Abstract
In the medical community, automatic epileptic seizure detection through electroencephalogram (EEG) signals is still a very challenging issue for medical professionals and also for the researchers. When measuring an EEG, huge amount of data are obtained with different categories. Therefore, EEG recording can be characterized as big data due to its high volume. Traditional methods are facing challenges to handle such Big Data as it exhibits non-stationarity, chaotic, voluminous, and volatile in nature. Motivated by this, we introduce a new idea for epilepsy detection using complex network statistical property by measuring different strengths of the edges in the natural visibility graph theory. We conducted 10-fold cross validation for evaluating the performance of our proposed methodology with support vector machine (SVM) and Discriminant Analysis (DA) families of classifiers. This study aims to investigate the effect of segmentation and non-segmentation of EEG signals in the detection of epilepsy disorder.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Around 50 million people in world-wide is effected by one of the most widely existing chronic neurological syndrome named as epilepsy [1]. In epilepsy, recurrent seizure attacks due to malfunctioning of the electrophysiological part of the brain come to pass at any stage of life. Electroencephalogram (EEG) is one of electro-physiological technique to study and measure the voltage fluctuation of the brain and helps in diagnose the epilepsy disorder as epilepsy leaves their signature in the EEG signals [2]. There are wide-ranging of existing methods from linear to non-linear for the detect epilepsy disorder [3]. However, these techniques do not preserve all characteristics of EEG time series data such as, non-stationarity, chaotic nature [4]. Hence there is an ever-increasing need to develop new techniques that can detect epilepsy disorder by preserving the relevant information and provide additional information about epileptic EEG signals.
In the current era, complex network and graph theory approach is becoming the emergent field to detect various brain disorders [5]. In 2010, Ahmadlou et al. [6] firstly applied visibility graph algorithm for the detection of Alzheimer disorder and obtained promising results. After that many researchers and clinicians applied visibility graph algorithm for the detection of epilepsy disorder [7, 8] but their proposed methods have some limitations as they have not considered an important fact that in network, the links exhibit different strengths and all the nodes of network are connected with each other on the basis of this strength. Therefore, by focusing the limitation of the existing (especially visibility graph) methods to detect the epilepsy disorder, our proposed technique has explore the idea of, the importance of edge weight in epilepsy detection through visibility graph by constructing weighted complex network. It is our believe that, this proposed methodology is really new and will be very useful in the field of epilepsy and other brain disorder detection.
1.1 Contribution and Organization of the Paper
In this paper, we perform several experiments to discriminate between different kinds of EEG signals and make the following contributions.
-
In this paper we developed new edge weight calculation method which helps to record the sudden changes happen in EEG signal for the duration of seizure activity. As during seizure activity the amplitude of EEG signals are too much fluctuating with time and our proposed method helps to recognize this fluctuation easily.
-
We investigate the effect of segmentation and non-segmentation process on EEG signals in detection of epilepsy disorder with our proposed methodology.
-
Several experiments performed for different classification problems and the outcomes results for all the test cases also suggests that our proposed technique is best appropriate to differentiate between different kinds of EEG signals. Moreover, it is quite promising for the classification of EEG signals of epileptic seizure activity set (E) and healthy person with eye open (A) with 100 % accuracy.
This remaining paper has been structured as: Sect. 2 comprises complete description of the proposed methodology. Section 3 presents the detailed discussion about the experimentation procedure and results. In Sect. 4, conclusions along with the future work have been mention.
2 Proposed Methodology
In this work, a novel algorithm based on new edge weight method for visibility graph is proposed to detect epilepsy disorder from EEG brain signals. The schematic diagram of proposed methodology is presented in Fig. 1. The approach is effective to distinguish between different EEG signals and epileptic EEG signals. The entire procedure of this methodology is composed of various sections: conversion of time series EEG signals into weighted complex network, statistical feature extraction of weighted complex network, classification of epileptic EEG signals from different kinds of EEG signals.
2.1 Conversion of Time Series Data into Complex Network
For mapping the time series data into weighted complex network we have used lucasa [9] visibility graph algorithm. Following steps are used for the construction of weighted complex network:
-
I.
Consider each sample point of a time series x(ti), i = 1,2,…….N of N sampling points as a node ni of graph G(N,E), where N represents the node set i.e. N = {ni},i = 1,2,…….N, and E = ei, i = 1,2,3,…………N, are the edges of graph.
-
II.
The edges between the nodes of the graph are determined on the basis of [9] the following Eq. (1).
where, ni, nj and nk are the nodes corresponds to the data sample points x(ti), x(tj) and x(tk) with the time events ti, tj and tk.
-
III.
In this study, we develop the below Eq. (2) to calculate the edge weight between two nodes:
where, wij is the edge weight between node i and j and also directional in nature from i to j. Also absolute value of edge weight has been considered in all cases.
-
IV.
Finally a weighted complex network has been constructed from the EEG time series data by utilizing the edges weight values.
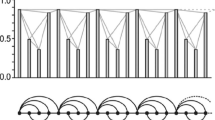
Figure 2 presents an example of the weighted complex network build upon the above four steps using EEG time series data sample points = {100, 124, 153, 185, 210, 220, 216, 222, 240, 265, 298, 330, 362, 381, 391}. The edge weight showing in this figure is constructed on the basis of our newly developed Eq. (2). The thickness of edges in the below figure is according to the edge weight values.
2.2 Feature Extraction
Feature extraction plays an important role for classification of EEG signal data. It helps to make the analysis process easier as extracted features compress the huge amount of EEG signal data into feature vector set by minimizing the loss of information in the original EEG signals. In this paper, we have extracted one statistical property of network named as average weighted degree of network as feature from weighted complex network. This statistical property helps to determine the underlying pattern of hidden information from brain EEG signals.
If a ANxN = {aij} is an adjacency matrix with N number of nodes is used to represent the weighted complex network then aij = 1 if there is an edge from node i to j otherwise it’s 0. According to [10] the weighted degree of the node i is the total weights of all the edges attached to node i which is represented by:
where, B(i) represents the neighborhood of node i and wij represents the weight of the edges between nodes i and j. And the average weighted degree of the network is the average mean of the total weights of the incident links on all the vertices in the network. It is important to note that due to sudden fluctuations in the epileptic EEG signals, the edge weight will show a discrepancy and different kinds of EEG signals exhibit different edge weight among their nodes and thus their resultant networks has different average weighted degree values.
2.3 Classification
In this paper, we have used two well-known supervised machine learning classification method named as Support Vector Machine (SVM) classifier with different kernel function and Discriminant Analysis classifier with discriminant type as linear and quadratic, for the evaluation of the performance of the proposed technique by utilizing the resulting feature extracted from feature extraction technique. As LDA and SVM is easy to implement and has fast prediction speed.
2.3.1 Support Vector Machine (SVM)
SVM is basically a binary classifier i.e. it can efficiently classify the data that belong to two different classes. SVM mechanism is based upon of finding the best hyperplane that separates the data of two different class of category [11]. To evaluate the performance of our proposed methodology for different test cases, we have employed the following three different kernel function in this paper.
-
I.
Linear kernel function:
-
II.
Polynomial kernel function with degree d:
-
III.
Radial basis kernel function with width σ:
2.3.2 Discriminant Analysis
In this paper, we have used two Discriminant Analysis (DA) classification methods named as Linear Discriminant Analysis (LDA) and Quadratic Discriminant Analysis (QDA). In case of LDA, each class has same covariance but the means vary whereas in case of QDA, the covariance and means parameters varies for each class. The detailed description about DA classifier are available in [12].
3 Experimental Results and Discussion
In this section, the proposed method is tested on the online available epileptic benchmark (http://epileptologiebonn.de/cms/front_content.php?idcat=193&lang=3) database: Bonn university epileptic EEG data. The whole database comprises of five EEG datasets (denoted as Set A–Set E). EEG signal in Set A and Set B were recorded from surface EEG recordings of five healthy volunteers with eyes open and eyes closed, respectively. Set C and Set D were collected in seizure-free intervals from five epileptic patients from the hippocampal formation of the opposite hemisphere of the brain and from within the epileptogenic zone, respectively. Set E contains the EEG records of five epileptic patients during seizure activity. Each channel of every set contains 4097 data sample points of 23.6 s. For detail description of this database please refer Andrzejak et al. [13]. The proposed technique is tested on the below four different classification problems named as test cases build upon this data set:
Test Case 1: Set A versus Set E
Test Case II: Set B versus Set E
Test Case III: Set C versus Set E
Test Case IV: Set D versus Set E
The proposed technique has been implemented with the help of MATLAB R2015b (version 8.6, 64 bit). In this research study, we wanted to examine the effect of segmentation of EEG signals in the detection of epilepsy disorder. In order to include more data, the segmentation of a signal can provide more meaningful information and can be considered as a part of the entire data set [7]. Moreover, this will also help to make computation task faster. By considering this information into account, the experimentation of the proposed technique has been conducted for the following two objectives:
-
1
First objective is to check the performance of our proposed method by considering whole data samples per channel i.e. by considering 4097 data sample points per channel. Here each channel is considered as independent samples. The implementation process of this method as discussed in Sect. 2.
-
2
Second objective is to check the performance of our proposed methodology with segmentation of each channel of EEG signals. During second approach, we divided each channel into four segment i.e. Seg1 = 1024, Seg2 = 1024, Seg3 = 1024, Seg4 = 1025 data sample points. Then these four segments are further used as a four independent samples. As in each data subset, there are 100 channels data with 4097 data points therefore after segmentation; we have 400 segments with 1024 data sample points. Afterward the proposed method (Sect. 2) is implemented on these segments.
To evaluate the performance of proposed epilepsy detection method and also to achieve more reliable results, k-fold cross validation method is applied on all the four test cases. In this paper, we have considered k = 10 i.e. in 10 fold cross validation, the feature sets are randomly partitioned into 10 groups. The classification model utilize 9 groups for training purposes and the remaining 10th group is used for testing. This whole procedure has been repeated 10 times. The performance of extracted feature vector sets is analyzed with the help of SVM and LDA families of classifiers. The results of different test cases after applying the proposed technique with LDA and QDA classifier is presented in Tables 1 and 2.
It is clear from both the tables, that the QDA is more efficient for our proposed methodology as compared to LDA. Moreover in case of QDA during segmented and non-segmented of EEG signals, the accuracy results are very close to each other except for test case 2, which is also showing only slight increase in case of non-segmentation. It can be seen that quadratic LDA classifier is demonstrating 100 % efficiency to distinguish between healthy (set A) and epileptic seizure activity (set E) EEG signals in both segmented and non-segmented process.
Our methodology is further analysed with other different kernel functions of SVM named as Linear, RBF and Polynomial kernel function. Table 3 illustrates the results after applying SVM classifier using linear kernel function on all the four test cases. Tables 4 and 5 lists the experimental results of all the four test cases with segmentation and without segmentation approach using SVM Rbf and SVM Polynomial kernel functions.
The experimental outcomes of Table 5 signifies that SVM Polynomial kernel function outperforms as compared to other SVM classifiers and achieve promising results with 100 % accuracy for distinguish between healthy person and epileptic seizure activity EEG signals. Moreover during segmented and non-segmented EEG signals, the accuracy results are very close to each other except for test case 2, which is also showing only slight increase in case of non-segmentation.
In order to provide a clear scenario, Fig. 3(a)–(c) presents a comparison of obtained performances for the different classifiers in both segmentation and non-segmentation process.
As can be observed from Fig. 3(a) SVM polynomial has higher accuracy and specificity classification performance as compared to other classifiers for all the four test cases problems whereas sensitivity of the LDA achieved higher performance results as compared to other classifiers. Table 6 presents the comparative analysis of the classification accuracy of the proposed method with different methods in the literature that perform experimentation on the same EEG data set and illustrate that the proposed methodology is more accurate for detection of epileptic seizure (set A vs set E) as compared to them. Moreover provide promising results for different test cases except for test case 3.
As for the duration of seizure activity, there is sudden change in neural discharge in the brain. Thus these consequences increase in variation of EEG signals and henceforth results with the sudden fluctuation in EEG signals. Due to this sudden fluctuation, the edge weight of the complex network build from this seizure EEG signals starts varying and helps to reveal the hidden information of brain functionality. This study also explores that different nodes of EEG weighted complex network interact with each other with different strengths. Therefore when complex network theory is used to detect epilepsy from brain EEG signals, the edge weight play an important role to detect the sudden fluctuation during seizure activity. This study also investigate that our proposed methodology produce very close results during segmentation and without segmentation approaches with SVM classifier. So there is no huge effect of segmentation and without segmentation of EEG signals on our proposed technique. Moreover segmentation approach takes fast computation time due to less amount of analysis data.
4 Conclusion
In this research study, we presented an efficient technique to detect epilepsy disorder from EEG brain signals. This methodology introduces new method to calculate the edge weight of complex network. We then constructed weighted complex network with the help of our newly developed edge weight method. Statistical property of complex network named as average weighted degree is used as extracted feature to compare the classification performance of SVM and DA families of classifiers. The outcomes of the experiments yield that in DA family, QDA provides higher accuracy performance results as compared to LDA but overall SVM polynomial is most promising classifier for our proposed methodology with higher performance results as compared to other classifiers. Moreover in this study, we investigate that, in case of segmentation and without segmentation approaches of EEG signals, the performance results for different test cases are very close to each and does not varies a lot. The pilot study in this paper has examined that the proposed methodology is best suitable to discriminate between different EEG signals. The classification accuracy performance result for ictal (set E) and normal healthy person EEG (set A) is achieved by100 % with SVM polynomial and quadratic discriminant analysis when considering both segmentation and without segmentation approach. Moreover the sensitivity performance for set A versus set E is 100 % with the family of SVM and LDA classifier. It is our believed that this research study will support the technicians to build a software system that will provides support for automatic detection of epileptic seizure. We are currently planning to extend this proposed methodology to detect other brain disorders through EEG signals and also to multi-class EEG classification in the epilepsy detection from brain signals.
References
Siuly, S., Li, Y.: Designing a robust feature extraction method based on optimum allocation and principal component analysis for epileptic EEG signal classification. Comput. Methods Programs Biomed. 119, 29–42 (2015)
Supriya, S., Siuly, S., Zhang, Y.: Automatic epilepsy detection from EEG introducing a new edge weight method in the complex network. Electron. Lett. (2016)
Donner, R., Small, M., Donges, J., Marwan, N., Zou, Y., Xiang, R., Kurths, J.: Recurrence-based time series analysis by means of complex network methods. Int. J. Bifurcat. Chaos 21, 1019–1046 (2011)
Campanharo, A., Sirer, M., Malmgren, R., Ramos, F., Amaral, L.: Duality between Time Series and Networks. PLoS ONE 6, e23378 (2011)
van Stam, C., Straaten, E.: The organization of physiological brain networks. Clin. Neurophysiol. 123, 1067–1087 (2012)
Ahmadlou, M., Adeli, H., Adeli, A.: New diagnostic EEG markers of the Alzheimer’s disease using visibility graph. J. Neural Transm. 117, 1099–1109 (2010)
Tang, X., Xia, L., Liao, Y., Liu, W., Peng, Y., Gao, T., Zeng, Y.: New approach to epileptic diagnosis using visibility graph of high-frequency signal. Clin. EEG Neurosci. 44, 150–156 (2013)
Ni, Y., Wang, Y., Yu, T., Li, X.: Analysis of epileptic seizures with complex network. Comput. Math. Methods Med. 2014, 1–6 (2014)
Lacasa, L., Luque, B., Ballesteros, F., Luque, J., Nuno, J.: From time series to complex networks: The visibility graph. Proc. Nat. Acad. Sci. 105, 4972–4975 (2008)
Antoniou, I., Tsompa, E.: Statistical analysis of weighted networks. Discrete Dyn. Nat. Soci. 2008, 1–16 (2008)
Andrew, A.: An Introduction to Support Vector Machines and Other Kernel‐based Learning Methods (2001)
Mikat, S., Fitscht, G., Weston, J., Scholkopft, B., Muller, K.-R.: Fisher discriminant analysis with kernels. Neural Net. Signal Proc. IX, 41–48 (1999)
Andrzejak, R., Lehnertz, K., Mormann, F., Rieke, C., David, P., Elger, C.: Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E 64, 61907 (2001)
Siuly, L.: Y., Wen, P.: Clustering technique-based least square support vector machine for EEG signal classification. Comput. Methods Programs Biomed. 104, 358–372 (2011)
Nicolaou, N., Georgiou, J.: Detection of epileptic electroencephalogram based on Permutation Entropy and support vector machines. Expert Syst. Appl. 39, 202–209 (2012)
Zhu, G., Li, Y., Wen, P.: Epileptic seizure detection in EEGs signals using a fast weighted horizontal visibility algorithm. Comput. Methods Programs Biomed. 115, 64–75 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Supriya, Siuly, Wang, H., Zhuo, G., Zhang, Y. (2016). Analyzing EEG Signal Data for Detection of Epileptic Seizure: Introducing Weight on Visibility Graph with Complex Network Feature. In: Cheema, M., Zhang, W., Chang, L. (eds) Databases Theory and Applications. ADC 2016. Lecture Notes in Computer Science(), vol 9877. Springer, Cham. https://doi.org/10.1007/978-3-319-46922-5_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-46922-5_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46921-8
Online ISBN: 978-3-319-46922-5
eBook Packages: Computer ScienceComputer Science (R0)