Abstract
Animal models have been an invaluable means to advance biomedical research as they provide experimental avenues for cellular and molecular investigations of disease pathology. The zebrafish (Danio rerio) is a good alternative to mammalian models that can be used to apply powerful genetic experimental methods normally used in invertebrates to answer questions about vertebrate development and disease. In the case of the kidney, the zebrafish has proven itself to be an applicable and versatile experimental system, mainly due to the simplicity of its pronephros, which contains two nephrons that possess conserved structural and physiological aspects with mammalian nephrons. Numerous genes that were not previously related to kidney conditions have now been linked to renal diseases by applying genetic screening with the zebrafish. In fact, a large collection of mutations that affect nephron formation and function were generated through phenotype-based forward screens. Complementary reverse genetic approaches have also been insightful, with methods spanning the use of antisense morpholino oligonucleotides to genome editing approaches such as the CRISPR/Cas9 system, to selectively knock down or knock out genes of interest to see if they produce kidney phenotypes. Acute kidney injury (AKI) has also been easily modeled in the zebrafish by injecting nephrotoxins, directly inducing damage through surgical intervention, or by generating transgenic lines that express compounds in a tissue-specific manner that when exposed to certain drugs promote an apoptotic response within cells. In this chapter, we provide an overview of these various approaches as well as discuss many of the contributions that have been achieved through the use of zebrafish to model kidney disease.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
1 Introduction
Animal models are the cornerstones of biomedical research. Over the last two decades, the zebrafish (Danio rerio) has received an unprecedented amount of attention within the scientific community (Pickart and Klee 2014). Although mammalian models have been a central means of studying human afflictions due to anatomical and evolutional similarities, there are also limiting factors with their applications. For example, rodent models are not always the optimal organisms to model certain conditions, due to factors such as inherent biological differences, the economical and space costs associated with the care and husbandry of the animals, or the molecular biology tools available for researching a particular question. A large degree of functional and genetic conservation between humans and nonmammalian organisms means that many diseases can be accurately modeled at the molecular level in a more cost-effective manner by using less complex organisms, such as flies or worms. With regard to this chapter, the genetic conservation between zebrafish and humans (Howe et al. 2013) has made them a premier model for efficacious basic and translational research and provided many prospects for novel ongoing studies (Lieschke and Currie 2007; Santoriello and Zon 2012).
For the purposes of kidney disease modeling, the zebrafish provides a particularly useful system to apply genetic approaches that have been traditionally used in the aforementioned invertebrate models to answer questions about cellular and developmental processes within vertebrates (Drummond 2005; Ebarasi et al. 2011). A suite of traits makes zebrafish amenable for organ development and disease studies in the embryo. For example, zebrafish development occurs ex utero, and the embryos are optically clear, enabling researchers to readily observe processes in real time within the context of the whole animal (Kimmel et al. 1995; Laale 1997). Most importantly, all common vertebrate organs are visible by 120–144 hours post-fertilization (hpf), with many apparent even earlier, at 24 hpf, such as the heart, eyes, and kidney (Rubenstein 2003). Thus, the development as well as the onset of metabolic functions for these common organs is not only fast but can be observed through the transparent embryos.
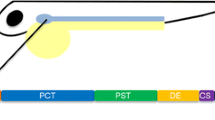
The zebrafish embryonic kidney, or pronephros, is composed of two blood-filtering glomeruli fused at the midline that are connected to two epithelial tubules (each divided into proximal and distal domains) and followed by paired bilateral pronephric ducts that join at the cloaca, which is where wastes exit the body (Drummond et al. 1998; Gerlach and Wingert 2013). The pronephros becomes functional beginning around 48 hpf (Drummond et al. 1998), and the structural composition of the zebrafish pronephros, as well as the segmentation pattern of the tubules, is highly conserved to that of the mammalian kidney (Wingert et al. 2007; Wingert and Davidson 2008, 2011). This high degree of structural conservation makes the zebrafish a useful counterpart to studies of mammalian kidney development (Fig. 3.1) (also discussed in Chap. 2).
Comparison between human nephron and zebrafish pronephros. There is a high degree of similarity between the nephron segmentation patterns of both organisms. Abbreviations in the human nephron: P podocytes, PCT proximal convoluted tubule, PST proximal straight tubule, TL thin limb, TAL thick ascending limb, MD macula densa, DCT distal convoluted tubule, CD collecting duct. Abbreviations in the zebrafish pronephros: P podocytes, N neck, PCT proximal convoluted tubule, PST proximal straight tubule, DE distal early, CS corpuscle of Stannius, DL distal late, PD pronephric duct, C cloaca
The adult kidney is also useful to model advanced developmental stages as well as renal afflictions that affect mature tissues. At around 12–14 days post-fertilization (dpf), the adult form, known as the mesonephros, begins to develop, with the progressive addition of new nephrons to the existing bilateral pair (Diep et al. 2015). When mesonephros generation is complete, the zebrafish possesses a more complex branched arrangement of nephron units within the kidney, but the nephrons show the same segmental composition (McCampbell et al. 2014, 2015). Mammals contain a fixed number of nephrons after birth and cannot regenerate or produce more after they are destroyed (Stocum 2012). Interestingly enough, in organisms such as the zebrafish or the goldfish, new nephrons keep being added to the existing blood-filtering array throughout the lifetime of the animal in response to a naturally increasing biomass (Reimschuessel et al. 1990; Reimschuessel and Williams 1995; Zhou et al. 2010; Diep et al. 2011). This phenomenon known as neonephrogenesis also occurs after kidney injury (Reimschuessel 2001). Because of the fundamental similarities between the mammalian and zebrafish kidney structures, the mesonephros can also be used to study the onset, progression, and recovery from kidney injury. Although complex in its anatomy, the mesonephros is relatively conducive to analysis when compared to arrangements of several thousand nephrons present in the rodent adult kidney (Kroeger and Wingert 2014).
In this chapter, we will discuss the versatility and advantages of the zebrafish pronephros and mesonephros kidney as models for renal disease, along with the various tools that have been implemented to study these systems in recent years.
2 Forward Genetic Approaches
One of the ways that zebrafish researchers can assess developmental dynamics is through the use of forward genetics, which involves identifying genes responsible for a specific phenotype. Because of the recently discussed advantages of zebrafish development, this organism can be subjected to large-scale forward genetic screens that produce a large number of mutants for analysis (Fig. 3.2).
Diagram of forward genetic screen approaches in the zebrafish model. F0 Generation: Mutagenized males are crossed with WT females to generate F1 families. F1 Generation: Heterozygotes are selected based on their phenotype and then outcrossed to expand the heterozygote population and generate F2 families. F2 Generation: Heterozygotes of the F2 generation are incrossed to bring the desired mutant phenotype to homozygosity. F3 Generation: Homozygous mutant is analyzed to identify the genetic cause of the observed mutant phenotype
2.1 Mutagenesis Screens
In a mutagenesis screen, adult males are exposed to an agent such as the chemical mutagen ethylnitrosourea (ENU), which induces random point mutations in their spermatogonial stem cells (Solnica-Krezel et al. 1994; Driever et al. 1996). The mutagenized males are then crossed with wild-type (WT) females to transmit the mutations onto their progeny. The F1 generation is then outcrossed with WT females in order to expand the heterozygote population, after which the resulting heterozygotes of the F2 generation are incrossed in order to produce homozygous mutants for analysis. Random mutagenesis screens can also be performed through exogenous DNA insertion. Although less efficient than chemical mutagenesis, the insertional mutagenesis method can greatly accelerate the identification of mutant genes (Amsterdam et al. 1999). This method involves inserting an exogenous DNA sequence, such as a retroviral vector, into the zebrafish genome to produce random mutations (Amsterdam and Hopkins 2006). The insertions can then serve as tags that can be used to identify the mutated gene in question. Both chemical and insertional mutagenesis screens can produce a multitude of mutants, where pertinent ones are then selected based on a phenotype of interest. Within the nephrology field, zebrafish mutants have been particularly useful for the study of cystic diseases.
Polycystic kidney disease (PKD) is a genetic condition characterized by the formation of multiple liquid-filled cysts within the kidney (Halvorson et al. 2010). This condition has been connected to mutations in the genes POLYCYSTIN-1 (PKD1) and POLYCYSTIN-2 (PKD2), as well as to overproliferation of the renal epithelium (Nadasy et al. 1995). PKD can be subdivided into two types: autosomal dominant (ADPKD) and autosomal recessive (ARPKD). ADPKD is one of the most common monogenic human disorders, affecting anywhere from 1/400 to 1/1000 live births, so the importance of understanding how this condition arises cannot be overstated. Using zebrafish mutants obtained through forward genetic mutagenesis screens, researchers have been able to develop various models of these conditions that have contributed to the understanding of disease onset and enabled further cell biological studies.
In a study by Drummond et al., researchers analyzed data obtained from a previously performed ENU mutagenesis screen for the purposes of identifying mutations affecting pronephric development (Drummond et al. 1998). Selection of mutants was based upon a common phenotype of fluid-filled cysts in place of the normal pronephric tubule. The group was able to identify 18 independent recessive mutations that affected all of the identifiable parts of the pronephros: the glomerulus, the tubules, and the duct. One of these identified mutants named double bubble showed glomerular expansion due to fluid buildup at around 40 hpf. By 56 hpf, the glomerulus was severely distorted and loose. Researchers stated that the aberrant glomerular morphology suggests that cyst formation was most likely due to early defects in glomerular structure, specifically with defects that affect filtration pressure. This study was able to link cyst formation with defective glomerulogenesis, providing some insight into the primary defects that can be responsible for cyst formation, which could lead to further insights about PKD pathogenesis.
A subsequent study used insertional mutagenesis to produce zebrafish embryos containing a similar polycystic renal phenotype (Sun et al. 2004). From this insertional mutagenesis screen, researchers were able to identify 12 genes whose disrupted expression causes cysts within the developing zebrafish kidney, one of which was pkd2, the zebrafish orthologue of human PKD2. Researchers found that although the pkd2 mutant produced by this screen did not exhibit kidney cysts before death, this was most likely due to maternal contribution of pkd2 transcript that carries the embryo through early development. Subsequent morpholino-mediated knockdown of pkd2 (method discussed in the next section), a process that targets both endogenous and maternally contributed transcripts, resulted in glomerular cyst formation, which speaks to the relevance these mutants have to human PKD.
Interestingly, three of the identified genes are homologues of genes encoding components of intraflagellar transport (IFT) particles, which are important in cilia formation. Cilia are microtubule-based organelles present on the cells in most tissues that serve a variety of metabolic and signaling functions. They have been shown to function as environmental sensors of mechanical stimuli (Vincensini et al. 2011) as well as signaling centers for various developmental pathways such as Hedgehog and Wnt (Lancaster and Gleeson 2009). Ciliopathies are conditions stemming from mutations in genes encoding and/or that process ciliary proteins, which result in ciliary dysfunction in the form of altered cilia structure or movement (Tobin and Beales 2008). Renal disease is a common aspect of ciliopathies, including, but not limited to, cyst formation, fusion of the lobules, and clubbing. For example, previous studies demonstrate that mutations in a human core intraflagellar transport gene, IFT80, can cause Jeune syndrome, a condition that among its characteristics features renal cysts (Beales et al. 2007). This work by Sun et al. (2004) similarly exemplifies how a forward genetic screen was useful to create zebrafish PKD models, and how mutants obtained through these screens can lead researchers to find previously unknown links between kidney cystogenesis and other cellular processes, providing a clearer picture of the genetic cause of PKD.
2.2 Future Directions
Various zebrafish mutants that resemble a number of human kidney-related conditions, as well as developmental defects, are still being discovered from the data produced by large-scale mutagenesis screens, and used to further elucidate the mechanisms of disease progression. Some such examples, the locke, switch hitter (swt), and kurly mutants (Brand et al. 1996; Haffter et al. 1996), which are characterized by a distinct “curly tail down” phenotype as well as cystic dilations of the pronephric tubules, were used to study the earlier cellular defects of cyst formation in the kidney. Sullivan-Brown et al. (2008) observed that locke, swt, kurly mutants develop tubular kidney cysts due to ciliary defects ranging from immotile cilia to cilia that beat irregularly. Analyzing the cilia of pkd2 morphants revealed that they beat in a coordinated fashion similar to that of WT embryos, which meant that the glomerular cysts resulting from pkd2 knockdown did not arise due to defects in cilia motility, but rather another, as of yet unknown mechanism. In doing this, researchers were able to observe a new mechanism for cyst formation that differed from what was seen in pkd2 morphants.
Combining the before-mentioned methods with the organism’s ex utero development, high fecundity, and the large number of embryos produced after mating, the zebrafish can be a powerful tool for in vivo drug discovery, looking for potential therapeutic agents that can ameliorate kidney conditions like PKD. Using a technique called “chemical genetics,” researchers expose embryos to a small molecule and then analyze the resulting phenotype within the context of a genetic pathway or disease of interest to assess the effects on gene expression or the disease phenotype.
For example, Cao et al. (2009) performed a chemical screen for compounds that ameliorated body curvature and laterality defects, aberrant phenotypes that are closely associated with kidney cyst formation, in previously identified zebrafish pkd2 and ift172 mutants. After applying the compounds to the mutant embryos, researchers were able to identify trichostatin A (TSA), a pan HDAC (histone deacetylase) inhibitor, as a compound that could modulate both body curvature and laterality defects. Additionally, they were able to demonstrate how this compound, as well as valproic acid (VPA), a Class I-specific HDAC inhibitor that is structurally unrelated to TSA that can also modulate body curvature and laterality defects, was able to suppress kidney cyst formation in pkd2 morphants. Through the use of a chemical genetic screen, Cao et al. (2009) were able to identify HDAC inhibitors as drug candidates for potential PKD treatments. Because of this and many other examples, forward genetic screens are a very promising approach that takes full advantage of the experimental versatility of this animal model.
3 Reverse Genetic Approaches
Although forward genetics is great at identifying multiple loci responsible for disease onset, random mutagenesis makes it hard to produce disease models for conditions that result from abnormal expression of specific genes because identification of mutants is based purely on the aberrant phenotype. Additionally, trying to identify the specific gene responsible for the observed mutant phenotype through methods like positional cloning is very work intensive and requires a large amount of effort. In cases like this, researchers turn to the use of reverse genetics, defined as the analysis of phenotypical consequences resulting from disruptions in genes of interest. Simply disrupt or mutate the gene, and then observe how the organism is affected.
3.1 Morpholino oligos
A standard reverse genetic technique performed on zebrafish is morpholino oligo-mediated gene knockdown. Morpholino oligos (MOs) are modified, antisense oligomer molecules that bind to a desired RNA transcript to prevent access of cell components to a target site on said transcript (Summerton 1999). MOs possess a nonionic backbone at physiological pH composed of 6-membered morpholine rings connected through phosphorodiamidate linkages. Because of the inherent nature of this chemical structure, MOs are highly soluble in aqueous solutions, highly stable in vivo, and resistant to a variety of enzymes and biological fluids (Janson and During 2006; Amantana and Iversen 2005; Hudziak et al. 2009). MOs work transiently in the embryo, where they act to block the ATG start site preventing translation, or they can block splice sites, which prevents processing of pre-mRNA into mature mRNA. Using this type of compound does not cause degradation of the RNA but prevents the biological activity of the RNA until it is degraded naturally. The process of using MOs involves microinjecting one-cell stage zebrafish embryos to deliver the oligonucleotide ultimately to all of the cells of the organism (Fig. 3.3a). This is possible because of the intrinsic high solubility of MOs, which allows them to diffuse readily throughout the one-cell stage embryo such that the resulting cells of the organism can subsequently inherit MO molecules. The embryos are then allowed to develop until a desired growth stage, at which point the specimen can be studied through any number of protocols to interrogate the phenotype.
Diagrams of reverse genetic techniques used in the zebrafish model. (a) Morpholino oligonucleotide is microinjected into the one-cell stage zebrafish embryo where it targets and binds to a specific mRNA strand. This prevents translation of the mRNA strand, drastically decreasing the amount of functional protein available. (b) CRISPR-Cas9 system. A guide RNA (gRNA) that specifically targets a gene of interest, as well as Cas9 endonuclease mRNA, is microinjected into the one-cell stage embryo. The gRNA binds to its target site, where it “guides” the binding of Cas9 which makes a double-strand cut in the DNA sequence. An insertion/deletion (indel) mutation is then produced through nonhomologous end joining
The binding affinity of the MO to a desired target sequence regulates its efficacy; however, MO concentration can be a limiting factor as it could become too diluted when development has progressed to a sufficient degree, making the MO-mediated knockdown a transient gene knockdown. Nevertheless, MO effects can be typically seen within the first three days of development but have also been reported after 5 dpf (Bill et al. 2009). To achieve genetic knockdowns over longer intervals, some researchers have supplemented the MO levels with more MO into individual or a small group of cells through techniques like electroporation which involves applying an electric field to cells in order to increase their permeabilization and introduce drugs or other molecules into the cell (Eisen and Smith 2008). Although not a complete gene knockout, this tool can still serve to substantially alter the normal expression of specific genes and can therefore provide a valuable approach to generate specific zebrafish models of disease.
One category of renal affliction that has benefited from the use of MOs has been ciliopathies. Researchers used MOs in zebrafish to generate models for this and other ciliopathies such as Jeune syndrome, Bardet–Biedl syndrome, Meckel syndrome, oro-facial-digital syndrome, and nephronophthisis, screen them for kidney phenotypes, and then identify compounds that might ameliorate said phenotypes. Representative gene candidates for all of these conditions were knocked down and each subsequent morphant developed kidney cysts, random body situs, and a downturned body axis, all features characteristic of ciliary mutants. These ciliopathy models were then used to assay the effects of rapamycin and roscovitine, two compounds that have been shown to retard or reverse the formation of kidney cysts (Tobin and Beales 2008). The individual compounds were able to rescue the renal edema caused by the cysts.
One drawback to using this method however is that, like most antisense technologies, MOs suffer from some off-target effects (Eisen and Smith 2008). A study by Kok et al. (2015) compares various mutant zebrafish lines generated through site-specific genome editing (discussed in next section) to published morpholino phenotypes. Researchers saw that out of the 20 genes they created mutant lines for, mutants for 10 of them did not recapitulate the morpholino phenotype, suggesting that these previously published morphant phenotypes were the result of morpholino off-target effects. In order to assess the off-target effects of an MO, appropriate controls are needed, and one of the best ways to do this is to compare the morphant phenotype to a mutant phenotype, should a mutation in the gene of interest be available. Validating the effects of one morpholino by analyzing the effects of another independent morpholino targeting the same gene to see if they are the same can also serve to assess off-target effects. Additionally, rescue studies can be performed to assess whether the morphant phenotype can be ameliorated through co-injection of the morpholino and a form of the targeted RNA (Eisen and Smith 2008). If the RNA for the affected gene is unable to rescue the phenotype, this suggests that the phenotype is most likely due to off-target effects (Eisen and Smith 2008). Ultimately, when the appropriate controls are applied, these works demonstrate the effectiveness of MOs as tools for renal disease modeling, as well as how the resulting models might be used for the development of therapeutics.
3.2 Genome Editing
MO knockdown may be an effective way to generate disease models in zebrafish; however, the nature of the “knockdown” allows for some transcripts to still be translated into functional protein. To create “knockout” models, in which expression of a desired gene is completely abrogated, zebrafish researchers have turned to genome editing approaches. One of the more promising and more recently developed techniques being used today is the CRISPR/Cas9 system. CRISPRs, an abbreviation for the term “Clustered Regularly Interspaced Short Palindromic Repeats,” are short segments of prokaryotic DNA that serve as part of the bacterial immune system to protect it from foreign DNA elements (Barrangou et al. 2007). By recognizing and binding to the foreign DNA sequences, they recruit CRISPR-associated protein 9 (Cas9), an RNA-guided endonuclease enzyme, to degrade DNA sequence the CRISPR is bound to. Scientists took advantage of this naturally occurring bacterial immune system to edit the genome of other animal models, such as the zebrafish (Cong et al. 2013). This type of genome editing begins by designing a guide RNA (gRNA) that will target a gene of interest and then “guide” the Cas9 protein to that location. Next, a cocktail of gRNA and Cas9 mRNA are microinjected into the one-cell stage zebrafish embryo, where the gRNA will bind to the desired target sequence and the Cas9 mRNA will be translated to produce the endonuclease enzyme. The Cas9 will then recognize the gRNA, bind to it, and produce a double-stranded break in the DNA. This break will then be repaired through nonhomologous end joining, which will result in random indel (insertion/deletion) mutations that can abrogate gene expression (Fig. 3.3b).
Using this system, researchers can successfully knock out genes that have been linked to various renal conditions (Anderson et al. 2015). Coding variants of human aplipoprotein L1 (APOL1) have been attributed to an increased risk of focal glomerular segmental sclerosis (FSGS) in African Americans; however, very little evidence exists as to the role of APOL1 in the kidney. Anderson et al. (2015) used the CRISPR/Cas9 system to create an apol1 knockout zebrafish mutant as a means of supplementing MO-mediated knockdown studies that try to analyze the role of this gene in renal function. Analysis of the mutant and morphant phenotypes revealed the presence of a pericardial edema at 3 dpf, indicative of glomerular filtration defects. Additionally, analysis of the organization and patterning of mutant and morphant glomeruli revealed an altered glomerular ultrastructure in both morphant and mutant embryos that was conducive to a poor glomerular filtration barrier. Using the CRISPR-Cas9 system, in conjunction with MO-mediated knockdown researchers were able to establish a solid, well-evidenced relationship between disease onset and variants of APOL1.
Like most molecular biology techniques, genome editing systems also have their drawbacks. A study by Rossi et al. (2015) revealed that mutants generated by TAL effector nucleases (TALENs), an alternative genome editing technique to the CRISPR-Cas9 system, could potentially be activating compensatory networks to shield against harmful mutations. Researchers generated mutants for egfl7, a gene that lacks obvious phenotypes when knocked out in mice and demonstrates severe vascular tube formation defects when knocked down in zebrafish, frog, and human cells, and then compared those mutants to egfl7 morphants. They observed that the severe vascular tube formation defects present in the morphants were either very mild or not present at all in the mutants. After confirming that the severe vascular phenotype in the morphants was not due to off-target effects, they proceeded to analyze both the transcriptome and proteome of morphant and mutant embryos and saw that emilin3a, emilin3b, and emilin2a transcripts and Emilin3a protein were upregulated in egfl7 mutants but not in morphant embryos. The emilin genes have an EMI domain, a key unit of Egfl7 function, and co-injection of emilin gene mRNA with egfl7 MO revealed that these transcripts could actually rescue egfl7 morphants. These results suggest that the upregulation of these genes within the egfl7 mutants could serve to compensate for the lack of egfl7. The possibility of these compensatory networks makes interpreting mutants generated via genome editing somewhat difficult which is why future studies should utilize both MO-mediated knockdown and mutants generated through genome editing techniques so as to compensate the drawbacks of each.
3.3 Future Directions
In addition to the MO and CRISPR/Cas9 systems, there are a number of other methods that are being used in zebrafish that have yet to be applied to the zebrafish renal field. One such example would be the Cre/loxP system, which has been widely used in mice to model diseases. Wang et al. (2008) demonstrated how efficient this system is to investigate genetic pathways in a tissue-specific manner by driving Cre recombinase under the control of the zebrafish lmo2 promoter, a tissue-specific promoter expressed in primitive hematopoietic stem cells as well as vascular endothelial cells. Not only were they able to show that Cre could effectively be expressed only in specific tissues, but they were also able to show that Cre expression faithfully recapitulated the endogenous lmo2 expression pattern. Additionally this system can be adapted to allow for temporal control of a transgene by expressing a modified version of Cre fused to a mutated hormone-binding domain of the estrogen receptor (CreER), which can only be activated in the presence of tamoxifen, a synthetic estrogen receptor ligand (Hans et al. 2009). This study serves to emphasize the usefulness of this method and denotes a possible way to study other, more late-onset renal diseases such as adult-onset PKD and renal cancer in the much simpler zebrafish renal system. A modified version of the CRISPR system can also be used for this purpose (inducible CRISPR or iCRISPR), by taking advantage of the Tetracycline-On system. This system uses a mutated version of the tetracycline repressor fused with virion protein 16 (VP16) called reverse tetracycline-controlled transactivator (rtTA) to activate expression of a transgene cassette when exposed to doxycycline, a member of the tetracycline class of antibiotics. Three expression cassettes are developed for this system: rtTA under the control of a constitutive promoter, the gRNA under the control of a ubiquitous promoter, and the Cas9 gene under the control of a tetracycline-dependent promoter. These tetracycline-dependent promoters consist of a tetracycline response element (TRE) upstream of a minimal promoter. When added, doxycycline binds with rtTA and changes the rtTA protein conformation, which now means that rtTA can bind to the TRE, effectively promoting expression of Cas9. The Cas9 then interacts with the gRNA molecules to produce targeted indel mutation (González et al. 2014; Dow et al. 2015). This system can also be modified for tissue-specific knockout by putting expression of rtTA under the control of a constitutively expressed, tissue-specific promoter like that of cadherin17, which is expressed exclusively in the zebrafish kidney from 24 hpf to adulthood.
An example of an alternate knockdown system would be the introduction of synthetic microRNA (miRNA) cassettes into the zebrafish genome to knockdown a desired gene (Dong et al. 2009; Giacomotto et al. 2015). RNA interference (RNAi) using synthetic small hairpin RNAs (shRNAs) has been shown to be an efficient method of posttranscriptional gene silencing in many model organisms (Dong et al. 2009). In combination with the miR-30 miRNA backbone, higher amounts of shRNAs can be produced. Dong et al. (2009) took the zebrafish homologues of miR-30 and miR-155 miRNA precursors (pri-miR-30e and pri-miR-155, respectively), cloned them into a pCS2 vector, and then replaced the endogenous miRNA stem-loop sequence with a 24 nucleotide long linker sequence containing two Bbs I restriction sites. This would allow insertion of a synthetic shRNA stem loop specific for a desired gene, effectively making a miRNA/shRNA hybrid construct (mir-shRNA). After determining that targeting the 3′ untranslated region (UTR) resulted in a more potent knockdown of reporter genes, researchers created miR-shRNA constructs for the genes chordin and α-catenin that, when injected into embryos, induced efficient knockdown of the two developmentally important genes. Transgenic zebrafish lines were also successfully created containing these miR-shRNA constructs under the control of the lineage-specific gata1 promoter, meaning that not only is this miRNA-mediated knockdown effect heritable but tissue specific as well. Giacomotto et al. (2015) also induced effective heritable gene knockdown of smn1 using this approach, effectively reproducing spinal muscular atrophy (SMA) in zebrafish, an autosomal recessive human disease that is characterized by motor neuron loss, progressive muscle weakness, and in severe cases death.
Notably, there has been some controversy regarding the use of RNAi-mediated knockdown. Zhao et al. (2008) demonstrated how injecting zebrafish embryos with small interrupting RNAs (siRNAs) specific for six3a and eri1 inhibited endogenous miR-430 processing, which resulted in many off-target effects such as tail truncations and distinct morphological features at the mesencephalic–mentencephalic boundary due to saturation of the miRNA pathway. However, recent studies have shown that generating transgenic lines that stably express integrated miRNA constructs for a desired gene of interest can circumvent these off-target effects, which speaks to the effectiveness of this knockdown method (Dong et al. 2009; Giacomotto et al. 2015).
Targeted Induced Local Lesions In Genomes, or TILLING, is probably one of the most recognized reverse genetic techniques currently in use across multiple fields. It involves screening DNA of randomly mutagenized fish for point mutations in a gene of interest. Once a mutation has been discovered, either the corresponding male is bred or its cryopreserved sperm are implanted in a female through in vitro fertilization until the mutation is brought to homozygosity (Draper et al. 2004). To date, there has been little to no data obtained using this technique pertaining to the study of renal disease; however, TILLING is still a good alternative to more common techniques, such as MOs or CRISPR, as a means to identify kidney mutants. These zebrafish research technologies are being developed and improved with each passing day, making said organism an increasingly attractive animal model for kidney disease.
4 Inducible Damage Models
Acute kidney injury (AKI) is a type of disorder that encompasses a wide range of factors, with a 50–70% mortality rate within intensive care patients, a rate that has not changed over the last few decades (Thadhani et al. 1996; Chertow et al. 2005). Treatments for AKI are extremely limited for patients with more severe cases, where renal replacement and dialysis therapies are the only treatments shown to be effective in humans (Thadhani et al. 1996). Mammalian models, such as rats or mice, of AKI are often difficult to interpret because of a plethora of challenges that include inaccessibility of the kidney and poor visualization of the renal tubules. Some alternative models like cell culture have come close but still cannot faithfully recapitulate the in vivo environment or produce adequate three-dimensional cultures in order to model injury.
Thus, researchers have turned to the zebrafish as a model system to study AKI. As previously stated, not only does the zebrafish kidney have conserved nephron structure and segmentation to that of mammalian nephrons, but their embryos and larvae are translucent, which makes observing changes along the entire length of the kidney under a microscope quite simple. Additionally, zebrafish have a relatively complex mesonephros that provides a convenient and useful adult setting for renal studies. Combined with the fact that they produce a large number of offspring when they breed, and that they have an incredible regenerative capacity after injury (McCampbell et al. 2015), zebrafish are uniquely qualified to help better elucidate the mechanisms of AKI progression and recovery.
4.1 Nephrotoxins
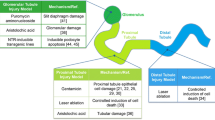
One of the ways that researchers model AKI in zebrafish is by injecting them with gentamicin and/or cisplatin, antibiotics that are also known nephotoxins (Fig. 3.4a). In a study led by Hentschel et al. (2004), the researchers characterized and quantified the effects of gentamicin injection in zebrafish larvae, as evidenced by histological and functional changes similar to aminoglycoside toxicity in mammals. Gentamicin injury was characterized by pronephric tubule flattening of the brush border, distension of the tubules and glomeruli, lysosomal phospholipidosis, accumulation of leukocytes, as well as debris in the tubular lumen. Similar effects were observed in cisplatin-injected zebrafish kidneys. These changes resulted in decreased renal function and an inability to maintain fluid homeostasis, resulting in the development of a pericardial edema.
Because of this ability to recapitulate AKI, nephrotoxins have also been used to characterize regeneration within the adult zebrafish kidney (Diep et al. 2011; McCampbell et al. 2015). In particular, McCampbell et al. (2015) analyzed and mapped the temporal progression of the cellular and molecular dynamics of regeneration after gentamicin injury, establishing a regeneration time course damage induction to complete regeneration of proximal epithelial nephron tubules.
4.2 Mechanically Induced Kidney Insults
Some drawbacks to using nephrotoxins on both larval and adult zebrafish are likely to include leakage of toxins out of the animal because solubilized liquid compounds are injected into an organism that lives in an aquatic environment. This may partially account for variability in the degree of AKI in fish exposed to the same dosage of antibiotic (Kamei et al. 2015). Consequently, there is no true way to quantify how much of the drug is actually affecting the fish kidney. As an alternative, inducing AKI through mechanical means can circumvent this difficulty. In a study done by Hellman et al. (2010), researchers mechanically obstructed the zebrafish kidney fluid flow for the purpose of understanding regulation of cilia function in response to injury by looking at foxj1a expression, an important transcription factor required for motile ciliogenesis. Researchers took anesthetized 12-month-old fish and made an incision at the level of the distal collecting system. Next, they used tweezers to pinch off the distal collecting system for 30 s which produced damage in the form of dilated nephron tubules. After which they proceeded to allow the fish to recover, along with providing antibiotics to prevent infection, for a period of 12 h overnight (Fig. 3.4b). Using this mechanical means of inducing kidney injury, they were able to determine that inducible foxj1a is essential for cilia beat rate maintenance over a 24-h time period, suggesting that enhanced cilia function may be an important, previously uncharacterized part of organ homeostasis.
Another type of mechanical insult used in the field today is laser-mediated cell ablation (Johnson et al. 2011). Palmyre et al. (2014) utilized focused violet (405 nm) laser photoablation on a 20–100 μm stretch of pronephric epithelium in transgenic zebrafish embryos expressing GFP under the control of kidney-specific promoters (Fig. 3.4c). The rationale behind the use of the 405 nm laser on GFP-expressing zebrafish is twofold: (1) the GFP would allow for a more focused and intense beam on a specific area, in this case the fluorescent tissue, and (2) GFP absorbs light around 405 nm, meaning that the fluorescent protein would serve as an energy sink to potentiate cell injury. Using this targeted and highly specific method for inducing injury in the zebrafish embryo, researchers were able to analyze the mechanisms of renal repair, noting how collective kidney epithelial cell migration is an early response to injury and how cell proliferation occurs after migration.
4.3 Spatial/Temporally Controlled Methods of Kidney Injury
One of the most significant advances in the last couple of years has been the development of a noninvasive, tissue-specific method of inducing kidney injury that is both quantifiable and able to recapitulate more specific and subtle forms of kidney injury, such as podocyte ablation, which leads to glomerulosclerosis. The injury models in question take advantage of bacterial nitroreductase (NTR) and its ability to convert the prodrug metronidazole (MTZ) into a cytotoxic metabolite (Zhou and Hildebrandt 2012; Huang et al. 2013). Researchers generated transgenic zebrafish expressing NTR fused to a fluorescent reporter under the control of the podocin promoter, so as to only induce injury in the podocytes. After which they added a specific amount of MTZ to the fish tanks, which was taken in by the fish through their gills (Fig. 3.4d). Using this system, the researchers were able to create animal models of glomerulosclerosis to characterize podocyte injury progression, as well as the mechanisms of recovery (Zhou and Hildebrandt 2012; Huang et al. 2013).
4.4 Future Directions
Zebrafish carry with them the immense possibility for answering many questions pertaining to AKI pathophysiology, as well as injury recovery and regeneration. Understanding both global and specific kidney damage using the methods described previously can help physicians provide better care for patients with AKI. Additionally, zebrafish have incredible regenerative capacity after AKI, completely recovering from damage 14–21 days after injury (McCampbell et al. 2015). This is of significant importance because even though mammals can regenerate their kidney tubule epithelium, they cannot regenerate nephrons after they are destroyed (Stocum 2012), by either disease or AKI. By understanding the cellular mechanisms of zebrafish kidney regeneration potential, cell therapies can be developed that can ameliorate kidney damage.
Another way zebrafish kidney damage models can be used to develop kidney therapies is when they are used in tandem with chemical genetic screens to find compounds that can ameliorate kidney damage, enhance kidney injury recovery, or post-injury fibrosis after AKI. de Groh et al. (2010) performed a chemical screen to identify compounds that would expand the renal progenitor cell field, explaining that compounds that expand renal progenitor cell number might also enhance recovery after injury. In this screen, they identified the HDAC inhibitor methyl-4-(phenylthio) butanoate (m4PTB) as a compound that expanded the renal progenitor cell field in a proliferation-dependent manner. Based on the results of this screen, Cianciolo et al. (2013) induced AKI in both zebrafish larvae and adult mice through gentamicin injection and ischemic reperfusion (IR) AKI, respectively, and then treated the organisms with m4PTB to see if this would affect their recovery. m4PTB treatment after AKI proved to accelerate recovery and survivorship in both zebrafish and mice, making this compound a potentially viable approach to treating AKI. These methods of inducible injury, combined with an incredible regenerative potential and a large molecular biology tool box, have made the zebrafish an incredible tool for disease study and therapy development.
5 Conclusion
From forward genetic screens to genome editing and inducible injuries approaches, this chapter has discussed various ways in which the zebrafish can be used to recapitulate kidney disease and has discussed how these can be used to further understand disease onset to provide better treatment, and hopefully create new therapies for patients with renal disease. The question of “how” zebrafish models of kidney disease and injury can further development of therapeutics, which is the ultimate goal of understanding any human disease, is one that is constantly being discussed however.
That being said, no disease model is perfect. There will be instances in which gene function will diverge between zebrafish and humans and others in which kidney physiology is too dissimilar in order to create an adequate disease phenotype, an example of which would be the lack of intermediate tubule segments in the zebrafish nephron. Nevertheless, the organism’s experimental versatility, limited in many ways only by a researcher’s imagination, is vast. The best way to take advantage of this animal model is to use it in conjunction with mammalian ones in order to get a clearer picture of the cellular processes taking place during kidney injury and disease. From a pet store novelty to an important model organism, the zebrafish has become an invaluable tool for scientific discovery.
References
Amantana A, Iversen PL (2005) Pharmacokinetics and biodistribution of phosphorodiamidate morpholino antisense oligomers. Curr Opin Pharmacol 5:550–555
Amsterdam A, Hopkins N (2006) Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet 22:473–478
Amsterdam A, Burgess S, Golling G et al (1999) A large-scale insertional mutagenesis screen in zebrafish. Genes Dev 13:2713–2724
Anderson BR, Howell DN, Soldano K et al (2015) In vivo modeling implicates APOL1 in nephropathy: evidence for dominant negative effects and epistasis under anemic stress. PLoS Genet 11:e1005349
Barrangou R, Fremaux C, Deveau H et al (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Beales PL, Balnd E, Tobin JL et al (2007) IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat Genet 39:727–729
Bill BR, Petzold AM, Clark KJ et al (2009) A primer for morpholino use in zebrafish. Zebrafish 6:69–77
Brand M, Heisenberg CP, Warga RM et al (1996) Mutations affecting development of the midline and general body shape during zebrafish embryogenesis. Development 123:129–142
Cao Y, Semanchik N, Lee SH et al (2009) Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc Natl Acad Sci 106:21819–21824
Chertow GM, Burdick E, Honour M et al (2005) Acute kidney injury, mortality, length of stay, and costs in hospitalized patients. J Am Soc Nephrol 16:3365–3370
Cianciolo CC, Skrypnyk NI, Brilli LL et al (2013) Histone deacetylase inhibitor enhances recovery after AKI. J Am Soc Nephrol 24:943–953
Cong L, Ran FA, Cox D et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
de Groh ED, Swanhart LM, Cosentino CC et al (2010) Inhibition of histone deacetylase expands the renal progenitor cell population. J Am Soc Nephrol 21:794–802
Diep CQ, Ma D, Deo RC et al (2011) Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature 470:95–100
Diep CQ, Peng Z, Ukah TK et al (2015) Development of the zebrafish mesonephros. Genesis 53:257–269
Dong M, Fu YF, Du TT et al (2009) Heritable and lineage-specific gene knockdown in zebrafish embryo. PLoS One 4:e6125
Dow LE, Fisher J, O’Rourke KP et al (2015) Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 33:390–394
Draper BW, McCallum CM, Stout JL et al (2004) A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol 77:91–112
Driever W, Solnica-Krezel L, Schier AF et al (1996) A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123:37–46
Drummond IA (2005) Kidney development and disease in the zebrafish. J Am Soc Nephrol 16:299–304
Drummond IA, Majumdar A, Hentschel H et al (1998) Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 125:4655–4667
Ebarasi L, Oddsson A, Hultenby K et al (2011) Zebrafish: a model system for the study of vertebrate renal development, function, and pathophysiology. Curr Opin Nephrol Hypertens 20:416–424
Eisen JS, Smith JC (2008) Controlling morpholino experiments: don’t stop making antisense. Development 135:1735–1743
Gerlach GF, Wingert RA (2013) Kidney organogenesis in the zebrafish: insights into vertebrate morphogenesis and regeneration. Wiley Interdiscip Rev Dev Biol 2:559–585
Giacomotto J, Rinkwitz S, Becker TS (2015) Effective heritable gene knockdown in zebrafish using synthetic microRNAs. Nat Commun 6:7378
González F, Zhu Z, Shi Z et al (2014) An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15:215–226
Haffter P, Granato M, Brand M et al (1996) The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123:1–36
Halvorson CR, Bremmer MS, Jacobs SC (2010) Polycystic kidney disease: inheritance, pathophysiology, prognosis, and treatment. Int J Nephrol Renovasc Dis 3:69–83
Hans S, Jaslin J, Freudenreich D et al (2009) Temporally–controlled site-specific recombination in zebrafish. PLoS One 4(2):e4640
Hellman NE, Liu Y, Merkel E et al (2010) The zebrafish foxj1a transcription factor regulates cilia function in response to injury and epithelial stretch. Proc Natl Acad Sci 107:18499–18504
Hentschel DM, Park KM, Cilenti L et al (2004) Acute renal failure in zebrafish-a novel system to study a complex disease. Am J Physiol Renal Physiol 288:F923–F929
Howe K, Clark MD, Torroja CF et al (2013) The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503
Huang J, McKee M, Huang HD et al (2013) A zebrafish model of conditional targeted podocyte ablation and regeneration. Kidney Int 83:1193–1200
Hudziak RM, Barofsky E, Barofsky DF et al (2009) Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev 6:267–272
Janson CG, During MJ (2006) Peptide nucleic acids, morpholinos and related antisense biomolecules. Morpholinos and PNAs compared. Landes Bioscience and Kluwer Academic, New York, pp. 89–113
Johnson CS, Holzemer NF, Wingert RA (2011) Laser ablation of the zebrafish pronephros to study renal epithelial regeneration. J Vis Exp 54:e2839
Kamei CN, Liu Y, Drummond IA (2015) Kidney regeneration in adult zebrafish by gentamicin induced injury. J Vis Exp 102:e51912
Kimmel CB, Ballard WW, Kimmel SR et al (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310
Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND (2015) Reverse genetic screening reveals poor correlation between morpholinoinduced and mutant phenotypes in zebrafish. Dev Cell 32:97–108
Kroeger PT Jr, Wingert RA (2014) Using zebrafish to study podocyte genesis during kidney development and regeneration. Genesis 52:771–792
Laale HW (1997) The biology and use of zebrafish. Brachydanio rerio in fisheries research. A literature review. J Fish Biol 10:121–173
Lancaster MA, Gleeson JG (2009) The primary cilium as a cellular signaling center: lessons from disease. Curr Opin Genet Dev 19:220–229
Lieschke GJ, Currie PD (2007) Animal models of human disease: zebrafish swim into view. Nat Rev Genet 8:353–367
McCampbell KK, Springer KN, Wingert RA (2014) Analysis of nephron composition and function in the adult zebrafish kidney. J Vis Exp 90:e51644
McCampbell KK, Springer K, Wingert RA (2015) Atlas of cellular dynamics during zebrafish adult kidney regeneration. Stem Cells Int 2015:547636
Nadasdy T, Laszik Z, Lajoie G et al (1995) Proliferative activity of cyst epithelium in human renal cystic diseases. J Am Soc Nephrol 5:1462–1468
Palmyre A, Lee J, Ryklin G et al (2014) Collective epithelial migration drives kidney repair after acute injury. PLoS One 9:e101304
Pickart MA, Klee EW (2014) Zebrafish approaches enhance the translational research tackle box. Transl Res 163:65–78
Reimschuessel R (2001) A fish model of renal regeneration and development. ILAR J 42:285–291
Reimschuessel R, Williams D (1995) Development of new nephrons in adult kidneys following gentamicin-induced nephrotoxicity. Ren Fail 17:101–106
Reimschuessel R, Bennett RO, May EB et al (1990) Renal tubular cell regeneration, cell proliferation and chronic nephrotoxicity in the goldfish Carassius auratus following exposure to a single sublethal dose of hexachlorobutadiene. Dis Aquat Org 8:211–224
Rossi A, Kontarakis Z, Gerri C et al (2015) Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524:230–233
Rubinstein AL (2003) Zebrafish: from disease modeling to drug discovery. Curr Opin Drug Discov Devel 6:218–223
Santoriello C, Zon LI (2012) Hooked! modeling human disease in zebrafish. J Clin Invest 122:2337–2343
Solnica-Krezel L, Schier AF, Driever W (1994) Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics 136:1401–1420
Stocum, DL (2012) Regeneration of digestive respiratory and urinary tissues. Regenerative biology and medicine. 2nd edn. Burlington: Elsevier Academic. Pg. 118
Sullivan-Brown J, Schottenfeld J, Okabe N et al (2008) Zebrafish mutations affecting cilia motility share similar cystic phenotypes and suggest a mechanism of cyst formation that differs from pkd2 morphants. Dev Biol 314:261–275
Summerton J (1999) Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta 1489:141–158
Sun Z, Amsterdam A, Pazour GJ et al (2004) A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131:4085–4093
Thadhani R, Pascual M, Bonventure JV (1996) Acute renal failure. N Engl J Med 334:1448–1460
Tobin JL, Beales PL (2008) Restoration of renal function in zebrafish models of ciliopathies. Pediatr Nephrol 23:2095–2099
Vincensini L, Blisnick T, Bastin P (2011) 1001 model organisms to study cilia and flagella. Biol Cell 103:109–130
Wang L, Zhang Y, Zhou T et al (2008) Functional characterization of Lmo2-Cre transgenic zebrafish. Dev Dyn 237:2139–2146
Wingert RA, Davidson AJ (2008) The zebrafish pronephros: a model to study nephron segmentation. Kidney Int 73:1120–1127
Wingert RA, Davidson AJ (2011) Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev Dyn 240:2011–2027
Wingert RA, Selleck R, Yu J et al (2007) The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet 3:1922–1938
Zhou W, Hildebrandt F (2012) Inducible podocyte injury and proteinuria in transgenic zebrafish. J Am Soc Nephrol 23:1039–1047
Zhou X, Fjose A, Larsen N et al (2008) Treatment with small interfering RNA affects the microRNA pathway and causes unspecific defects in zebrafish embryos. FEBS J 275:2177–2184
Zhou W, Boucher RC, Bollig F et al (2010) Characterization of mesonephric development and regeneration using transgenic zebrafish. Am J Physiol Renal Physiol 299:F1040–F1047
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Morales, E.E., Wingert, R.A. (2017). Zebrafish as a Model of Kidney Disease. In: Miller, R. (eds) Kidney Development and Disease. Results and Problems in Cell Differentiation, vol 60. Springer, Cham. https://doi.org/10.1007/978-3-319-51436-9_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-51436-9_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-51435-2
Online ISBN: 978-3-319-51436-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)