, the peak was registered in August, 10 weeks later than in the past 9 years [6].
7 Ecology
Influenza A viruses can be found in various animals such as ducks, chickens, pigs, whales, horses, seals, and dogs. Influenza B viruses circulate only among humans. All known subtypes of influenza A virus were detected in birds, with the exception of H18N11 and H17N10 subtypes, which were only found in bats. Wild birds are the primary natural reservoir for all subtypes of influenza A viruses and are thought to be the source of influenza A viruses in all other animals. Argentina had been searched for evidences of influenza A infection in Antarctica through antibody and genome detection in different migratory bird species. Between December 2001and May 2004, sera and cloacal swabs were collected in different locations. Antibody against the A(H3N2), A(H1N1), A(H9N2), A(H5N1), and A(H7N2) subtypes and sequences of the M gene was detected in migratory birds in Antarctica, showing that these species had been in contact with influenza A virus during their lifetime. These findings were obtained also in young birds, born in Antarctica, and would suggest the local acquisition of the infection [2, 4].
As part of the ongoing efforts on animal influenza surveillance in Argentina, Xu et al. [35] described the isolation of an H9N2 virus from a wild aquatic bird (Netta peposaca), named as A/rosy-billed pochard/Argentina/CIP051–559/2007 (H9N2). Phylogenetic analysis of the HA gene revealed that the 559/H9N2 virus maintained an independent evolutionary pathway and shared a sister-group relationship with North American viruses, suggesting a common ancestor. The rest of the genome segments are clustered with viruses from South America . Experimental inoculation of the 559/H9N2 in chickens and quail revealed efficient replication and transmission only in quail. These viruses could easily jump to other bird species, thus highlighting the potential threat posed to local poultry. This study increases local understanding of H9N2 viruses in nature [35].
When a highly pathogenic virus (HPAI) appears, it produces severe symptoms and almost 100% mortality within 2 days. Since 2002, outbreaks of HPAI have occurred in the Americas: in Chile A(H7N3) 2002, U.S. (Texas) A(H5N2) 2004, 21 U.S. states A(H5) 2014–2015, and Canada (H7N3) 2004 were identified. In each of these outbreaks, a precursor virus of low pathogenicity mutated to become highly pathogenic after circulating in poultry [29].
The understanding of the global ecology of avian influenza A viruses is impeded by historically low levels of viral surveillance in Latin America. Through sampling and whole-genome sequencing of 31 avian influenza viruses from wild birds in Peru, ten HA subtypes (H1–H4, H6–H7, H10–H13) and eight N subtypes (N1–N3, N5–N9) were identified. The majority of those Peruvian avian influenza viruses were closely related to avian influenza viruses found in North America. However, unusual reassortants, including a H13 virus containing a PA segment related to extremely divergent Argentinean viruses, suggest that substantial avian diversity circulates undetected throughout South America [23].
In recent years, there has been extensive surveillance of the virus in aquatic birds in the Northern Hemisphere; however, in contrast, only a few studies have been attempted to detect avian influenza viruses in wild birds in South America. There are major flyways connecting South America to Central and North America, whereas avian migration routes between South America and the remaining continents are uncommon. As a result, it has been hypothesized that South American avian influenza strains would be more closely related to the strains from North America than to those from other regions in the world. The full genome of three avian influenza subtype H11N9 isolates obtained from ruddy turnstones (Arenaria interpres) on the Amazon coast of Brazil was studied. For all gene segments, all three strains consistently clustered together within evolutionary lineages of avian influenza viruses that had been previously described from aquatic birds in North America. In particular, the H11N9 isolates were remarkably closely related to avian influenza strains from shorebirds sampled at the Delaware Bay region, on the northeastern coast of the U.S. There was also evidence of genetic similarity to avian influenza strains from ducks and teals from the interior U.S. and Canada [16].
In 2002, the Chilean poultry industry was afflicted with a highly pathogenic avian influenza strain, which created economic loss and triggered the establishment of a surveillance program in wild birds. This effort consisted of periodic samplings of sick or suspicious animals found along the coast and analyses with standardized techniques for the detection of influenza A virus. The detection of three avian influenza strains (H13N2, H5N9, H13N9) in gulls from Chile between 2007 and 2009 showed highest similarities to viruses detected in wild birds from North America. These results suggest a dissemination route for influenza viruses along the coasts of the Americas [20].
Swine influenza virus (SIV) is enzootic in most regions with dense porcine populations. This disease is common in North and South America, Europe, Asia, and also in Africa. Although the viral subtypes found in the U.S. and Europe are the same, they are actually different strains. In Argentina, swine origin viruses have been identified in producing farm populations. An Argentinean study demonstrated the circulation of influenza A(H3N2) viruses in 19 pig farms between 2000 and 2002; the seroprevalence rate was 16.5% [5]. In sera collected in 2002, Piñeyro et al. [27] demonstrated the seroprevalence was 38.46% to 100% against H1N1 and 7.69% to 100% for H3N2 in 13 Argentinean swine.
In November 2008, an outbreak of respiratory disease in pigs consistent with SIV infection was detected in Argentina. Phylogenetic analysis revealed that the virus isolated shared nucleotide identities of 96–98% with A(H3N2) viruses that circulated in humans from 2000 to 2003. Sera collected from experimental inoculated animals mainly cross-reacted with noncontemporary human-origin H3N2 influenza viruses [8].
In 2009, A(H1N1)pdm09 transmission from human to pig was confirmed. In Argentina, seroepidemiological analyses performed in 17 pig farms showed that ≈41% of pigs had antibodies against A(H1N1) and A(H3N2) subtypes. Vaccines against swine influenza viruses were not licensed in Argentina in that period. In June–July 2009, an outbreak caused by influenza A(H1N1)pdm09 virus occurred on a pig farm. The virus was genetically related to the pandemic strain isolated in humans, and no evidence of further reassortment was confirmed [25]. Equine influenza occurs in almost all countries with a significant number of horses.
In July 2006, horses from various regions of Chile presented with fever, serious nasal discharge, dry cough, anorexia, and depression. The virus was identified as equine influenza virus H3N8. After performing sequencing of the H, N, and NP genes, important differences with the Santiago/85 isolate was observed, with a closer relationship to North American isolates, especially to the Florida lineage, and to Argentinean isolates from the 1990s [22].
There is no evidence that canine influenza virus is currently circulating outside the U.S. However, occasional infections with equine influenza virus are seen among dogs in other regions. In the UK, an equine virus H3N8 was responsible for outbreaks of respiratory disease in dog kennels in 2002 and 2003, and also in Australia in 2007. H3N2 viruses have been reported in dogs only in Korea.
8 Prophylaxis
In uncomplicated cases, bed rest with adequate hydration is the treatment of choice for most adolescents and young adult patients. Salicylates must be avoided in children of 18 years or younger because of the association to Reyes syndrome. Antibiotic treatment should be reserved for the treatment of secondary bacterial pneumonia. More severe cases or infections at high risk of complications can be treated with antiviral drugs. Two classes of antiviral drugs are available for the prevention and treatment of influenza: (a) the neuraminidase inhibitors (NAIs) zanamivir, oseltamivir, peramivir, and laninamivir, which are active against both influenza A and influenza B, and (b) the adamantanes, amantadine, and rimantadine, which are only active against influenza A. Before licensing of the NAIs in 1999, the adamantanes were the only drugs used for the treatment and prevention of influenza. These drugs target the virus M2 ion channel protein, involved in virus uncoating in the endosome . However, because of central nervous system complications in the elderly and lack of efficacy against influenza B, these agents were not widely employed. Additionally, since 2000, many viruses have acquired the substitutions L26F, L/V27A, A30T, S31 N, or G34E in the M2 gene conferring resistance, including the current human A(H3N2) and A(H1N1)pdm09 viruses and avian influenza A(H5N1) and A(H7N9) viruses, which have caused sporadic human infections. Resistance initially emerged in China, possibly related to the ready availability of the adamantanes in over-the-counter medications and use in poultry feed.
The second group of antivirals described here is called “NA inhibitor” (NAI) because these antiviral drugs bind to NA influenza virus and inhibit the enzymatic activity of this protein [21].
Antiviral agents may be prescribed as treatment to potentially shorten the duration and decrease the severity of influenza infection. Antivirals may also be prescribed for chemoprophylaxis to prevent/attenuate a potential influenza infection following contact with an infected individual or in vulnerable individuals during a community outbreak (e.g., nursing homes). When used as treatment, initiation of antiviral agents should not be delayed and ideally should be started within 48 h of the onset of symptoms.
When NA proteins change, the NAI can lose its ability to bind to and inhibit the function of the viral NA proteins, resulting in NAI resistance (non-susceptibility). A particular genetic change named the “H275Y” mutation is known to confer oseltamivir resistance in 2009 A(H1N1)pdm09 viruses. (The H275Y mutation is a substitution of histidine for tyrosine at position 275 in the NA.) This substitution prevents oseltamivir from inhibiting NA activity and allows the mutated virus to spread to healthy cells, which results in the drug not working as well. Although instances of antiviral drug resistance among influenza viruses have occurred during antiviral therapy, resistant influenza strains have also spread widely in the absence of such drug pressure. Viruses resistant to oseltamivir could not disseminate properly in the human population , which could explain the low frequency of resistant virus detected in the entire world. In a study performed in Central and South America in 2005–2008, the M2 and NA genes were sequenced, and resistance was inferred by comparison with published sequences and known resistant mutations. The results obtained indicated resistance to adamantanes in the majority of the A(H3N2) isolates, but in only one isolate of the influenza A(H1N1) viruses, and resistance to NAIs began to be detected in 2008 A(H1N1) isolates. Also, none of the influenza B viruses analyzed was resistant to NAIs. [13]. To spread in community settings, H275Y mutants must contain additional mutations. Oseltamivir-resistant A(H1N1)pdm09 strains with compensatory mutations were identified in different states of Brazil and in different countries [19]. Most of the influenza viruses tested during past seasons (2014, 2015) continued to be susceptible to the antiviral drugs recommended, although resistance to the adamantanes class of antiviral drugs among A/H3N2 and A/H1N1 viruses remains widespread [15]. In Argentina, in 2009, the first A(H1N1)pdm09 virus resistant to oseltamivir was detected in a 3-year-old child under treatment who had received an unrelated bone marrow transplantation and who had developed an upper respiratory tract infection with a long period of viral excretion. Between 2011 and 2015, in Argentina, using two rapid genotypic screenings, the H275Y substitution was found in 25 of 2216 influenza A(H1N1)pdm09 viruses tested and the E119V change in 1 of 1515 A(H3N2) viruses studied. Most of the viruses carrying the substitutions were collected from patients at risk without oseltamivir therapy . The resistant A(H1N1)pdm09 viruses showed the compensatory changes V241I and N369K. Oseltamivir appears to increase the chance of survival in people infected with H5N1 Asian lineage. These viruses are resistant to adamantanes and rarely to oseltamivir and zanamivir.
9 Prevention
Currently, the influenza vaccine, annually revised, includes three influenza virus strains: influenza A(H1N1)pdm09, A(H3N2), and influenza B. The vaccine includes seasonal strains and does not protect against the virus influenza C. According to the Centers for Disease Control and Prevention (CDC) , influenza vaccine is the best way to protect people against influenza and prevent its spread. This vaccine can also reduce the severity of the disease if a person contracts a strain of the influenza that is not perfectly antigenically matched with the strain included in the vaccine formula.
About 2 weeks following vaccination are needed for antibodies to protect against the virus. Influenza vaccine will not protect against infections and diseases caused by other viruses that can also cause symptoms similar to those of influenza. This vaccine is available in the Southern Hemisphere countries during the autumn months (March and April), before influenza season starts. It contains the H and N viral surface proteins that have been previously grown in eggs, and it is administered subcutaneously. There are two different types of inactivated influenza vaccines, trivalent and quadruple . Trivalent vaccines protect against two influenza A viruses (H1N1 and H3N2) and an influenza B virus. Quadruple vaccines protect against two influenza A viruses and two influenza B viruses (Yamagata and Victoria lineages). Influenza vaccine has to be applied every season because the body’s immune response from vaccination declines over time, so an annual vaccine is needed for optimal protection and also because influenza viruses are constantly changing. The vaccine formula is reviewed each year and sometimes updated to keep up with changing influenza viruses. The selection of the strains included in the vaccine is based on the analysis of the strains circulating during the previous winter season in the countries of the Northern Hemisphere and the Southern Hemisphere carried out by an expert group coordinated by WHO.
Until 1998, for many years, WHO had held a consultation meeting once a year, in mid-February, to formulate a recommendation for the composition of inactivated influenza vaccines intended for the following winter. Epidemics of influenza occur at different times of the year in different parts of the world. Consequently, it was appropriate for WHO to review the recommendation twice a year. Since October 1998, a recommendation every February continues, which relates to the composition of vaccines intended for use for the following winter in the Northern Hemisphere (November to April), and a second recommendation made each September was implemented that relates to vaccines which will be used for the following winter in the Southern Hemisphere (May to October) [1]. To determine if the locally circulating strains are antigenically closely related to the vaccine strains administered, Argentinean strains were compared with the reference influenza viruses isolated from May 1994 to December 1997. Nasopharyngeal aspirates and nasopharyngeal swabs collected from hospitalized children and adults, respectively, with acute lower respiratory tract infection were tested. In this study, it has been shown that influenza A(H3N2) circulating in Argentina during the past 4 years matched partially with the antigens present in the vaccines administered during the 1994–1997 period. These antigenic variants sometimes circulated late in the year (October 1994 and 1997), initiating during the following influenza season and becoming prevalent: they were present in the vaccine formula administered in the Southern Hemisphere 2 years later [28]. Influenza vaccines can be administered from 6 months of age and also to people considered to belong to at-risk groups, such as pregnant women in the second or third trimester and people suffering from chronic respiratory diseases; or those with cardiac, metabolic, and immune compromise; or those who are morbidly obese. Healthcare workers who provide community services should consider vaccination to minimize disruption of essential activities during influenza outbreaks.
In addition to the trivalent and quadruple inactivated vaccine, other vaccines are currently available. In 2003, a live attenuated influenza virus vaccine was approved for use in the U.S. This vaccine is approved only for healthy individuals from 5 to 49 years old. In 2007, it was also approved for healthy children aged 24 to 59 months. It is administered nasally and provides mucosal, humoral, and cell-mediated immunity. The strain included, which is cold adapted, can grow in the upper respiratory tract where the temperature is lower than in the lower tract. It is attenuated by multiple changes in different segments of the genome. Because it is a live virus vaccine, when administered intranasally (as an aerosol) it generates a response in IgA and IgM/IgG.
Because the virus is grown in eggs, vaccine application is contraindicated in persons with hypersensitivity to any component thereof.
References
Anonymous (1998) Recommendation for the composition of influenza virus vaccines for use in 1999. Wkly Epidemiol Rec 73:305–308
Baumeister ELG, Pontoriero A et al (2004) Serological evidences of influenza A virus infection in Antarctica migratory birds. In: Options for the control of influenza. International congress series V. Elsevier, San Diego, pp 737–740
Baumeister E, Palacios G, Cisterna D et al (2010) Molecular characterization of severe and mild cases of influenza A (H1N1) 2009 strain from Argentina. Medicina (B Aires) 70:518–523
Baumeister EG, Salvador R, Vigo G et al (2005) Molecular evidences of the influenza A virus presence in migratory Antarctic birds. In: VII Congreso Argentino de Virología Libro de resúmenes. Sociedad Argentina de Virología, Ciudad de Buenos Aires
Baumeister EG, Perfumo C et al (2007) Influenza subtype H3 circulation in fattening pigs of Argentina. In: Proceedings, options for the control of influenza VI. International Medical Press 2008., London, p 333. ISBN 978-1-901-769-15-6 https://isirv.org/site/images/stories/publication/isirv_Options_VI_Proceedings_Book.pdf
Benedetti E, Daniels RS, Pontoriero A et al (2016) Influenza virus surveillance in Argentina during the 2012 season: antigenic characterization, genetic analysis and antiviral susceptibility. Epidemiol Infect 144:751–767
Born PS, Siqueira MM, Faria NR et al (2016) Phylodynamics of influenza A(H3N2) in South America, 1999–2012. Infect Genet Evol 43:312–320
Cappuccio JA, Pena L, Dibarbora M et al (2011) Outbreak of swine influenza in Argentina reveals a non-contemporary human H3N2 virus highly transmissible among pigs. J Gen Virol 92:2871–2878
CDC (2014) How the flu virus can change: “drift” and “shift”. http://www.cdc.gov/flu/about/viruses/change.htm
Comas V, Moratorio G, Sonora M et al (2015) Phylogenetic analysis of the neuraminidase gene of pandemic H1N1 influenza A virus circulating in the South American region. Virus Res 197:1–7
Escalera-Zamudio M, Nelson MI, Cobian Guemes AG et al (2014) Molecular epidemiology of influenza A/H3N2 viruses circulating in Mexico from 2003 to 2012. PLoS One 9:e102453
Espinola EE, Amarilla AA, Martinez M et al (2014) Influenza A H1N1pdm 2009 virus in Paraguay: nucleotide point mutations in hemagglutinin and neuraminidase genes are not associated with drug resistance. Open Virol J 8:9–13
Garcia J, Sovero M, Torres AL et al (2009) Antiviral resistance in influenza viruses circulating in Central and South America based on the detection of established genetic markers. Influenza Other Respir Viruses 3:69–74
Guan Y, Vijaykrishna D, Bahl J et al (2010) The emergence of pandemic influenza viruses. Protein Cell 1:9–13
Hurt AC, Besselaar TG, Daniels RS et al (2016) Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2014–2015. Antivir Res 132:178–185
Hurtado R, Fabrizio T, Vanstreels RE et al (2015) Molecular characterization of subtype H11N9 avian influenza virus isolated from shorebirds in Brazil. PLoS One 10:e0145627
King Amq AM, Carstens EB, Lefkowitz EJ (2012) Virus taxonomy. Ninth report of the International Committee on taxonomy of viruses. Elsevier Academic Press, San Diego. www.trevorwilliams.info/ictv_iridoviridae_2012.pdf
Libster R, Bugna J, Coviello S et al (2010) Pediatric hospitalizations associated with 2009 pandemic influenza A (H1N1) in Argentina. N Engl J Med 362:45–55
Lopes E, Souza TM, Fintelman-Rodrigues N, Resende PC et al (2015) Oseltamivir-resistant influenza A(H1N1)pdm2009 strains found in Brazil are endowed with permissive mutations, which compensate the loss of fitness imposed by antiviral resistance. Mem Inst Oswaldo Cruz 110:101–105
Mathieu C, Moreno V, Pedersen J et al (2015) Avian influenza in wild birds from Chile, 2007–2009. Virus Res 199:42–45
Mckimm-Breschkin JL, Fry AM (2016) Meeting report: 4th ISIRV antiviral group conference: novel antiviral therapies for influenza and other respiratory viruses. Antivir Res 129:21–38
Muller I, Pinto E, Santibanez MC et al (2009) Isolation and characterization of the equine influenza virus causing the 2006 outbreak in Chile. Vet Microbiol 137:172–177
Nelson MI, Pollett S, Ghersi B et al (2016) The genetic diversity of influenza A viruses in wild birds in Peru. PLoS One 11:e0146059
Neumann G, Kawaoka Y (2011) The first influenza pandemic of the new millennium. Influenza Other Respir Viruses 5:157–166
Pereda A, Cappuccio J, Quiroga MA et al (2010) Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg Infect Dis 16:304–307
Periolo N, Avaro M, Czech A et al (2015) Pregnant women infected with pandemic influenza A(H1N1)pdm09 virus showed differential immune response correlated with disease severity. J Clin Virol 64:52–58
Pineyro PE, Baumeister E, Cappuccio JA et al (2010) Seroprevalence of the swine influenza virus in fattening pigs in Argentina in the 2002 season: evaluation by hemagglutination-inhibition and ELISA tests. Rev Argent Microbiol 42:98–101
Savy VL, Baumeister EG, Pontoriero AV (1999) Antigenic relationship between influenza A (H3N2) strains circulating in Argentina and vaccine strains. Medicina (B Aires) 59:225–230
Senne DA, Suarez DL, Stallnecht DE et al (2006) Ecology and epidemiology of avian influenza in North and South America. Dev Biol (Basel) 124:37–44
Shinya K, Ebina M, Yamada S et al (2006) Avian flu: influenza virus receptors in the human airway. Nature (Lond) 440:435–436
Tong S, Li Y, Rivailler P et al (2012) A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274
Tong S, Zhu X, Li Y et al (2013) New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657
Webster RG, Bean WJ, Gorman OT et al (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179
Webster RG, Govorkova EA (2014) Continuing challenges in influenza. Ann N Y Acad Sci 1323:115–139
Xu K, Ferreri L, Rimondi A et al (2012) Isolation and characterization of an H9N2 influenza virus isolated in Argentina. Virus Res 168:41–47
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Baumeister, E.G., Pontoriero, A.V. (2017). Influenza Viruses, Biology, Epidemiology, and Control. In: Ludert, J., Pujol, F., Arbiza, J. (eds) Human Virology in Latin America. Springer, Cham. https://doi.org/10.1007/978-3-319-54567-7_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-54567-7_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-54566-0
Online ISBN: 978-3-319-54567-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)