Abstract
The human hippocampus is a complex structure consisting of multiple anatomically and functionally distinct subfields. Obtaining subfield-specific measures from in vivo MRI is challenging, and can benefit from a detailed 3D anatomical reference. This paper builds a computational atlas of the hippocampus from high-resolution ex vivo MRI of 26 specimens using groupwise deformable registration. A surface-based approach based on the explicit segmentation and geometric modeling of hippocampal layers is used to initialize deformable registration of ex vivo MRI scans. This initialization improves of groupwise registration quality, as measured in terms of similarity metrics and qualitatively. The resulting atlas, which also includes annotations mapped from histology, is a unique resource for describing variability in hippocampal anatomy.
This work is supported by NIH grants R01 AG037376, R01 EB017255, and R01 AG040271. Specimens obtained from NDRI and UPenn brain banks.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
The hippocampus, located in the medial temporal lobe (MTL), is a major component of the declarative memory system, and is of acute interest in many brain disorders. It is a complex anatomical structure formed by multiple layers in a “swiss roll” configuration. The outer layers include subfields Cornu Ammonis (CA) 1–3 and subiculum, and the inner layer includes the dentate gyrus (DG), divided into the inner hilus region and the outer DG proper. Hippocampal subfields are hypothesized to be selectively affected by Alzheimer’s disease, aging, epilepsy, and other disorders [10], and to play different roles in normal memory.
Given their differential involvement in disease and memory, there has been increased interest in using in vivo MRI to interrogate hippocampal subfields [5]. However, even with highly optimized parameters, in vivo MRI provides only limited ability to distinguish hippocampal subfields. For instance, T2-weighted MRI with high resolution in the plane parallel to the main axis of the hippocampus makes it possible to see a dark layers separating the CA and subiculum from the DG; but boundaries between CA subfields, CA3 and DG, and CA1 and subiculum have to be inferred, typically based on heuristic rules [5].
Some authors have proposed to use ex vivo MRI, which can provide dramatically better resolution and contrast, to inform and guide the analysis of in vivo MRI. In [15], an atlas of the hippocampus was created from five specimens using groupwise image registration, and subfield distributions from the atlas were mapped into the in vivo MRI space using shape-based interpolation. More recently, in [7], an atlas was constructed by applying groupwise registration to multi-label manual segmentations of 15 ex vivo MRI scans, and used as a shape and intensity prior for in vivo MRI segmentation.
In addition to MRI, ex vivo specimens can be processed histologically, and rich information from histology can be mapped into the MRI space. This makes it possible to define anatomical boundaries in the MRI space based on the cytoarchitectural features used by anatomists to define subfields, rather than based on heuristic rules used in the MRI literature [1]. Furthermore, histology can be used to quantify pathology, e.g., tau neurofibrillary tangles in Alzheimer’s disease. Mapping such pathological information into an atlas of the hippocampal region can provide a statistical characterization of the distribution of pathology in this region, and in turn, serve as valuable resource for analysis and interpretation of in vivo MRI (or PET) studies.
The current paper develops the largest ex vivo computational atlas of the hippocampus to date, and the first such atlas to combine information from high-resolution MRI and dense serial histology. The algorithmic contribution of the paper lies in the use of a surface-based registration scheme to initialize pairwise registration of ex vivo MRI scans, leading to improved groupwise registration.
2 Methods and Results
2.1 Ex Vivo Imaging
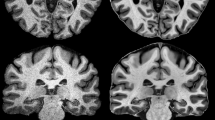
Intact ex vivo brain bank specimens of the hippocampus (n = 26, 14 left / 12 right, 10 left/right pairs from same subject) were scanned after >21 days formalin fixation at 9.4 tesla at \(0.2 \times 0.2 \times 0.2\,\mathrm {mm}^3\) or similar resolution using a multi-slice spin echo sequence described in [15] (see Fig. 1 for example). A subset of specimens (n = 8) was processed histologically, by cutting tissue into 1 cm blocks, paraffin embedding, slicing at \(\sim 200\,{\mu }\mathrm {m}\) intervals, staining using the Kluver-Barrera method, and optical scanning at \(0.5 \times 0.5\,{\mu \mathrm{m}}^2\) resolution.
2.2 Overview of the Two-Stage Registration Approach
As shown below in Fig. 3, direct image registration between specimens does not align structures in the hippocampus well. The proposed two-stage approach is based on the segmentation of two anatomical structures: the whole hippocampus, and the myelinated layers of CA and DG (strata radiatum and lacunosomoleculare; SRLM) that separate these two structures and appear dark in MRI. Laying between CA and DG, and spanning the whole length of the hippocampus, the SRLM forms a natural “skeleton” of the structure (figuratively speaking, distinct from the medial axis of the hippocampus). To make our atlas, SRLM and whole hippocampus were segmented manually, but we show that semi-automatic segmentation is feasible. For each specimen, we solve the Laplace equation to obtain a half-way surface between the hippocampal boundary and SRLM. Surface correspondences between half-way surfaces of different specimens are obtained by solving the Laplace equation between each half-way surface and the tightest fitting ellipsoid. These correspondences are then propagated to the entire image volume and used to initialize groupwise volumetric image registration between specimens. The sections below detail the different steps in this approach.
2.3 Ex Vivo MRI Segmentation
One author segmented the hippocampus and SRLM in all specimens (Fig. 1). Given the size of data, fully manual segmentation was impractical, and instead we manually edited segmentations generated semi-automatically (hippocampus: multi-atlas segmentation [13] using “rough” segmentations drawn in earlier work; SRLM: random forest [3] classifier applied to intensity processed with sheetness filter [6]). Overall, manual editing was extensive – over 8 h per specimen.
To establish the feasibility of automating segmentation in future work, we implemented a multi-atlas automatic pipeline for hippocampus and SRLM segmentation. This pipeline requires the user to label the outer hippocampal boundary every 20th coronal slice of the target image, which is interpolated and used for initial affine registration. Then each atlas is registered to the target image using diffeomorphic deformable registration with the normalized cross-correlation metric [2]. Consensus hippocampus and SRLM labels are obtained using the joint label fusion (JLF) algorithm [13]. This is repeated 5 times, with each iteration’s affine atlas-target registrations bootstrapped by the results of the previous iteration’s joint label fusion. This pipeline was applied in a leave-one-out framework. The average Dice coefficient between the (pseudo) manual segmentation and JLF segmentation was \(0.80\pm 0.08\) for SRLM and \(0.94\pm 0.02\) for the hippocampus. Unfortunately, there is no prior data on the reliability of manual or automatic segmentation of SRLM in ex vivo MRI to put these numbers into context. The most comparable high-resolution in vivo MRI study [14] reports intra-rater Dice coefficient 0.71 for SRLM and 0.91 for whole hippocampus. Overall, our Dice coefficients can be considered high, particularly since SRLM is a thin structure. This suggests that semi-automatic segmentation may be used in place of extensive manual workup in the future, as new specimens are added to the atlas.
2.4 Initial Shape-Based Normalization
The hippocampus occupies a different location in each specimen, and before intensity-based image registration can be performed, initial alignment of the hippocampi is required. In [15], a strategy based on the continuous medial representation (cm-rep) was proposed. The cm-rep is a deformable model that approximates the hippocampus and imposes a shape-based coordinate system on its interior (the coordinate system follows the medial axis of the deformable model). Each MRI scan can then be warped into the space of the cm-rep template, such that the outer boundaries and the medial axes of the hippocampi in different specimens approximately align. This step is not necessary per se for the proposed surface-initialized registration strategy, but to facilitate its comparison with [15], cm-rep normalization was applied as a preprocessing step.
Surface-based correspondence approach. A. Hippocampal and SRLM boundaries and the halfway (mid-potential) surface \(L_i^0\) of one subject. B. Superior and inferior views of halfway surface \(L_i^0\), rendered within its minimum volume enclosing ellipsoid (MVEE), and colored by its spherical parameterization angles \(\theta \) and \(\phi \). C. Spherical coordinate parameterization of the halfway surface in 4 specimens. D. Parameterization of the image domain by potential \(\rho \) and spherical coordinates \(\theta \) and \(\phi \). (Color figure online)
2.5 Surface Registration
Surface registration in our method leverages special properties of harmonic functions, and is closely related to prior work on cortical thickness estimation [8]. For subject i, let \(\mathcal{S}_i\) and \(\mathcal{H}_i\) denote the SRLM and hippocampus segmentation, respectively. Let \(\varOmega \) be the image domain after initial normalization. Since SRLM is fully contained in the hippocampus label, \(\mathcal{S}_i \subset \mathcal{H}_i \subset \varOmega \). Surface registration begins with solving the Laplace equation on \(\varOmega \), with the boundaries of \(\mathcal{S}_i\), \(\mathcal{H}_i\) and \(\varOmega \) modeled as equipotential surfaces. We find \(\rho : \varOmega \rightarrow \mathbb {R}\) that solves
Assuming \(\partial \mathcal{S}_i\) and \(\partial \mathcal{H}_i\) have spherical topology, the shells \(L^t_i = \{x: \rho (x) = t\}\) for \(t \in (-1,1)\) are a family of nested spherical surfaces that smoothly span the region between \(\partial \mathcal{S}_i\) and \(\partial \mathcal{H}_i\). We define the mid-potential surface \(L^0_i\) as the “halfway surface” between SRLM and hippocampus for subject i. Shown in Fig. 2A, this surface captures the geometric characteristics of both SRLM and overall hippocampus, which, we believe, makes it the ideal target for finding geometric correspondences between specimens.
To establish such correspondences, for each subject i, we construct a bijective mapping \(f : L^0_i \rightarrow S^2\) from the halfway surface to the unit sphere \(S^2\), based on diffeomorphic potential gradient flow mapping between \(L^0_i\) and its minimum volume enclosing ellipsoid (MVEE), denoted \(E_i\). \(E_i\) is the tri-axial ellipsoid of minimum volume and arbitrary spatial orientation that entirely encloses \(L^0_i\). It is uniquely defined and may be seen as a tight, quadric approximation of \(L^0_i\) that, being the affine image of \(S^2\), has a trivial spherical parameterization. The MVEE is computed using Khachiyan’s method [12]. Next, we solve the Laplace equation on the image domain, with \(L^0_i\) and \(E_i\) as equipotential surfaces:
The field \(\nabla \tau (x)\) is then integrated along its non-intersecting gradient field lines (or streamlines) from \(L^0_i\) to \(E_i\). Each point \(x \in L^0_i\) is assigned the spherical coordinate \((\theta , \phi )\) of the point in \(E_i\) at which the streamline traced at x terminates. This yields a coordinate map \(\theta (x), \phi (x)\) on \(L^0_i\) (Fig. 2B). We note that this approach is analogous to the spherical shape parameterization strategy in [4], except that [4] used a spherical enclosing boundary (rather than ellipsoid).
Correspondences obtained using gradient flow to the MVEE do not take into account local surface features like curvature, but rather the overall hippocampal shape. As shown for four specimens in Fig. 2C, they appear sensible upon visual inspection. Since our method does not “lock in” these correspondences but uses them as an initial point for groupwise intensity registration (see Sect. 2.6), such approximate correspondence is suitable for our purposes.
Lastly, the spherical coordinates \((\theta , \phi )\) for each subject i are propagated from \(L^0_i\) to the entire image volume \(\varOmega \). This is done by tracing the streamlines of \(\nabla \rho \) from each voxel center \(y \in \varOmega \) to the endpoint \(x_y \in L^0_i\). Voxel y is then assigned a triple of coordinates \(\{\rho (y), \theta (x_y), \phi (x_y)\}\), where \(\rho (y)\) can be interpreted as the “depth” coordinate (e.g. \(0< \rho < 1\) means y is between \(L^0_i\) and \(\mathcal{H}_i\)). An example of the resulting image of \(\rho , \theta , \phi \) coordinates is plotted in Fig. 2D.
2.6 Groupwise Intensity Registration
Deformable image registration [2] was performed in a groupwise unbiased framework proposed in [9]. Given input images \(Z_1 \ldots Z_k\) and initial groupwise template \(T_0\) (initialized as the voxel-wise average of \(Z_1 \ldots Z_k\)), this iterative approach alternates between performing diffeomorphic registration between \(Z_1 \ldots Z_k\) and \(T_p\) and updating \(T_{p+1}\) as the average of images \(Z_1 \ldots Z_k\) warped to \(T_p\). Groupwise registration was applied in three settings:(1) directly to the MRI intensity of the ex vivo scans after initial cm-rep normalization; (2) to the coordinate maps \(\rho , \theta , \phi \); (3) to the MRI intensity of ex vivo scans after applying warps obtained by groupwise registration of coordinate maps \(\rho , \theta , \phi \) in Method 2. Method 1 is the “reference” intensity-based approach used in [15]. Method 2 is a shape-based method that disregards MRI intensity. Method 3 is the proposed “hybrid” method. The normalized cross-correlation (NCC) metric was used for MRI registration, and the mean square difference metric was used for coordinate map registration; otherwise registration parameters were the same. Template-building ran for 5 iterations. Fig. 3 shows the templates obtained by these three methods. The hybrid method yields the sharpest template.
Table 1 reports mean pairwise agreement between specimens warped into the space of the three templates. Average Dice coefficient for SRLM and hippocampus masks is highest for the shape-based method, followed closely by the hybrid method. This is to be expected, since \(\rho , \theta , \phi \) maps are derived directly from SRLM and hippocampus segmentations. The key observation in Table 1 is that the template created with the hybrid method has greater average intensity similarity between pairs of MRI scans, as measured by the NCC metric. This suggests that initialization by surface correspondences yields a better groupwise MRI registration result, although further confirmation of this using expert-placed anatomical landmarks is needed.
2.7 Mapping Histology to Atlas Space
Interactive software HistoloZee was used to manually label hippocampal subfields in individual histology slices (Fig. 4A) and to reconstruct histology stacks in 3D with MRI as a reference (Fig. 4B). The software tool allows interactive in-plane translation, rotation, scaling, shearing of histology slices, adjustment of histology slice z-spacing, and 3D rigid transformation of the MRI volume. Histology slices were matched in this way to intermediate ex vivo MRI scans of the 1 cm blocks sectioned for histology. The intermediate block MRI scans were registered to the intact specimen MRI scans. These transformations were composed with the transformations from groupwise registration to map histology segmentations of 8 specimens into MRI template space. Figure 4C shows the mean distribution of histologically-derived subfield labels in the MRI template space.
A. Example histology slice, with manual segmentation, and overlay on the corresponding MRI slice. B. Serial histology stacks, reconstructed and aligned to the intact specimen MRI, through the intermediate stage of aligning to 1 cm tissue block MRI. C. Distribution of histologically-derived subfield labels in MRI template space obtained by averaging aligned annotations from 8 specimens.
3 Discussion
Contribution. Our ex vivo hippocampus atlas, constructed from 26 MRI specimen scans and 8 dense serial histology datasets, is the largest and most complex such atlas, to our knowledge. In contrast to the large (n = 15) atlas in [7], our atlas incorporates histology, which, according to [11], is the most accurate way to identify subfield boundaries. There are many potential uses of this atlas, including as a prior for in vivo segmentation [7], as an anatomical reference space for analysis of functional MRI data [15], and as a way to relate cytoarchitectonic aspects of hippocampal anatomy to MRI-observable macroscopic features.
Limitations. The surface-volume registration approach was built and evaluated using pseudo-manual segmentation, which is acceptable for the one-time purpose of creating this unique atlas, but problematic for extending the atlas to new specimens. The accuracy of automatic segmentation of SRLM and hippocampus reported in Sect. 2.3 is encouraging, but it remains to be shown that the gains from the surface-based initialization will persist when applied to automatic segmentations. Relatedly, the evaluation of templates in Table 1 is biased in the sense that Dice agreement in same structures that are used to establish correspondence is reported. This does not preclude us from reaching the main conclusion – that leveraging shape helps improve intensity match (NCC metric), but in the future, the use of expert-placed landmarks could help evaluate the method more extensively. Lastly, the proposed groupwise registration strategy did not combine matching of \((\rho ,\theta ,\phi )\) maps and MRI intensity in the same optimization, but rather performed the two types of registration sequentially. Joint optimization of shape and intensity matching may lead to even better templates.
References
Adler, D.H., Pluta, J., Kadivar, S., Craige, C., Gee, J.C., Avants, B.B., Yushkevich, P.A.: Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage 84, 505–523 (2014)
Avants, B., Epstein, C., Grossman, M., Gee, J.: Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008)
Breiman, L.: Random forests. Mach. Learn. 45(1), 5–32 (2001)
Chung, M.K., Worsley, K.J., Nacewicz, B.M., Dalton, K.M., Davidson, R.J.: General multivariate linear modeling of surface shapes using surfstat. Neuroimage 53(2), 491–505 (2010)
de Flores, R., La Joie, R., Chételat, G.: Structural imaging of hippocampal subfields in healthy aging and Alzheimer’s disease. Neuroscience 309, 29–50 (2015)
Frangi, A.F., Niessen, W.J., Vincken, K.L., Viergever, M.A.: Multiscale vessel enhancement filtering. In: Wells, W.M., Colchester, A., Delp, S. (eds.) MICCAI 1998. LNCS, vol. 1496, pp. 130–137. Springer, Heidelberg (1998). doi:10.1007/BFb0056195
Iglesias, J.E., Augustinack, J.C., Nguyen, K., Player, C.M., Player, A., Wright, M., Roy, N., Frosch, M.P., McKee, A.C., Wald, L.L., Fischl, B., Van Leemput, K.: Alzheimer’s disease neuroimaging initiative: a computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage 115, 117–137 (2015)
Jones, S.E., Buchbinder, B.R., Aharon, I.: Three-dimensional mapping of cortical thickness using laplace’s equation. Hum. Brain Mapp. 11(1), 12–32 (2000)
Joshi, S., Davis, B., Jomier, M., Gerig, G.: Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage 23(Suppl 1), S151–S160 (2004)
Small, S., Schobel, S., Buxton, R., Witter, M., Barnes, C.: A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 12(10), 585–601 (2011)
van Strien, N.M., Widerøe, M., van de Berg, W.D.J., Uylings, H.B.M.: Imaging hippocampal subregions with in vivo MRI: advances and limitations. Nat. Rev. Neurosci. 13(1), 70 (2012)
Todd, M.J., Yıldırım, E.A.: On khachiyan’s algorithm for the computation of minimum-volume enclosing ellipsoids. Discrete Appl. Math. 155(13), 1731–1744 (2007)
Wang, H., Suh, J.W., Das, S.R., Pluta, J., Craige, C., Yushkevich, P.A.: Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 611–623 (2013)
Winterburn, J.L., Pruessner, J.C., Chavez, S., Schira, M.M., Lobaugh, N.J., Voineskos, A.N., Chakravarty, M.M.: A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. Neuroimage 74, 254–265 (2013)
Yushkevich, P.A., Avants, B.B., Pluta, J., Das, S., Minkoff, D., Mechanic-Hamilton, D., Glynn, S., Pickup, S., Liu, W., Gee, J.C., Grossman, M., Detre, J.A.: A high-resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 t. Neuroimage 44(2), 385–398 (2009)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Adler, D.H. et al. (2016). Probabilistic Atlas of the Human Hippocampus Combining Ex Vivo MRI and Histology. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention - MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9902. Springer, Cham. https://doi.org/10.1007/978-3-319-46726-9_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-46726-9_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46725-2
Online ISBN: 978-3-319-46726-9
eBook Packages: Computer ScienceComputer Science (R0)