Abstract
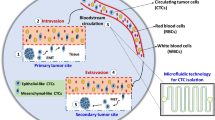
Cancer is a major cause of mortality worldwide, with a disease burden estimated to grow over the coming decades. Circulating tumor cells (CTCs) are rare cancer cells released from the primary or metastatic tumors and transported though the peripheral circulatory system to their specific secondary locations. The presence of CTCs in the a cancer patient’s blood has been used as a prognostic biomarker, with lower CTC count correlating with greater overall survival [1]. In spite of its clinical potential, the isolation and detection of CTCs has been a challenging task due to its rare presence amongst other blood cells (as low as 1–10 CTCs per billions of blood cells) and variability in terms of both morphological and biochemical markers. Recent developments of microfluidics technology have paved the way for better isolation and characterization of CTCs due to several advantages such as lower sample volume, higher sensitivity and throughput and lesser production cost [2, 3]. In this chapter, various CTC isolation devices are classified under two major categories: microfluidics and conventional macro-scale devices, as illustrated in Fig. 1. We will be discussing both label-free methods and antibody-dependent methods for CTC isolation, and will provide discussion and future perspectives on the advantages and drawbacks of both these techniques on potential clinical applications. Advancement in these technologies for CTCs and associated components, such as exosomes, led to an unraveling of tumor variation, ranging histology, molecular, proteomic and functional heterogeneity, which will be discussed in the subsequent sections.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Cancer is a major cause of mortality worldwide, with a disease burden estimated to grow over the coming decades. Circulating tumor cells (CTCs) are rare cancer cells released from the primary or metastatic tumors and transported though the peripheral circulatory system to their specific secondary locations. The presence of CTCs in a cancer patient’s blood has been used as a prognostic biomarker, with lower CTC count correlating with greater overall survival [1]. In spite of its clinical potential, the isolation and detection of CTCs has been a challenging task due to its rare presence amongst other blood cells (as low as 1–10 CTCs per billions of blood cells) and variability in terms of both morphological and biochemical markers. Recent developments of microfluidics technology have paved the way for better isolation and characterization of CTCs due to several advantages such as lower sample volume, higher sensitivity and throughput and lesser production cost [2, 3]. In this chapter, various CTC isolation devices are classified under two major categories: microfluidics and conventional macro-scale devices, as illustrated in Fig. 1. We will be discussing both label-free methods and antibody-dependent methods for CTC isolation, and will provide discussion and future perspectives on the advantages and drawbacks of both these techniques on potential clinical applications. Advancement in these technologies for CTCs and associated components, such as exosomes, led to an unraveling of tumor variation, ranging histology, molecular, proteomic and functional heterogeneity, which will be discussed in the subsequent sections.
Schematic classifications of various circulating tumor cell (CTC) isolation technologies. CTC isolation platforms can be classified into two major classes: microfluidics and conventional macro scale devices. The microfluidic devices can be further sub-divided into label-free (microfiltration, deterministic lateral displacement (DLD) devices and inertial microfluidics), antibody-based (anti-EpCAM coated channels, affinity-based magnetic beads and adhesion-based techniques) and hybrid techniques (combining DLD with magnetophoresis or anti-EpCAM coated channels) that uses the advantages of both label-free and antibody-based methods. Macro scale techniques can also be further classified into label-free (density gradient centrifugation and micro filtration membranes) and antibody-based (CellSearch system, MagSweeper system) platforms
CTCs are heterogeneous in terms of the morphology and surface expression of various biomarkers. Therefore, it is an uphill task to isolate these rare cells from clinical samples in the presence of billions of other hematologic cells. Recent technological advancements observed that CTCs differ from blood cells in various biophysical properties (such as size, adhesion and stiffness) and cell surface receptor expressions (such as Epithelial cell adhesion molecule (EpCAM) and Cytokeratin (CK)) [4, 5]. Current microfluidics techniques and conventional methods can target such distinct properties of the CTCs and achieve high isolation efficiency and throughput along with greater cell viability for downstream single-cell analysis.
2 Microfluidics Devices
Over the past decade, microfluidics technologies have been extensively utilized for study of human disease such as cancer. Microfluidic systems normally leverage on the disparities in the intrinsic properties of the different cell populations (i.e., size, deformability, surface charge, density, etc.) to achieve separations. Isolation and characterization of CTCs using microfluidic systems has been a flourishing area of research which can be broadly categorized to label-free and antibody-based approaches (see Fig. 1).
2.1 Label-Free Technologies
Differences in the biophysical properties between the CTCs and blood cells, such as size, deformability, magnetic susceptibility and electrical behaviors have been exploited to develop label-free sorting of CTCs. The key advantages of this method compared to the antibody-based devices are the collection of a complete pool of CTC population consisting of both EpCAM positive and negative cells and greater compatibility to a wide range of assays that require viable unlabeled cells. Size and deformability-based sorting of CTCs can be achieved by using microfiltration devices and a membrane pore size of around 8 μm has been proved optimal for CTCs capturing [6]. The size of the CTCs varies between 6 and 30 μm and is usually greater than normal hematologic cells [7]. 3D membrane micro filter consisting of two-layers of membrane has been developed with the upper membrane pore size diameter of 9 μm and the pores are aligned centrally to the smaller pores (8 μm) on the basal layer [8]. When blood samples are passed through the device, the CTCs are captured in the upper membrane while other blood cells pass through the gap between the two membranes. This device has an isolation efficiency of 86 % and processing throughput of 3.75 ml/min. Recently, a novel design of the 3D membrane micro filter is introduced with 5 times greater upper membrane pore diameter than that of the lower membrane pores (8 μm) [9]. The CTCs are captured between the gap of the two membranes and the cell can be analyzed by separating the two membranes. Isolation efficiency ranging from 78 to 83 % can be obtained using this device with higher cell viability (greater than 70 %). In order to mitigate the pressure buildup problem during membrane filtration, cross-flow conformation in parallel direction to the filtration membrane was introduced [10]. Membrane microfilters developed using this principle can obtain a capture efficiency of 98 % for MCF-7 cells spiked into blood samples. In another interesting study, weir-shaped structures are used as barrier across the filtration chip to trap most of the CTCs while the blood cells can pass through the narrow opening at the upper portion of the barrier [11]. This device has an isolation efficiency of >95 % and processing speed of 20 ml/h. Microfiltration devices can also be integrated with conical shaped holes to achieve highly efficient cell capture (96 %) at 0.2 ml/min processing rate, as illustrated in Fig. 2a [10]. Clearbridge Biomedics has commercialized a microfluidics device, the CTChip® with an array of crescent-shaped structures composed of three closely spaced micropillars that can trap the CTCs from clinical samples with high throughput and without channel clogging from the cell debris [15, 16]. This device can isolate single and double CTCs and the viable cells can be retrieved and cultured by reversing the flow direction. ISET® (Rarecells, Paris, France) and ScreenCell® are two commercialized systems for sized-based isolation of CTCs that provide cost-effective and high-throughput enrichment of fixed and viable CTCs respectively [17, 18]. The major advantages of the microfiltration devices are its simple design and its ability to obtain the CTCs from the whole blood in a single pass while maintaining its cellular integrity for detection and further downstream analysis. Although size based enrichment of CTCs provides a high-throughput, label-free technique, it has some drawbacks; such as the cells are subjected to high mechanical stress that can alter their normal function [19] and there is a size overlap between the CTCs and leukocytes that can increase the probability of isolating more contaminating cells [20].
Diagrammatic representation of CTC isolation devices. (a) Integration of microfiltration with conical holes for size-based capturing of CTCs. The sample is processed through inlet 1 and it passes through the filter and subsequently gets collected by constant pulling from a syringe pump at outlet 2 (top right). Representation of the microfluidics platform processing clinical samples (middle right). Image of 9 mm diameter micro-filter (bottom right). Scanning electron micrograph (SEM) of conical holes (scale 40 μm). Reproduced with permission from [10]. (b) Representation of multiplexed spiral biochip with two inlets for clinical samples and sheath fluid respectively. The CTCs are sorted due to the action of inertial lift and Dean drag forces and are segregated towards the inner wall of the microfluidics channel (A–A) whereas the WBCs and platelets are concentrated towards the outer wall (B–B). Reproduced with permission from [12]. (c) Working principle of Thermoresponsive NanoVelcro substrate for capturing CTCs with biotinylated anti-EpCAM antibodies at 37 °C and subsequent release of the captured CTCs at 4 °C due to temperature-dependent conformational changes of the polymer brushes that changes the availability of anti-EpCAM antibodies on the surface. Reproduced with permission from [13]. (d) Schematic of the working principle of Gilupi nanodetector system for capturing CTCs from the in-vivo environment. The gold-plated medical steel wire is coated with hydrogel (indicated in brown) and functionalized with anti-EpCAM antibodies (indicated in red) and is inserted into the patient vein to capture EpCAM positive CTCs. Reproduced with permission from [14] (Color figure online)
Dielectrophoresis (DEP) phenomena have also been used for CTC isolation based on the differential motion of the cells when exposed to a non-uniform electric field. Cells can exhibit attractive and repulsive behaviors depending on their size and dielectric polarizability under a non-uniform electric field [21]. Compared to normal cells, CTCs have greater surface area and higher capacitance per unit area that provides them a unique dielectric property and thereby affecting their motion in presence of an electric field. Contactless DEP (cDEP) is developed with greater sensitivity and eliminating the drawbacks of traditional DEP such as high cost, air bubble production, electrode delamination and culture contamination [22]. This device reports an isolation efficiency of 64.5 % for carcinoma cells from concentrated RBC solution. ApoCell laboratories have commercialized ApoStream device in 2010 and validation study using breast cancer cells spiked into clinical sample has observed an isolation efficiency of 86.6 % with higher viability (97.6 %) [23]. The main advantage of DEP devices is that the effect of therapeutic agents could be determined by monitoring the differences in the frequency responses of the cells under the action of various chemotherapeutic agents.
Deterministic lateral displacement (DLD) is another label-free method used for sorting CTCs in microfluidic devices. A novel DLD platform with 58 μm triangular micropost and 42 μm gap is used to capture CTCs at high efficiency (>85 %) and throughput (10 ml/min) [24]. Inertial microfluidics is also used for isolating CTCs by combining inertial lift forces and pinched flow dynamics to focus particles in their preferential equilibrium position along a microfluidic channel. Inertial microfluidics can be combined with microvortex particle capturing to isolate CTCs in a high-throughput, clogging free manner [25]. Apart from straight channels, spiral microfluidics can be used to achieve inertial sorting by combining inertial and Dean drag forces. Our group developed a spiral microfluidics chip with rectangular cross section to isolate CTCs (varying from 5 to 88 CTCs/ml of blood) from 20 metastatic lung cancer patients [26]. Strong inertial lift forces focus the bigger CTCs towards the inner wall while the smaller hematologic cells move towards the outer wall. Due to greater channel dimensions (500 μm width and 160 μm height) and increased flow rate (3 ml/h), clogging of the microchannels by the cell debris is also mitigated, thereby increasing the performance and sensitivity of the device. More recently, we further developed our spiral microfluidics chip with trapezoidal cross-section that can separate the CTCs from metastatic breast and lung cancer clinical samples in an ultra-high throughput manner (7.5 ml within 8 min) [27]. Our group subsequently developed multiplexed spiral microfluidics chip for ultra-high throughput sorting of viable clinical CTC (12–1275 CTCs/ml for breast cancer patients and 10–1535 CTCs/ml for lung cancer patients) for further downstream single-cell characterization such as fluorescence in-situ hybridization (FISH) and proteomics analysis, as represented in Fig. 2b [12]. In another study, contraction-expansion array microfluidic channels is used to enrich cancer cells spiked into blood suspension with 99.5 % efficiency [28]. In negative selection methods, the CTCs are untouched; however the RBCs are lysed and the WBCs are magnetically removed using specific markers such as CD45, CD61 [29].
2.2 Antibody Based Technologies
Antibody mediated CTC isolation techniques are dependent on the specific binding of the cell surface receptors with the antibody bound matrix. The matrix could be of mainly two types, such as magnetic beads or functionalized microfluidics channel. The two commonly used markers for CTC isolation and detection are EpCAM and different subtype of CK. However, due to the occurrence of epithelial to mesenchymal transition (EMT), all CTCs do not express these markers and therefore these techniques fail to collect some subpopulations of the CTCs [30, 31]. CTCs can be selectively labeled with antibody-tagged magnetic microbeads (diameter: 0.5–5 μm) or nanoparticles (diameter: 50–250 nm) and sorted in a non-uniform magnetic field, whereby the labeled CTCs migrate to a region of higher magnetic field and get trapped with an isolation efficiency of 86 % [32, 33]. A straight microchannel with many square indentation on its sidewalls can be used to pull and trap the EpCAM tagged CTCs towards the sidewalls under the influence of an external magnetic field [34]. Fluxion Biosciences have commercialized a microfluidics device, IsoFlux™, which can trap magnetically labeled CTCs when passed through a microchannel with uniform magnetic field. Other promising strategies to further develop the enrichment of EpCAM-positive CTCs in a highly sensitive and efficient manner includes 3D nanostructured substrate or nano “Fly Paper” Technology [35] and a Velcro-like microfluidics platform with isolation efficiency ranging from 40 to 70 %, as depicted in Fig. 2c [13].
Adhesion-based CTC isolation techniques depend on the binding affinity of CTCs to a surface whose biochemical (using antibody coated surface) and structural properties (using nano topographical features) can be modified to favor suitable adhesion. This technique can be performed either in static [36] or dynamic flow modes [37], while the later one is more sensitive due to the greater interaction between the cells and the surface and prevention of non-specific adhesion due to fluid shear forces. Using 3D structures such as microposts inside a microfluidics channel (the CTC-Chip), the effective surface area can be enhanced and the collision frequency between the cell and the EpCAM functionalized surface can be increased [38]. The CTC-Chip has recovery rate of 60 % and the recovery efficiency does not depend on the different expression of EpCAM by the CTCs due to the greater collision frequency. Geometrically enhanced differential immunocapture (GEDI) chip is developed to further increase the effective collision frequency between the CTCs and the antibody-coated microstructure to enhance the isolation efficiency (~90 %) [37]. In another study, herringbone-chip was discovered with anti-EpCAM coated herringbones structures to increase the collision between the CTCs and antibody functionalized PDMS channels [39]. This device has an isolation efficiency of 91.8 % with 95 % cell viability. Recently, geometrically enhanced mixing (GEM) chip is introduced with a further improved herringbone micromixers to enhance the throughput and isolation purity in antibody-dependent devices [40]. CTCs are detected using this device in 17 out of the 18 pancreatic cancer patients studied and the CTC number correlates with the tumor size in three advanced stage patients during the period of the treatment. Adhesion-based CTC isolation techniques using micropost arrays has been commercialized by OnQChip™ (On-Q-ity, MA, USA) and the CEE™ chip (Biocept Laboratories, CA, USA). Graphene oxide sheets can be functionalized with anti-EpCAM antibodies and adsorbed on gold patterned substrates for efficient and sensitive enrichment of CTCs [41]. The increased surface area and biocompatibility of graphene oxide enables greater loading capacity of anti-EpCAM antibodies on its surface and the isolated cells can be cultured on the gold substrates for further downstream characterization such as RT-PCR and drug testing.
Different techniques can be combined to create a better hybrid platform for effective enrichment of CTCs with higher throughput. The CTC-iChip uses both antigen dependent (magnetophoresis) and independent (DLD with inertial focusing) strategies to isolate CTCs with greater sensitive (0.5 CTCs per ml) from clinical samples with lower CTC numbers [42]. Another study combined DLD with EpCAM functionalized isolation chambers to enrich CTCs with greater efficiency (90 %) and throughput (9.6 ml/min) [43]. CTCs are initially sorted from other blood cells in the DLD compartment comprising of triangular microposts and subsequently they are captured in an EpCAM functionalized chambers with fishbone conformation to enhance the isolation capacity.
3 Conventional Macro Scale Devices
3.1 Label-Free Technologies
Density gradient centrifugation is a label-free technique using centrifugal forces for sorting cells from blood based on the difference in their sedimentation coefficient. When a clinical sample is subjected to density gradient centrifugation, the denser RBC and neutrophils settles at the base of the tube whereas the CTCs, plasma and mononucleated cells are collected above the buffy coat. In another study using silicon-blending oil as a floatation media, cancer cells are identified in 53 % and 33 % of gastrointestinal tract cancer and breast cancer patients respectively [44]. OncoQuick centrifugation system has been developed with a porous membrane within the centrifugation tube to prevent the mixing of the separation media with the clinical sample before centrifugation. This system has a greater CTC isolation efficiency compared to the Ficoll density gradient centrifugation [45]. OncoQuick has been used for the enrichment of CTCs from metastatic breast cancer patients and it can successfully detect CTCs in 69.2 % of the clinical samples [46]. RBCs and WBCs can be cross-linked to form rosettes using a cocktail of antibodies and RosetteSep™ (STEMCELL Tech., BC, Canada) commercialized this technique. On application of centrifugal forces these rosettes can be efficiently separated from the CTCs due to their greater densities [47]. In another study, the use of sieve material as a filter with pore size of 4.5 μm could separate 100 % of the HeLa cells spiked into blood and detect cancer cells in 19 out of the 50 cancer patient specimen [48].
3.2 Antibody Based Technologies
Notable achievement in the isolation techniques of CTCs has been accomplished by the introduction of FDA (Food and Drug Administration)–approved method, the CellSearch system (Veridex) [49, 50]. CellSearch system consists of a semi-automated platform for capturing EpCAM positive CTCs using immunomagnetic cell sorting and the cells are subsequently identified by antibody staining of various subtypes of cytokeratins. This system was validated for CTC detection in metastatic breast cancer patients and it was reported that CTC assessment could provide critical prognostic information such as overall survival and progression-free survival [51–53]. However, CellSearch system can only successfully isolate EpCAM positive sub-population of the CTCs and the isolated CTCs are permanently labeled with antibodies that limit their downstream characterization. Illumina Inc. (CA, USA) commercialized MagSweeper system, which consists of a robotic arm with a magnetic rod that continuously shifts the region of high magnetic field within a multiwell plate [54]. The speed and trajectory of the rod movement has been standardized to favor the adsorption of EpCAM-labeled CTCs on the rod and prevents non-specific adsorption by other non-target cells. This system can perform at a high throughput (9 ml/h) and higher isolation efficiency (~81 %). Flow cytometry technologies such as fluorescence-activated cell sorting (FACS) can be utilized to simultaneously detect several fluorescently labeled antibodies and thereby increasing CTC isolation purity [55]. However, this technique can not provide greater sensitivity for CTC isolation due to the varied expression pattern of different antigens on the CTCs depending on the types and stages of cancer [56]. Anti-EpCAM coated detachable microbeads can be used to enlarge the size of the CTCs before passaging the whole blood sample through the microfiltration device (8 μm pore size) [57]. Leukocytes and other blood cells can pass through the filter while the larger CTCs remain trapped on the filter membrane. This device reports a capture efficiency of 89 % and isolated 1–31 CTCs/ml of clinical samples. CTCs can also be isolated directly from the in vivo environment using the Gilupi nanodetector® system, as shown in Fig. 2D [14]. The nanodetector is coated with anti-EpCAM antibodies and inserted in the peripheral arm vein for 30 min to collect larger number of CTCs.
4 Characterization of Cancer Cell Heterogeneity
Tumors demonstrate huge heterogeneity in morphology, immuno-phenotype, and genotype [58]. Novel therapeutic approaches for BRAF mutant cutaneous melanoma [59] and ALK rearranged NSCLC [60], in the recent decades have been unable to increase the survival of patients with advanced stage cancer. This is largely attributed to tumor heterogeneity and the rise of drug tolerant or resistant subtypes under the influence of ineffective drug therapy [61]. Similarly, CTCs have been found to demonstrate a similar range of heterogeneity across multiple parameters, including morphology, histology and proteomics (Fig. 3).
Varied manifestation of CTC heterogeneity. (a) Pleomorphism of CTC morphology obtained from the same patient demonstrated with immunofluorescence staining (left) and Papanicolau stain (right) [62]. (b) Immunostaining of various forms of cytokeratin reveals heterogeneity in protein marker expression. Representative images of minority cohort are provided in boxed images (bottom left). Scale bar, 20 μm [63]. (c) Molecular FISH analysis carried out with clinical human CTCs from an NSCLC patient using Vysis ALK Break Apart FISH probe. Varied frequency of separation of the original gene fusion signal (arrows) is detected in cells obtained from the same sample [12]. Scale bar: 16 mm [12]. (d) Antibody coated membranes demonstrates active protein secretion of CK19 (green) and MUC1 (red) from viable CTCs respectively [64] (Color figure online)
4.1 Histology and Morphology
Observations of tumor histology are often correlated with disease severity [65], and have been found to exhibit strong correlations to genotype [66]. Generally, malignant cancer cells are identified as cells with a round or oval nucleus, and a high nuclear to cytoplasmic (N/C) ratio [67]. However, cancer subtypes display distinct morphology variations, such as the established case of small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) [68].
Recent analysis with specific cancer subpopulations [62, 69, 70], such as disseminated tumor cells (DTCs) and CTCs revealed extensive intertumor histological variation (within the same tumor type among individuals) [71]. CTCs are derived from the blood samples of cancer patients and selectively isolated by targeting protein expression [49], negative selection [29], physical properties [26, 42], or by means of expansion [8, 63, 72]. CTCs can be isolated on a routine basis via a non-invasive process of blood withdrawal [73], allowing monitoring of therapy-associated changes in protein expression, a process which could illuminate the mechanisms that facilitate drug resistance development. Proteomics profiling of CTCs has revealed some of the extent of heterogeneity present. Results suggest the role of multiple pathways, such as the process of EMT transition [74], which is involved in the metastatic cascade [75, 76]. CTC size and other physical parameters may also vary significantly [77], which limits the sensitivity of CTC enrichment devices. These variations can be observed within the cells obtained from tumors of the same patient. Upcoming high throughput spectral imaging techniques will allow rapid processing of single-cell images, providing dynamic insights to cancer cell morphological changes under stimulus or drug exposure (see Fig. 3a) [78, 79].
4.2 Proteomics and Genetic Profiling
Although some histology aspects of cancer cells correspond to genotype, intertumor variation in protein expression has been identified in tumors classified under similar subtypes [80]. Depending on the microenvironment [81], as well as tissue organization and composition [82], cancer cells may be exposed to varying stimulus which induces different rates of oncogene or tumor suppressor gene expressions. For example, accumulation of mutations in the adenomatous polyposis coli (APC) gene may lead to abnormal production of APC protein, an alteration associated with increased colorectal cancer risk [83]. Cancer cells presented with the APC mutation also has increased resistance towards certain drug strategies [84].
The varied protein expression often confers cancer cells with favorable characteristics for growth and survival. For example, the amplified production of human epidermal growth factor receptor 2 (HER2) has been suggested to induce cluster formation in membranes, leading to heightened susceptibility to tumorigenesis [85]. Processing and comparison of large databases have led to resources that provide quantitative values for EMT phenotypes, based on various protein expressions [86]. These dynamics have hindered attempts to completely map the extent of cancer heterogeneity [87]. Our inability to fully comprehend cancer progression often generates conflicting hypotheses and reduces our ability to generate effective treatment strategies.
4.2.1 Understanding CTC Proteomics for Diagnostic and Prognostic Relevance
Tumor-associated proteins have been regularly identified to characterize cancer for administering precise therapeutic treatment. The golden standard involves extraction of these proteins from tumor biopsies [88]. However, few proteins demonstrate actual clinical utility due to cancer heterogeneity [89], except for few instances such as the prostate-specific antigen (PSA). CTC has been broadly defined in a similar way as DTCs [90], namely being positive for epithelial markers (e.g. EpCAM and CKs) and negative for leukocyte markers. However, the proteomics of CTCs are extremely varied (Fig. 3b), and may even not reflect the phenotype observed from the tumor(s) of the same patient. Hence, researchers have been looking for unique specific markers.
To better identify the full CTC spectrum, researchers have started to explore tumour-associated proteins detectable in the serum. Although the field of serum proteomics is in its infancy, technological advances are surfacing which will aid in identifying novel markers. The serum proteome of tumors is a rich source for the analysis of cancer cell activity and interaction with the tumor niche [91]. Circulating tumor DNA (ctDNA), which are short fragments of 150–200 bp DNA, are released from apoptotic CTCs and are now a major interest for diagnostic purposes [92]. ctDNA constitutes about 3–93 % of the total cell-free DNA (cfDNA) and ctDNA can be used as a liquid biopsy biomarker for cancer treatment by analyzing somatic mutations, chromosomal aberrations and loss of heterozygosity [93]. Recently, de novo multiplexed identification of ctDNA mutation is performed from different types of tumor (SHIVA trial) and 28 of 29 mutations detected in the tissue biopsy can be identified in the ctDNA from 27 cancer patients. This signifies the potential of ctDNA as a biomarker [94]. DEP microelectrodes can be used for the isolation of ctDNA from whole blood; however, this system is unsuitable for separation of small fragments of apoptotic DNA (<0.01 μm) and the use of DEP microelectrodes also suffers from cross-sample contamination issues and higher fabrication cost [95, 96]. In another study, digital PCR is used to define malignant growth in various types of cancer such as pancreatic, ovarian, colorectal and they can identify ctDNAs in greater than 75 % of the patients [97]. Additionally, next generation sequencing (NGS) techniques has also been used for sequencing ctDNA and to monitor the response of patients to anti-cancer treatments [98]. ctDNA retains the spectrum of gene mutations found in tumours, and has the same ability to capture mutation patterns as per tumor biopsies [94]. A supposed advantage of utilizing ctDNA is the heightened abundance of material over CTCs, as well as the ease of detection over capturing CTCs [97]. Most importantly, the technology advancement in DNA profiling has enabled sensitive detection of a small amount of genetic material, in fact less than the amount of genetic material within a single CTC [93].
MicroRNAs (miRs) are small (20–22 bases), single stranded fragments of nucleic acid that can negatively regulate gene expression and many tumors have a characteristic miR-based tissue profiles. The miRs can be released into the microenvironment due to an active process or via passive secretion from a dying cancer cell; however, some miRs are not secreted from tumor cells [99]. Circulating miRs can be identified from serum and plasma of clinical samples and different types of solid tumors such as breast, colon, gastric can be distinguished based on miR profiles [100]. miRs from cancer cells [101] may also be isolated from plasma or found within microvesicles (100–1000 nm) [102], exosomes [103] and oncosomes (1–10 μm) (Fig. 4). Exosomes are cell-derived nanovesicles (30–100 nm) that are secreted into tumor microenvironment and play a critical role in cell-cell communication. Exosomes contains genetic information and signaling molecules including proteins, lipids, mRNA, miR and dsDNA that help in the formation of the premetastatic niche at the secondary tumor site [104, 105]. Exosomes can be extracted from the body fluids using conventional methods such as ultracentrifugation and combination of sucrose gradient ultracentrifugation with ultrafiltration centrifugation. However, the former method is much simpler in terms of sample preparation (requires 4–5 h) and has greater isolation efficiency (5–25 %) [106]. Recently, Exo-Quick™ (System Biosciences) and Exo-spin™ (Cell Guidance System) commercialized the isolation kits for exosomes in a cost effective and user-friendly manner. Exosomes can also be isolated by targeting antibody-immobilized magnetic beads against specific antigens that are expressed on their surface such as Alix, annexin, EpCAM, Rab5, CD63, CD81, CD82, CD9 [107]. This technique has greater isolation efficiency than conventional methods; however, it fails to collect the entire population, as some of the exosomes do not express the targeted protein on its surface. Microfluidics isolation of exosomes has been developed using ExoChip, which uses fluorescent imaging to quantify the exosomes that are captured on anti-CD63 coated chips [108]. Interestingly, this study observed a greater number of exosomes in cancer patients (2.34 ± 0.31 fold higher) compared to healthy individuals, which indicates the significance of exosomes as a circulating biomarker. Microfiltration devices (with membrane pore size of <500 nm) have also been used for exosome sorting from whole blood in a highly efficient manner [109]. This device can be combined with electrophoresis and pressure driven flow to further increase the enrichment efficiency. Label-free identification of exosomes can be carried out using surface plasmon resonance (SPR) sensor chip, the nano-plasmonic exosome (nPLEX) [110]. The nPLEX chip can be functionalized with various exosomal antibodies and can potentially identify 12 exosomal markers within 30 min.
Utilizing CTC components for diagnostic and prognostic relevance. (a) Intact CTCs are the primary targets for isolation and enrichment for further analysis and utility. A representative image of enriched CTCs (white arrow) is provided (Green: cytokeratin; Red: CD45; Blue: Nuclei stain, Hoechst). Scale bar: 20 mm [12]. (b) An example of a scatter plot demonstrating the differential signal intensities of miRNAs isolated from serum samples of cancer patients and healthy controls [101]. (c–e) Extracellular vesicles can be classified into three categories based on size—microvesicles, exosomes and large oncosomes. Oncosomes appear to display unique properties and could serve as an important target for cancer characterization (Color figure online)
Research for novel markers in glioma has been extensive for the recent years, and several new markers such as CXCR4 [111] and haptoglobin [91] has been discovered. However, the heterogeneity of cancer complicates the validation of any new candidate markers, and extensive single-cell analysis will be required. Some clinicians are also keen to utilize the serum markers from CTCs, namely exosomes and ctDNA, as diagnostic markers for cancer. For example, the mutant EGFRvIII protein, found in exosomes [112], has been demonstrated as a potential therapeutic marker. Markers from ctDNA, such as miR181 [113], can also be a source for diagnostic means, if the half-life of ctDNA is established [114]. To overcome the problem of information loss due to DNA degradation, extracellular vesicles (EVs) may also be extracted from blood serum. EVs are vesicles that bud off from membranes or secreted from cells for intercellular communication [115], and the transported components are protected from degradation. In the latter instance, these vesicles form exosomes [116].
Another alternative to overcoming the low genetic material available from rare cancer cell subpopulations, such as CTCs, is via single-cell analysis (SCA). Advanced systems, such as the microfluidic image cytometry (MIC) [117] and mass cytometry platform (CyTOF) [118], are available. They can simultaneously separate, process and analysis information from large panels of protein expression, such as the Akt kinase [119, 120].
4.3 Molecular Profiling
Ideally, CTCs can help to reflect the complete tumor spectrum, if the technological limitations impeding enrichment and analysis of rare cells are overcome. Recent advances now allow the simultaneous monitoring of multiple gene expression in parallel. Sensitive transcriptomic tools, such as the single-cell PCR based approach (SINCE-PCR) [121], serve to obtain quantitative and qualitative information which can be beneficial in both scientific and clinical settings. Single-cell analysis can also be carried out with fluorescence in-situ hybridization (FISH) (Fig. 3c).
Thus far, the identification of CTCs has been made complicated due to the lack of specificity of known markers. In fact, most existing molecular subtyping methods do not distinguish cancer cells from normal stroma tissue. Varied histopathological sub-types [122] exhibit different sensitivity of markers as well, and some markers may be common across several cancer types [123, 124]. Variation also exists across tumors from the same patient [125, 126], and some specific cancer types only have traces of molecular similarity with other cancer types [127]. This hinders the accuracy of tumor profiling, since the small amount of clinical samples may be insufficient to reveal rare mutations or intrinsic changes that occurred from response to therapeutic treatment [128]. Such extent of tumor heterogeneity may contribute to differential response to anti-cancer treatment [93], due to incomplete tumor profiling or adaptation to evade treatment.
The rise in advanced technologies, such as the Ensemble Decision Aliquot Ranking (eDAR), is now revealing vast heterogeneity in CTC gene expression, both within and across different patients [129]. An integrative approach to enrich, recover and characterize CTC exomes enabled extensive profiling of prostate CTCs. These profiles revealed a match to a portion of the tumor tissue from the same patient [130]. The spectrum of mutations varied significantly from cell lines, highlighting the issue of translational relevance of immortalized cancer cell phenotypes [131]. Extensive profiling also enables mapping of clonality. Preliminary studies suggest that the cells from the original tumor and metastases are phenotypically distinct, but presence of non-metastatic intermediate phenotypes which are found to persist in both the original tumor and its metastases [132]. Screening of various nuclei at the single-cell level also revealed extensive heterogeneity for KRAS, PIK3CA and other gene expressions [133–135]. These molecular variations may be the reason for a high failure in various anti-cancer regimes, due to an inaccurate diagnosis, or because the drugs are unable to target all cancer subtypes [93]. Detailed single-cell profiling of tumors may reveal novel mechanisms of tumor progression, such as gene amplification (e.g. MET) or chromosomal rearrangements [e.g. ALK (anaplastic lymphoma kinase)] [136].
4.4 Lipidomics Profiling
Lipidomics is a relatively new area of research, and is now increasingly utilized due to development of novel technologies such as the time-of-flight secondary-ion mass spectrometry (TOF-SIMS) [137]. Detection of key enzymes involved in lipid metabolism, such as stearoyl-CoA desaturase 1, may also provide a signature for monitoring lipid levels, even in single cells [138].
4.5 Functional and Metabolic Heterogeneity
Generally, the EMT spectrum of CTCs demonstrates both intrapatient and interpatient heterogeneity. Researchers have also showed that not all phenotypes within the EMT spectrum may be present; with some being fully epithelial-like or mesenchymal-like [139]. More recently, it has been hypothesized that single-cell metabolomics [140] can be a unique factor for the identification of cancer cells from blood cells. These studies may be aided by the novel integrated systems, such as the nanostructure-initiator mass spectrometry (NIMS), which will serve to reveal cancer cells that may have a higher metastatic potential [141]. However, functional and metabolic studies of CTCs are highly limited by the method of CTC enrichment, which is often stressful to the primary cancer cells and compromise cellular viability.
4.5.1 Heterogeneity in Cultured CTCs
The functional properties of CTCs can be investigated with viable CTCs. However, most CTC enrichment techniques are laborious and require harsh procedures which compromise CTC viability. Analysis of the spatial-temporal characteristics of CTCs may be possible by the establishment of short-term CTC cultures with in vitro tumorsphere assays [142], structured microwell assays [63], or long-term CTC cell lines spontaneously immortalized after >6 months in culture [72, 143]. Secretion capabilities of CTCs from various cancer types can also be investigated with a membrane-based assay that provides quantification of proteins [64]. The proliferative capabilities of CTCs vary depending on marker expression [142], as well as response to therapeutic treatment [159]. More specifically, upar+/int β1 − CTC subsets are shown to expand better than other combinational subsets of CTCs, and longer treatment duration correlates with lower proliferative potential. Upar−/int β1 − CTC subsets also appeared to be more mesenchymal and displayed delayed clustering.
CTC may also demonstrate varied capabilities for adhesion, albeit most studies report low adhesive capabilities of CTCs. These low adhesive properties may reflect their heightened potential for extravasation and migration. Interestingly, it has been described that low adhesive CTCs demonstrate high plasticity and may eventually adhere to form spheroids under suitable 3D gel conditions [142]. Understandably, the more mesenchymal phenotypes also demonstrate higher motility and invasiveness. Analysis of these factors and other functional phenotypes, such as the formation of invadopodia [142], may reveal valuable information of the metastatic cascade occurring in vivo.
4.5.2 Cancer Stem Cells
Technological advances have enabled isolation of rare cancer subtypes with unique properties. Cancer stem cells (CSCs) are cells present in the tumor that demonstrates heightened tumorigenic potential. They are found to demonstrate distinct protein signatures, for example, breast CSCs are recognized as CD44+/CD24− [144] or aldehyde dehydrogenase 1+ (ALDH1+) cells [12, 145]. Recently, other pluripotency factors, such as POUF51 (OCT4), have been associated with the CSC subtype [146], supporting their potential role in metastasis. CSCs are often speculated to be associated with worsened patient prognosis [147], as they are found with anti-cancer drug resistant or tolerant traits [148]. Despite the distinctive characteristics of CSCs, the clinical relevance of this rare sub-population is still unclear, and further functional analysis may help to resolve this question.
5 Discussions and Conclusion
Compared to conventional macro scale devices, microfluidics technologies have provided several advantages in the field of CTC isolation such as higher throughput, greater sensitivity, portability, and ease of operation [149]. Additionally, this technique enables manipulation of fluid and particle motion at a micron-scale that is critical for CTC enrichment considering their rare occurrence in the blood. The antibody based isolation techniques provides some inherent benefits including higher specificity, lesser leukocyte contamination and enriching viable CTCs due to minimum sample handling procedure. However, these devices suffer from low throughput (1–3 ml/h) and are dependent on the expression of specific antigen markers. Due to the occurrence of EMT, some subpopulations of CTCs might not express the epithelial markers such as EpCAM and this might lead to the loss in capture efficiency [150, 151]. Label-free approaches can overcome these drawbacks of antibody-dependent methods by isolating CTCs based on their physical properties including size, deformability and dielectric properties in an unbiased manner independent of EpCAM expression. However, these techniques are limited by the heterogeneity of the cancer cells in terms of their size (CTC size varies from 6 to 30 μm) and thereby resulting in contamination from other blood cells and decreasing the isolation purity [152].
Overall, CTC isolation devices have made rapid progress over the last decade; however, till date there are few FDA approved methods (the CellSearch system and cobas EGFR Mutation Test) [153] for clinical diagnosis. Therefore, development of universal optimal protocol for examining the performances of these different devices is essential. Recent advancement of microfluidics technology has paved the way for solving some of the initial problems associated with conventional macroscale systems such as lower throughput, purity and sensitivity. The development of hybrid systems such as CTC-iChip that uses the advantages of both antibody-dependent and label-free approaches might overcome the limitations associated with both these systems individually. Additionally, negative selection devices that rely on the depletion of leukocytes may be an alternative unbiased approach for CTC enumeration. In future, we envision the development of an integrated, fully automated CTC enrichment platform that can isolate the CTCs from a simple blood test, characterize their biochemical and biophysical properties and identify the potential targets for chemotherapeutic administration.
Cancer manifests as a heterogeneous disease in several different ways [154], and detailed profiling of this manifestation is essential to develop personalized drug therapeutic strategies [155]. Biopsies of tumors are generally undesirable due to the invasive nature and possibility of dissociation of cancer cells during the surgical process. Hence CTCs represent an alternative source of cancer cells obtained via blood withdrawal, so termed as liquid biopsy [155]. The CTCs may also provide a better profile for the tumors, as they are shed from a range of sites as compared to a single biopsied region.
Several questions remain which impede the clinical utility of CTCs. The relevance of CTCs in influencing therapeutic strategies is in question since it is not known how CTCs persist within the circulation and contribute to metastasis. CTC lifespan has also not been conclusively defined, and it is speculated that this may vary from a few hours to years [156, 157]. The persistence of CTCs in blood fuels the conjecture that these rare cells may play a role in tumor relapse. Overall, the clinical relevance of CTCs will be determined by their resemblance to the original tumor phenotype, their transition in blood and their functional dynamics with time. CTCs and associated markers (e.g. microRNAs) could help to rectify the incomplete tumor profile. The characterization of CTC components may shed light for a single pathway [158] or on the patient conditions at a point of time during treatment, but monitoring of a patient’s condition will require serial sampling or culturing of CTCs which could reveal dynamics of an individual’s cancer progression.
References
Allen JE, El-Deiry WS (2010) Circulating tumor cells and colorectal cancer. Curr Colorectal Cancer Rep 6(4):212–220
Majid EW, Lim CT (2013) Microfluidic platforms for human disease cell mechanics studies. In: Buehler MJ, Ballarini R (eds) Materiomics: multiscale mechanics of biological materials and structures. Springer, New York, pp 107–119
Chaudhuri PK et al (2016) Microfluidics for research and applications in oncology. Analyst 141(2):504–524
Low WS, Wan Abu Bakar WA (2015) Benchtop technologies for circulating tumor cells separation based on biophysical properties. BioMed Res Int 2015:239362
Alix-Panabières C, Pantel K (2014) Technologies for detection of circulating tumor cells: facts and vision. Lab Chip 14(1):57–62
Zabaglo L et al (2003) Cell filtration-laser scanning cytometry for the characterisation of circulating breast cancer cells. Cytometry A 55(2):102–108
Allard WJ et al (2004) Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 10(20):6897–6904
Zheng S et al (2011) 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices 13(1):203–213
Zhou MD et al (2014) Separable bilayer microfiltration device for viable label-free enrichment of circulating tumour cells. Sci Rep 4:7392
Adams DL et al (2014) The systematic study of circulating tumor cell isolation using lithographic microfilters. RSC Adv 4(9):4334–4342
Chung J et al (2012) Microfluidic cell sorter (μFCS) for on-chip capture and analysis of single cells. Adv Healthcare Mater 1(4):432–436
Khoo BL et al (2014) Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PLoS One 9(7), e99409
Hou S et al (2013) Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv Mater 25(11):1547–1551
Saucedo-Zeni N et al (2012) A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol 41(4):1241–1250
Tan SJ et al (2009) Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microdevices 11(4):883–892
Tan SJ et al (2010) Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens Bioelectron 26(4):1701–1705
Desitter I et al (2011) A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res 31(2):427–441
Vona G et al (2000) Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol 156(1):57–63
Kuo JS et al (2010) Deformability considerations in filtration of biological cells. Lab Chip 10(7):837–842
Alunni-Fabbroni M, Sandri MT (2010) Circulating tumour cells in clinical practice: methods of detection and possible characterization. Methods 50(4):289–297
Shim S et al (2013) Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics 7(1):011807
Huang C-T et al (2012) Selectively concentrating cervical carcinoma cells from red blood cells utilizing dielectrophoresis with circular ITO electrodes in stepping electric fields. J Med Biol Eng 33(1):51–58
Gupta V et al (2012) ApoStream™, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 6(2):024133
Loutherback K et al (2012) Deterministic separation of cancer cells from blood at 10 mL/min. AIP Adv 2(4):042107
Sollier E et al (2014) Size-selective collection of circulating tumor cells using Vortex technology. Lab Chip 14(1):63–77
Hou HW et al (2013) Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep 3
Warkiani ME et al (2014) Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip 14(1):128–137
Lee MG et al (2013) Label-free cancer cell separation from human whole blood using inertial microfluidics at low shear stress. Anal Chem 85(13):6213–6218
Zborowski M, Chalmers JJ (2011) Rare cell separation and analysis by magnetic sorting. Anal Chem 83(21):8050–8056
Fehm T et al (2009) Detection and characterization of circulating tumor cells in blood of primary breast cancer patients by RT-PCR and comparison to status of bone marrow disseminated cells. Breast Cancer Res 11(4):R59
Punnoose EA et al (2010) Molecular biomarker analyses using circulating tumor cells. PLoS One 5(9), e12517
Hoshino K et al (2011) Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 11(20):3449–3457
Pamme N (2012) On-chip bioanalysis with magnetic particles. Curr Opin Chem Biol 16(3):436–443
Kang JH et al (2012) A combined micromagnetic-microfluidic device for rapid capture and culture of rare circulating tumor cells. Lab Chip 12(12):2175–2181
Wang S, Owens GE, Tseng HR (2011) Nano “fly paper” technology for the capture of circulating tumor cells. Methods Mol Biol 726:141–150
Lu J et al (2010) Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients. Int J Cancer 126(3):669–683
Smith JP et al (2012) Microfluidic transport in microdevices for rare cell capture. Electrophoresis 33(21):3133–3142
Nagrath S et al (2007) Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450(7173):1235–1239
Stott SL et al (2010) Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci 107(43):18392–18397
Sheng W et al (2014) Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip 14(1):89–98
Yoon HJ et al (2013) Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nanotechnol 8(10):735–741
Ozkumur E et al (2013) Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 5(179):179ra47
Liu Z et al (2013) High throughput capture of circulating tumor cells using an integrated microfluidic system. Biosens Bioelectron 47:113–119
Seal S (1959) Silicone flotation: a simple quantitative method for the isolation of free-floating cancer cells from the blood. Cancer 12(3):590–595
Gertler R et al (2003) Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. In: Allgayer H, Heiss M (eds) Molecular staging of cancer. Springer, Berlin, pp 149–155
Königsberg R et al (2011) Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol 50(5):700–710
Naume B et al (2004) Detection of isolated tumor cells in peripheral blood and in BM: evaluation of a new enrichment method. Cytotherapy 6(3):244–252
Seal S (1964) A sieve for the isolation of cancer cells and other large cells from the blood. Cancer 17(5):637–642
Riethdorf S et al (2007) Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the Cell Search system. Clin Cancer Res 13(3):920–928
Diamandis EP (2002) Tumor markers: physiology, pathobiology, technology, and clinical applications. American Association for Clinical Chemistry, Washington
Cristofanilli M et al (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351(8):781–791
Cristofanilli M et al (2005) Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol 23(7):1420–1430
Giordano A et al (2013) Establishment and validation of circulating tumor cell-based prognostic nomograms in first-line metastatic breast cancer patients. Clin Cancer Res 19(6):1596–1602
Talasaz AH et al (2009) Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci 106(10):3970–3975
Cruz I et al (2005) Evaluation of multiparameter flow cytometry for the detection of breast cancer tumor cells in blood samples. Am J Clin Pathol 123(1):66–74
Yu M et al (2011) Circulating tumor cells: approaches to isolation and characterization. J Cell Biol 192(3):373–382
Lee HJ et al (2013) Efficient isolation and accurate in situ analysis of circulating tumor cells using detachable beads and a high-pore-density filter. Angew Chem Int Ed 52(32):8337–8340
McCarthy N (2014) The cancer kaleidoscope. Nat Rev Cancer 14:151–152
Chapman PB et al (2011) Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364(26):2507–2516
Kwak EL et al (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363(18):1693–1703
Singh AK et al (2015) Tumor heterogeneity and cancer stem cell paradigm: updates in concept, controversies and clinical relevance. Int J Cancer 136(9):1991–2000
Marrinucci D et al (2010) Cytomorphology of circulating colorectal tumor cells: a small case series. J Oncol 2010:861341. doi:10.1155/2010/861341
Khoo BL et al (2016c) Liquid biopsy and therapeutic response: circulating tumor cell cultures for evaluation of anticancer treatment. Sci Adv 2, e1600274
Alix-Panabieres C et al (2009) Full-length cytokeratin-19 is released by human tumor cells: a potential role in metastatic progression of breast cancer. Breast Cancer Res 11(3):R39
Weichselbaum RR, Hellman S (2011) Oligometastases revisited. Nat Rev Clin Oncol 8(6):378–382
Soslow RA et al (2012) Morphologic patterns associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod Pathol 25(4):625–636
Chen S et al (2012) Recent advances in morphological cell image analysis. Comput Math Methods Med 2012:101536
Gazdar AF et al (1985) Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 45(6):2924–2930
van de Stolpe A et al (2011) Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res 71(18):5955–5960
Leversha MA et al (2009) Fluorescence in situ hybridization analysis of circulating tumor cells in metastatic prostate cancer. Clin Cancer Res 15(6):2091–2097
Min JW et al (2015) Identification of distinct tumor subpopulations in lung adenocarcinoma via single-cell RNA-seq. PLoS One 10(8), e0135817
Zhang L et al (2013) The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med 5(180):180ra48
Pantel K, Alix-Panabieres C (2013) Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res 73(21):6384–6388
Thiery JP (2002) Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2(6):442–454
Ting DT et al (2014) Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep 8(6):1905–1918
Khoo BL et al (2016) Genesis of circulating tumor cells through epithelial–mesenchymal transition as a mechanism for distant dissemination. In: Circulating tumor cells. Springer, New York, pp 139–182
Marrinucci D et al (2007) Case study of the morphologic variation of circulating tumor cells. Hum Pathol 38(3):514–519
Goda K et al (2012) High-throughput single-microparticle imaging flow analyzer. Proc Natl Acad Sci U S A 109(29):11630–11635
Goda K, Tsia KK, Jalali B (2009) Serial time-encoded amplified imaging for real-time observation of fast dynamic phenomena. Nature 458(7242):1145–1149
Navin N et al (2011) Tumour evolution inferred by single-cell sequencing. Nature 472:90–96
Bissell MJ, Hines WC (2011) Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 17(3):320–329
Michor F, Weaver VM (2014) Understanding tissue context influences on intratumour heterogeneity. Nat Cell Biol 16(4):301–302
Network CGA (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487(7407):330–337
Brown JM, Attardi LD (2005) The role of apoptosis in cancer development and treatment response. Nat Rev Cancer 5(3):231–237
Nagy P et al (1999) Activation-dependent clustering of the erbB2 receptor tyrosine kinase detected by scanning near-field optical microscopy. J Cell Sci 112(Pt 11):1733–1741
Tan TZ et al (2014) Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med 6(10):1279–1293
Pantel K, Brakenhoff RH (2004) Dissecting the metastatic cascade. Nat Rev Cancer 4(6):448–456
Basik M et al (2013) Biopsies: next-generation biospecimens for tailoring therapy. Nat Rev Clin Oncol 10:437–450
Mimeault M, Batra SK (2014) Molecular biomarkers of cancer stem/progenitor cells associated with progression, metastases, and treatment resistance of aggressive cancers. Cancer Epidemiol Biomarkers Prev 23(2):234–254
Borgen E et al (1999) Standardization of the immunocytochemical detection of cancer cells in BM and blood: I. Establishment of objective criteria for the evaluation of immunostained cells. Cytotherapy 1(5):377–388
Gollapalli K et al (2012) Investigation of serum proteome alterations in human glioblastoma multiforme. Proteomics 12(14):2378–2390
Francis G, Stein S (2015) Circulating cell-free tumour DNA in the management of cancer. Int J Mol Sci 16(6):14122–14142
Diaz LA Jr et al (2012) The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486(7404):537–540
Lebofsky R et al (2015) Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 9(4):783–790
Sonnenberg A et al (2013) Dielectrophoretic isolation and detection of cfc-DNA nanoparticulate biomarkers and virus from blood. Electrophoresis 34(7):1076–1084
McCanna JP, Sonnenberg A, Heller MJ (2014) Low level epifluorescent detection of nanoparticles and DNA on dielectrophoretic microarrays. J Biophotonics 7(11–12):863–873
Bettegowda C et al (2014) Detection of circulating tumor DNA in early-and late-stage human malignancies. Sci Transl Med 6(224):224ra24
Diaz LA Jr et al (2013) Insights into therapeutic resistance from whole-genome analyses of circulating tumor DNA. Oncotarget 4(10):1856
Chan M et al (2013) Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res 19(16):4477–4487
Chen X et al (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18(10):997–1006
Dong L et al (2014) miRNA microarray reveals specific expression in the peripheral blood of glioblastoma patients. Int J Oncol 45(2):746–756
Noerholm M et al (2012) RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer 12:22
Manterola L et al (2014) A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol 16(4):520–527
Peinado H, Lavotshkin S, Lyden D (2011) The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol 21(2):139–146
Costa-Silva B et al (2015) Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 17(6):816–826
Zhao L et al (2015) The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett 356(2):339–346
Chen C et al (2010) Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 10(4):505–511
Kanwar SS et al (2014) Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 14(11):1891–1900
Davies RT et al (2012) Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip 12(24):5202–5210
Im H et al (2014) Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol 32(5):490–495
Popescu ID et al (2014) Potential serum biomarkers for glioblastoma diagnostic assessed by proteomic approaches. Proc Natl Acad Sci U S A 12(1):47
Skog J et al (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10(12):1470–1476
Zhang W et al (2012) miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol 14(6):712–719
Sozzi G et al (2014) Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol 32(8):768–773
Belting M, Wittrup A (2008) Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. J Cell Biol 183(7):1187–1191
Minciacchi VR, Freeman MR, Di Vizio D (2015) Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol 40:41–51
Sun J et al (2010) A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res 70(15):6128–6138
Amir el-AD et al (2013) viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31(6):545–552
Vivanco I, Sawyers CL (2002) The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2(7):489–501
Khoo BL et al (2016b) Single‐cell profiling approaches to probing tumor heterogeneity. Int J Cancer 139(2):243–255
Dalerba P et al (2011) Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 29(12):1120–1127
Jerome Marson V et al (2004) Expression of TTF-1 and cytokeratins in primary and secondary epithelial lung tumours: correlation with histological type and grade. Histopathology 45(2):125–134
Xu B et al (2010) Expression of thyroid transcription factor-1 in colorectal carcinoma. Appl Immunohistochem Mol Morphol 18(3):244–249
Ordonez NG (2012) Value of thyroid transcription factor-1 immunostaining in tumor diagnosis: a review and update. Appl Immunohistochem Mol Morphol 20(5):429–444
Lee JY et al (2015) Tumor evolution and intratumor heterogeneity of an epithelial ovarian cancer investigated using next-generation sequencing. BMC Cancer 15:85
Sottoriva A et al (2013) Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 110(10):4009–4014
Hoadley KA et al (2014) Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158(4):929–944
Punnoose EA et al (2012) Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 18(8):2391–2401
Schiro PG et al (2012) Sensitive and high-throughput isolation of rare cells from peripheral blood with ensemble-decision aliquot ranking. Angew Chem Int Ed Engl 51(19):4618–4622
Lohr JG et al (2014) Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 32(5):479–484
Lacroix M (2007) Persistent use of “false” cell lines. Int J Cancer 122(1):1–4
Cummings EB (2003) Streaming dielectrophoresis for continuous-flow microfluidic devices. IEEE Eng Med Biol Mag 22(6):75–84
Baldus SE et al (2010) Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 16:790
Kosmidou V et al (2014) Tumor heterogeneity revealed by KRAS, BRAF, and PIK3CA pyrosequencing: KRAS and PIK3CA intratumor mutation profile differences and their therapeutic implications. Hum Mutat 35(3):329–340
Deng G et al (2014) Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer 14:456
Cooper WA et al (2013) Molecular biology of lung cancer. J Thorac Dis 5(Suppl 5):S479–S490
Ide Y et al (2014) Single cell lipidomics of SKBR-3 breast cancer cells by using time-of-flight secondary-ion mass spectrometry. Surf Interface Anal. doi:10.1002/sia.5523
Denbigh JL, Lockyer NP (2014) ToF-SIMS as tool for profiling lipids in cancer and other diseases. Mater Sci Technol. doi:10.1179/1743284714Y.0000000648(0)
Joosse SA, Gorges TM, Pantel K (2015) Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med 7(1):1–11
Nemes P et al (2012) Single-cell metabolomics: changes in the metabolome of freshly isolated and cultured neurons. ACS Chem Neurosci 3:782
O’Brien PJ et al (2013) Monitoring metabolic responses to chemotherapy in single cells and tumors using nanostructure-initiator mass spectrometry (NIMS) imaging. Cancer Metab 1(1):4
Vishnoi M et al (2015) The isolation and characterization of CTC subsets related to breast cancer dormancy. Sci Rep 5:17533
Yu M et al (2014) Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345(6193):216–220
Prince ME et al (2007) Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A 104(3):973–978
Charafe-Jauffret E et al (2010) Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res 16(1):45–55
Ennen M et al (2014) Single-cell gene expression signatures reveal melanoma cell heterogeneity. Oncogene. doi:10.1038/onc.2014.262
Tinhofer I et al (2014) Cancer stem cell characteristics of circulating tumor cells. Int J Radiat Biol 90(8):622–627
Seymour T, Nowak A, Kakulas F (2015) Targeting aggressive cancer stem cells in glioblastoma. Front Oncol 5:159
Whitesides GM (2006) The origins and the future of microfluidics. Nature 442(7101):368–373
Alix-Panabières C, Pantel K (2014) Challenges in circulating tumour cell research. Nat Rev Cancer 14(9):623–631
de Wit S et al (2015) The detection of EpCAM+ and EpCAM− circulating tumor cells. Sci Rep 5
Sun Y-F et al (2011) Circulating tumor cells: advances in detection methods, biological issues, and clinical relevance. J Cancer Res Clin Oncol 137(8):1151–1173
Brown P (2016) The Cobas(R) EGFR Mutation Test v2 assay. Future Oncol 12:451–452
Larsen JE, Minna JD (2011) Molecular biology of lung cancer: clinical implications. Clin Chest Med 32(4):703–740
Krebs MG et al (2014) Molecular analysis of circulating tumour cells—biology and biomarkers. Nat Rev Clin Oncol 11(3):129–144
Plaks V, Koopman CD, Werb Z (2013) Cancer. Circulating tumor cells. Science 341(6151):1186–1188
Meng S et al (2004) Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res 10(24):8152–8162
Kros JM et al (2015) Circulating glioma biomarkers. Neuro Oncol 17(3):343–360
Khoo, BL et al (2016) Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Science Advances 2(e1600274).
Acknowledgments
B.L.K. acknowledges support from the National Research Foundation (NRF), Prime Minister’s Office, Singapore, under CREATE, Singapore-MIT Alliance for Research and Technology (SMART) BioSystems and Micromechanics (BioSyM) IRG and MBI. P.K.C. acknowledges the support from the Mechanobiology Institute (MBI) for the Graduate Scholarship. We thank Mr. Wong Chun Xi (MBInfo) for helping with the illustrations.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Khoo, B.L., Chaudhuri, P.K., Lim, C.T., Warkiani, M.E. (2017). Advancing Techniques and Insights in Circulating Tumor Cell (CTC) Research. In: Aref, A., Barbie, D. (eds) Ex Vivo Engineering of the Tumor Microenvironment. Cancer Drug Discovery and Development. Humana Press, Cham. https://doi.org/10.1007/978-3-319-45397-2_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-45397-2_5
Published:
Publisher Name: Humana Press, Cham
Print ISBN: 978-3-319-45395-8
Online ISBN: 978-3-319-45397-2
eBook Packages: MedicineMedicine (R0)