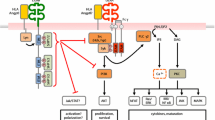
Graphical Abstract

Abstract
Immune cells express several adhesion G protein-coupled receptors (aGPCRs), including the ADGRE subfamily members EMR1 (F4/80, ADGRE1), EMR2 (ADGRE2), EMR3 (ADGRE3), EMR4 (FIRE, ADGRE4), and CD97 (ADGRE5), the ADGRB subfamily member BAI1 (ADGRB1), and the ADGRG subfamily members GPR56 (ADGRG1), GPR97 (Pb99, ADGRG3), and GPR114 (ADGRG5). Expression of these molecules in hematopoietic stem and progenitor cells, monocytes/macrophages (Mφs), dendritic cells, granulocytes, and lymphocytes depends on lineage diversification and maturation, making them suitable markers for individual leukocyte subsets (e.g., F4/80 on mouse Mφs). Recent studies revealed intriguing activities of aGPCRs in tolerance induction (EMR1), granulopoiesis (CD97), engulfment of apoptotic cells and bacteria (BAI1), hematopoietic stem cell formation (GPR56), and control of cytotoxicity (GPR56). Here, we review these findings and discuss their biological and translational implications.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Adhesion GPCRs in the Immune System
Though regulating numerous cellular processes, G protein-coupled receptors (GPCRs) in the immune system are mostly known for their ability to steer cell migration and homing in response to chemokine gradients [1]. However, leukocytes express many more GPCRs, including members of the adhesion family. In fact, the first aGPCRs that have been described—F4/80 and CD97—are molecules expressed on mouse macrophages (Mφs) and human activated T cells, respectively [2, 3]. Since then, ADGRE, ADGRB, and ADGRG subfamily members on hematopoietic cells have been identified and extensively studied, both in vitro and in vivo. Here, we review current ideas about the role of these molecules in relation to immune cell development and function and discuss the biological and translational implications of these findings.
2 EGF-TM7/ADGRE Subfamily
The five members of the ADGRE subfamily, also known as EGF-TM7 receptors , are encoded by two gene clusters on chromosome 19p13.3 (EMR1 and EMR4) and chromosome 19p13.1 (EMR2, EMR3, and CD97) [4–6]. EGF-TM7 receptors possess N-terminal epidermal growth factor (EGF)-like domains encoded by a single exon. Alternative splicing of these exons in EMR1, EMR2, and CD97 gives rise to isoforms with a different EGF-domain composition. Expression of EMR1–4 is restricted to myeloid cells. In contrast, CD97 is found on virtually all hematopoietic cells as well as on many non-hematopoietic cell types (Fig. 1).
Expression of adhesion GPCRs in immune cells. Summary of available published data on the expression of aGPCRs in hematopoietic cells in (a) human and (b) mice, obtained by transcriptional profiling (green squares) and protein analysis techniques (purple squares). For additional information and references, see the descriptions of individual aGPCRs at www.guidetopharmacology.org/
2.1 EMR1 (F4/80, ADGRE1)
The F4/80 antigen (Ag) was initially described as a novel cell surface marker of mouse Mφs by Jon Austyn and Siamon Gordon in 1981, following the development of the F4/80 monoclonal antibody (mAb) in rats against isolated thioglycollate-elicited mouse peritoneal Mφs [2]. Subsequent experiments established the limited expression pattern of F4/80 in myeloid cells, including monocytes, most tissue Mφs, and some dendritic cells (DCs) [7, 8]. Eosinophils also express the F4/80 Ag, whose expression is enhanced after parasitic infection [9]. No F4/80 reactivity was detected in other cells of the myeloid lineage, such as neutrophilic or basophilic granulocytes and osteoclasts, in cells of the lymphoid lineage, and in non-hematopoietic cells. Due to its robust reactivity and specificity in restricted Mφ subsets, the F4/80 Ag has been extensively used as a tissue Mφ marker in mice ever since [10–12]. The F4/80 Ag is detected as early as embryonic day 8/9 in Mφs in the aorta–gonad–mesonephros region and yolk sac, in fetal livers, and in splenic red pulp and bone marrow before birth [13]. In the adult mouse, F4/80 is weak on circulating monocytes, but is highly expressed on sinusoidal Mϕs in the liver (Kupffer cells), spleen, and adrenal glands and on almost all tissue extravascular Mϕs. F4/80 has also been used to identify tumor-associated Mφs [14].
F4/80 was identified as the murine ortholog of human EMR1 through genomic and phylogenetic analyses [4–6]. F4/80 and EMR1 possess seven and six N-terminal EGF-like domains, respectively [15–17]. They are the only members of the ADGRE subfamily that are not proteolytically processed due to the presence of an atypical GPCR proteolysis site (GPS) sequence. Indeed, both F4/80 and EMR1 are expressed as single-chain polypeptide receptor molecules. Notably, EMR1 is not expressed in Mφs, but instead is a specific marker of human eosinophilic granulocytes [18, 19]. Therefore, it is believed that F4/80 and EMR1 are structural orthologs that possess different cell-type- and species-specific expression patterns and functions.
Several approaches, including Ab treatment and gene targeting, have been used to investigate the physiological function(s) of F4/80 ex vivo and in vivo. Using the F4/80 mAb in an in vitro model of facultative intracellular pathogen infection, Warschkau et al. demonstrated a role for F4/80 in interferon (IFN)-γ production by severe combined immunodeficiency (SCID) mouse splenic cells in response to heat-killed Listeria monocytogenes [20]. IFN-γ is critical for the control of bacterial growth during L. monocytogenes infection in vivo. It was shown that heat-killed L. monocytogenes-stimulated Mϕs secreted tumor necrosis factor (TNF) and interleukin (IL)-12, which activated natural killer (NK) cells to produce IFN-γ. It was concluded that F4/80 is critical in the Mφ–NK cell interaction during L. monocytogenes infection, because the F4/80 mAb treatment downregulated TNF, IL-12, and IFN-γ production [20]. The F4/80 molecule was believed to interact with a potential cellular ligand on NK cell, which delivered a stimulatory signal for cytokine release.
Two strains of F4/80 knockout mice were generated using different gene targeting approaches; one involved the production of F4/80–Cre knock-in mice, where the first coding exon of F4/80 was replaced with the cDNA of the Cre recombinase, while the other used a conventional method by deleting the first coding exon [21, 22]. Interestingly, the F4/80-deficient animals were healthy and phenotypically normal. No obvious defect in the maturation and development of tissue Mϕs was noted in the mutant mice either. This indicated that F4/80 is not required for the differentiation and development of cells of the monocyte–Mφ lineage. In contrast to what was found in vitro, the F4/80-deficient animals show normal responses to infectious challenge with L. monocytogenes, suggesting a minimum role for F4/80 in the immune responses against L. monocytogenes infection in vivo [22].
In another study, the F4/80-deficient mice were employed to confirm the role of F4/80 in the induction of peripheral immune tolerance [21]. Indeed, the F4/80-deficient mice did not develop anterior chamber-associated immune deviation (ACAID) because the animals failed to generate functional Ag-specific efferent CD8+ regulatory T (Treg) cells. Similarly, it was shown that efferent Treg cells were only produced when the APCs are F4/80+ in an in vitro ACAID model using both the F4/80 mAb and F4/80-deficient APC. Furthermore, the F4/80-deficient animals were found unable to produce an efficient immune tolerance response in a low-dose oral tolerance model. Importantly, reconstitution of the F4/80-deficient animals with the F4/80+ APCs restored their ability to induce ACAID and low-dose oral tolerance [21]. Therefore, the F4/80 molecule is required for the generation of Ag-specific efferent Treg cells [23]. The molecular mechanism mediated by the F4/80 receptor to induce peripheral immune tolerance remains elusive, but a putative cellular ligand of F4/80 on interacting immune cells is a likely candidate.
2.2 EMR2 (ADGRE2)
EMR2 seems to be a chimera of CD97 and another EGF-TM7 receptor, EMR3, as a result of a relatively recent evolution, comprising gene conversion and duplication [24]. Indeed, EMR2 shares ~97 % amino acid sequence identity in the EGF-like domains with CD97 and ~90 % identity in the seven-transmembrane (7TM ) region with EMR3, respectively [6, 24–26]. This suggests that EMR2 might interact with similar cellular ligand(s) as CD97 while sharing with EMR3 comparable signaling activities. The EMR2 gene is lost in rodents, suggesting a specific need for EMR2 in the immune reactions of human [24].
Unlike F4/80 and human EMR1 in Mφs and eosinophils, EMR2 is more widely expressed in myeloid cells, including monocytes, Mφs, DCs, and granulocytes, suggesting a role for EMR2 in these innate immune cells [27]. Resting or activated lymphocytes do not express any detectable EMR2. CD16+ blood monocytes, Mφs, and BDCA-3+ myeloid DCs express the highest levels of EMR2. Interestingly, EMR2 expression is upregulated during the differentiation and maturation of Mφs in vitro, but is downregulated during DC maturation [28]. Lipopolysaccharide (LPS ) treatment upregulated EMR2 expression in monocytes and Mφs in an IL-10-dependent manner. In tissues, EMR2 expression was detected in certain Mφ subpopulations of the lung, skin, spleen, tonsil, and placenta, but not in those of the liver and kidney. In inflamed tissues, EMR2 staining was found in subpopulations of Mφs and neutrophils [28]. Recently, high levels of EMR2 were detected in foamy Mϕs in atherosclerotic vessels and in Gaucher cells in the spleen. On the contrary, multiple sclerosis brain foam cells express little if any EMR2 [29].
The full-length EMR2 protein contains a total of five EGF-like motifs. Alternative splicing results in four isoforms: EMR2(EGF1,2), EMR2(EGF1,2,5), EMR2(EGF1,2,3,5), and EMR2(EGF1–5) [25]. EMR2(EGF1–5) shares with CD97(EGF1–5) a glycosaminoglycan ligand, chondroitin sulfate B (dermatan sulfate), while EMR2(EGF1,2,5) binds to CD55 with a tenfold weaker affinity than does CD97(EGF1,2,5) [30, 31]. These findings suggest possible overlapping, but also specific functions for EMR2 and CD97.
Studies using the mAb 2A1, which binds downstream of the EGF domains, have demonstrated a role for EMR2 in potentiating the activation and recruitment of human neutrophils [32]. Specifically, ligation of EMR2 by 2A1 enhanced neutrophil adherence and migration under both static and shear stress conditions. In addition, ligation and activation of EMR2 was shown to work synergistically with a number of pro-inflammatory mediators in potentiating neutrophil respiratory burst and degranulation [32]. Patients suffering from noninfectious systemic inflammatory response syndrome (SIRS) and sepsis often die through multiple organ failure, caused by increased numbers and hyperactivation of neutrophils. A more recent study revealed higher EMR2 expression in neutrophils of SIRS patients and a positive correlation between the percentage of EMR2+ neutrophils and the occurrence of organ failure. In fact, EMR2 in neutrophil was considered a potential biomarker for SIRS [33]. Huang et al. have investigated the potential role of EMR2 in Mφ activation. They demonstrated that cross-linking of EMR2 in Mφs by 2A1 induced the translocation and colocalization of N-terminal fragment (NTF ) and the C-terminal fragment (CTF) within lipid rafts. This in turn led to receptor signaling and production of inflammatory cytokines, such as IL-8 and TNF [34]. Therefore, EMR2 seems to act as an activation receptor in myeloid cells, including neutrophils and Mφs.
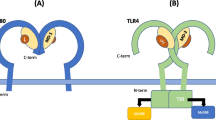
Notably, a search for genetic variants causing the autoinflammatory disorder vibratory urticaria recently identified a missense substitution 26 amino acids upstream from the GPS in EMR2 [35] (see also [36, 37]). This C492Y variant destabilized the association between the NTF and the CTF, such that shear stress that is incurred during vibration disrupted their noncovalent attachment. The remaining CTF elicited greater degranulation from transfected LAD2 mast cells, which is consistent with numerous observations indicating enhanced signaling activity of the CTF of aGPCRs upon removal of the NTF (Fig. 2a) (see also [38, 39]).
Functions of adhesion GPCRs in immune cells . (a) Left: A mAb binding at the NTF of EMR2 underneath the EGF-domain region potentiates the activation and recruitment of human neutrophils and Mφ. Right: A mutation near the GPS of EMR2 facilitates sheer stress-induced dissociation of the NTF from the CTF upon ligation by antibody or glycosaminoglycan (GAG). The remaining CTF enhances vibration-induced degranulation of mast cells in patients suffering from vibratory urticaria. (b) Left: CD97 triggers different activities. Mice lacking either CD97 or CD55 display a comparable phenotype with about twofold more circulating granulocytes. Augmented numbers of Gr-1-positive cells in cell cycle in the bone marrow indicate a higher granulopoietic activity. In addition, the NTF of CD97 can activate T cells by engaging CD55. Right: In the circulation, CD97 expression is constantly downregulated by contact with CD55 on blood and stromal cells, possible to restrict CD97–CD55-mediated cell adhesion to tissue sites. (c) BAI1 on Mφ binds PtdSer on apoptotic cells and LPS on gram-negative bacteria via the TSRs in its extracellular domain. BAI1 interacts with the N‐terminal domain of ELMO via an alpha‐helical domain in its intracellular tail. ELMO recruits Dock180 and promotes guanine nucleotide exchange factor activity. The GEF activity of the ELMO–Dock180 complex promotes Rac1 association with GTP and thereby mediates actin cytoskeletal reorganization during engulfment and ROS production. (d) GPR56 on NK cells forms a complex with the tetraspanin CD81 to inhibit cytotoxicity. Expression of GPR56 is induced by the transcription factor Hobit and terminated upon physiological activation through shedding of the NTF and transcriptional downregulation
2.3 EMR3 (ADGRE3 )
EMR3 was first identified in a DNA database, based on its high similarity with EMR2. cDNA cloning revealed that EMR3, due to concerted evolution of the encoding genes, shares ~90 % identity in the 7TM region with EMR2 [26]. EMR3 possesses two N-terminal EGF-like domains that may recognize a ligand on human Mφs and activated neutrophils [26]. Flow cytometric analysis revealed that all types of myeloid cells express EMR3 [40]. In peripheral blood, the highest expression of EMR3 was found on polymorphonuclear granulocytes. Moreover, mature CD16+ monocytes express high levels of EMR3, while CD16− monocytes and myeloid DCs are EMR3dim/low. Lymphocytes and plasmacytoid DCs lack EMR3. Interestingly, CD34+CD33−/CD38− committed hematopoietic stem cells and CD34+CD33+/CD38+ progenitors in bone marrow do not express EMR3, which is only upregulated during late granulopoiesis. Accordingly, EMR3 can be used to monitor granulocyte maturation [41]. In rodents, the EMR3 gene has been lost together with gene for EMR2 [24].
2.4 EMR4 (FIRE, ADGRE4)
Like EMR1, EMR4 was first identified in mouse, where it was called F4/80-like receptor (FIRE) [42]. FIRE has two N-terminal EGF-like domains and is expressed exclusively in myeloid cells, including CD8− DCs, but not by CD8+ DCs, monocytes, or Mφs [42, 43]. Immunohistochemical analysis revealed that FIRE+ cells reside predominantly in the red pulp and in the marginal zone of the spleen; yet, very few FIRE+ cells are seen in the white pulp. FIRE is downregulated during DC and Mφ activation, although the latter is not conclusive [43]; also the distribution of EMR4 on tissue Mφ warrants further investigation. A ligand for FIRE, present on the B lymphoma cell line A20, remains to be identified [43].
Human EMR4 is not expressed at the cell surface. Due to a one-nucleotide deletion in exon 8, translation of EMR4 would result in a truncated 232-amino acid protein, lacking the entire 7TM region [44]. Whether human cells express a soluble secreted fragment of EMR4 is not clear; transcripts are present in BDCA-1+ DCs and monocytes, but protein expression remains to be demonstrated [45]. Of note, the deletion disabling EMR4 surface expression is not found in nonhuman primates, including chimpanzees, suggesting that the gene became nonfunctional only after human speciation, about 5 million years ago [44]. Thus, EMR4 accounts for a genetic difference between humans and primates related to immunity.
2.5 CD97 (ADGRE5)
CD97 is a prototypical member of the EGF-TM7 subfamily of aGPCRs [46]. Evolutionary conserved, the gene encoding CD97 is found in all vertebrate genomes analyzed so far. Moreover, the structure and cellular distribution of CD97 are similar in human and mouse, indicating a conserved biological role. Initially, CD97 has been described as an activation-dependent Ag on lymphocytes [3]. Later, it became clear that hematopoietic stem and progenitor cells (HSPCs) and all leukocytes in human and mouse express CD97+, with highest levels found in myeloid cells [47, 48]. Moreover, many normal and malignant non-hematopoietic cell types express CD97 [49, 50].
Like EMR2, CD97 possesses up to five N-terminal EGF-like domains. Alternative splicing gives rise to three isoforms in human, namely CD97(EGF1,2,5), CD97(EGF1,2,3,5), and CD97(EGF1–5) [51]. CD97 isoforms expressed in the mouse are CD97(EGF1,2,4), CD97(EGF1,2,3,4), and CD97(EGF1,2,X,3,4), with X indicating a 45-amino acids sequence that does not correspond to any known protein module [52, 53]. EGF-like domains 1 and 2 interact with CD55 (decay-accelerating factor), a membrane-based regulator of complement activation [54, 55], also deposited on collagen fibers [56]. EGF-like domain 4, which is present only in the largest isoform, binds chondroitin sulfate B, a glycosaminoglycan that is found abundantly on cell surfaces and extracellular matrix [31, 57]. An RGD (Arg-Gly-Asp) motif 3′ to the EGF-domain region of human CD97 is bound by the integrin α5β1 and, possibly, αvβ3 [58]. Finally, through the GPCR autoproteolysis-inducing (GAIN) domain, CD97 interacts with the stromal and T-cell Ag Thy-1 (CD90) [59]. Thus, CD97 interacts with at least four matricellular ligands.
Based on its molecular structure and ligand interactions, a role for CD97 in leukocyte adhesion and/or trafficking has been proposed [54]. To investigate this assumption, mAbs directed against individual EGF domains were generated, interfering with either CD55 or chondroitin sulfate B binding. The functional consequences of targeting CD97 and blocking its ligand interactions were studied using several mouse inflammation models, each representing different aspects of innate and adaptive immunity. CD97 mAbs inhibited the accumulation of neutrophils at sites of inflammation, thereby affecting antibacterial host defense, inflammatory disorders, and stem cell mobilization from bone marrow [60–62]. Unexpectedly, they did so independent of the ligand interaction they interfered with. Moreover, the tested mAbs had no impact on Ag-specific (adaptive) responses, such as delayed-type hypersensitivity or experimental autoimmune encephalomyelitis [63]. Comparison of the consequences of Ab treatment and gene targeting implied that CD97 mAbs actively inhibit the innate response, presumably at the level of granulocyte and/or Mφ recruitment to sites of inflammation. Detailed investigation showed that CD97 mAbs deplete polymorphonuclear granulocytes in bone marrow and blood, which involved Fc receptors, but not complement activation, and was associated with an increase in serum levels of TNF and other pro-inflammatory cytokines [64]. Of note, depletion of granulocytes by CD97 mAbs requires acute inflammation, suggesting a mechanism of conditional, Ab-mediated granulocytopenia.
Two independently developed CD97-deficient mice showed no overt phenotype, except for a mild granulocytosis that increased under inflammatory conditions [50, 65]. A comparable phenotype with about twofold more circulating granulocytes was found in mice deficient for the CD97 ligand CD55 [66]. Augmented numbers of Gr-1-positive cells in cell cycle in the bone marrow indicated a higher granulopoietic activity in mice lacking either CD97 or CD55 (Fig. 2b). Concomitant with the increase in blood granulocyte numbers, CD55 knockout mice, challenged with the respiratory pathogen Streptococcus pneumoniae, developed less bacteremia and died later after infection [66], while CD97 knockout mice displayed an improved immune response toward acute infection with L. monocytogenes [65]. Moreover, possibly related to the interaction of CD55 with CD97, amelioration of collagen-induced arthritis and a trend toward less severe K/BxN serum transfer arthritis was found in mice that lack CD97 or CD55 [67].
Cellular and molecular assays have proved the interaction between CD97 and its various ligands in vitro. For example, soluble CD97 can cross-link CD55 on T cells in vitro, thereby increasing CD4+ T-cell proliferation, activation, and IL-10 and granulocyte–macrophage colony-stimulating factor (GM-CSF) secretion [68]. Yet, it has been difficult to prove ligand interactions of CD97 in vivo. Conclusive evidence was obtained when leukocytes from CD55-deficient mice were shown to express significantly increased levels of cell surface CD97 that normalized after transfer into wild-type mice because of contact with CD55 on both leukocytes and stromal cells [69]. Downregulation of both CD97 subunits occurred within minutes after first contact with CD55 in vivo and was strictly dependent on shear stress in the circulation. Of note, de novo ligation of CD97 did not activate signaling molecules constitutively engaged by CD97 in cancer cells, suggesting that CD55 downregulates CD97 surface expression on circulating leukocytes by a process that requires physical forces, but may not induce receptor signaling (Fig. 2b).
3 BAI/ADGRB Subfamily
The members of the ADGRB subfamily, commonly referred to as brain-specific angiogenesis inhibitors (BAIs), possess a large NTF that contains, next to the GAIN domain, a single hormone receptor motif (HRM) and 4–5 thrombospondin type 1 repeat (TSR) domains [70–72]. Moreover, BAIs contain a long C-terminal tail, downstream of the 7TM region, which comprises the PDZ-binding motif QTEV (Gln-Thr-Glu-Val) for intracellular signal transduction. BAIs were initially known for their ability to inhibit angiogenesis and tumor formation. Later studies unraveled key roles in the clearance of apoptotic cells and bacteria and in the regulation of immune responses. Finally, BAIs have been involved in synaptogenesis and dendritic spine formation. Below, we focus on the roles of BAI family proteins in cell clearance and immune regulation.
3.1 BAI1 (ADGRB1 )
BAI1 (human chromosome location at 8q24) was first cloned in 1997 as a target of the tumor suppressor gene p53 [73]; yet, it is not clear whether the expression of BAI1 indeed is dependent on p53 [74]. Abundantly found in the brain on neurons, astrocytes, and microglia, BAI1 is also expressed in other tissues (bone marrow, spleen, testis, and colon) and in distinct cell types (Mφs, skeletal muscle myoblasts) [70, 72] (Fig. 1). The TSR region of BAI1 comprises five repeats, which can bind phosphatidylserine (PtdSer) and LPS [75, 76]. Moreover, an N-terminal RGD motif can interact with the integrin α5β1 that regulates extracellular matrix attachment and motility [77]. Autoproteolysis of BAI1 at the GPS or behind the first TSR generates soluble ectodomains with antiangiogenic properties, known as vasculostatin 120 (kDa) and vasculostatin 40 (kDa) [78–80].
Park et al. identified BAI1 as an engulfment receptor for apoptotic cells that can both, bind the corpses and signal intracellularly in the phagocyte to facilitate their internalization [75]. BAI1 was found to function as a phagocytic receptor both, in Mφs and nonprofessional phagocytes, such as fibroblasts and epithelial cells. In embryonic zebra fish brains, BAI1 mediates engulfment of dying neurons by microglia [81]. BAI1 directly binds PtdSer, which is exposed on the outer leaflet of the plasma membrane of cells undergoing apoptosis, via the TSR regions, as determined by a combination of biochemical approaches [75]. Upon PtdSer binding, BAI1 transmits signals via the downstream intracellular engulfment molecule ELMO1 (engulfment and cell motility 1). The specific interaction occurs via an α-helical motif within the cytoplasmic tail of BAI1 and the N-terminal region of ELMO1 [75]. In fact, all three ELMO family members (ELMO1, ELMO2, and ELMO3) can bind to the cytoplasmic tail of BAI1. ELMO1 acts as a guanine nucleotide exchange factor for Rac and activates Rac, which promotes actin cytoskeletal rearrangements, necessary to mediate the phagocytosis of apoptotic cells (Fig. 2c). When phagocytes engulf apoptotic cells, they release anti-inflammatory mediators, such as tumor growth factor (TGF)β, IL-10, and prostaglandin E2, and these, in turn, reduce the levels of pro-inflammatory TNF in the local tissue. BAI1-mediated apoptotic cell clearance has been implicated in anti-inflammatory response.
Upon stimulation with apoptotic cells, BAI1 signaling in Mφs leads to upregulation of ABCA1 (ATP-binding cassette transporter 1) [82]. ABCA1 is a large transmembrane transporter that mediates cholesterol efflux from Mφs and is a key player for high-density lipoprotein (HDL) biogenesis. Genetic loss of BAI1 prevented ABCA1 upregulation in response to apoptotic cells and reduced serum levels of total cholesterol, HDL, low-density lipoprotein (LDL), and triglycerides [82]. Furthermore, transgenic mice that overexpressed BAI1 had an improved HDL/LDL ratio [82]. HDL has potent anti-inflammatory functions. Therefore, BAI1 in Mφs can regulate lipid homeostasis and anti-inflammatory responses via mediating apoptotic cell clearance.
A more recent study revealed a novel role for BAI1 in colonic epithelial cells [83]. Lee et al. showed that healthy epithelial cells mediate the engulfment of their apoptotic neighbors that arise due to acute injury, and this BAI1-mediated apoptotic cell removal beneficially influenced the level of tissue inflammation. Genetic deficiency of BAI1 caused increased numbers of uncleared apoptotic cells, greater inflammatory colon regions, and higher inflammatory cytokine levels (IL-1α and TNF) in the dextran sulfate sodium-induced colitis model. Conversely, transgenic overexpression of BAI1 in colonic epithelial cells resulted in a reduction of uncleared apoptotic cells, smaller inflammatory colon regions, and lower inflammatory cytokine levels. Furthermore, transgenic mice expressing a mutant version of BAI1 that cannot signal failed to protect mice suggesting that BAI1 signaling (BAI1–ELMO) is required for the beneficial effects in experimental colitis.
In addition to its role as a PtdSer receptor , BAI1 mediates engulfment of gram-negative bacteria [76]. The TSR region of BAI1 directly interacts with the LPS inner core of gram-negative bacteria. BAI1-mediated engulfment of bacteria by Mφs through this interaction induced production of the pro-inflammatory cytokine TNF [76]. Furthermore, a recent study revealed that BAI1 mediates reactive oxygen species (ROS) production and bacterial clearance in vivo [84]. BAI1-deficient mice showed impaired bacterial clearance and were more susceptible to peritoneal infection. These studies suggest that BAI1 can act as a pattern recognition receptor that mediates bacterial clearance through microbicidal activity and local inflammation (Fig. 2c). An interesting question that arises from these observations is how the signaling downstream of BAI1 remains anti-inflammatory during apoptotic cell engulfment while being pro-inflammatory during bacterial engulfment; whether this is mediated through other receptors that interact with BAI1 during apoptotic cell recognition or a different set of intracellular signaling molecules that might be attracted to the tail of BAI1 during these recognition events remains to be determined.
Besides its immune-related activities, BAI1 possesses other unique functions. As a PtdSer receptor, BAI1 induces myoblast fusion into multinucleated myofibers in response to apoptotic or PtdSer-exposing myoblasts [85]. Moreover, BAI1 regulates spinogenesis and synaptogenesis [86]. Finally, in a study of BAI1-deficient mice, BAI1 was found to regulate spatial learning and memory as well as synaptic plasticity [87].
3.2 BAI2 (ADGRB2) and BAI3 (ADGRB3)
BAI2 (human chromosome location at 1p35) and BAI3 (human chromosome location at 6q12) were identified as homologs of BAI1 [88]. Although expression patterns are not well characterized, BAI2 and BAI3 are found in many tissues, including the brain, heart, skeletal muscle, intestine, and thymus [70, 72]. Both receptors possess four TSR motifs, and although immune-related functions have not been reported yet, BAI3 possesses the motif necessary for binding ELMO proteins that allows BAI1 to engulf apoptotic cells [89]. Moreover, similar to BAI1, BAI3 has been implicated in skeletal myoblast fusion and muscle fiber formation [90].
4 ADGRG Subfamily
The ADGRG subfamily member GPR56 is one of the best-studied aGPCRs, well known for its causal involvement in neuronal and cortical development [91]. The gene encoding GPR56 clusters with the genes for GPR97 and GPR114 on chromosome 16q21, suggesting a common evolutionary origin. In contrast to GPR56, which is expressed in many different cell types, very little is known about GPR97 and GPR114, which both seem to have a rather restricted cellular distribution [92]. All three genes are expressed in leukocytes (Fig. 1).
4.1 GPR56 (ADGRG1)
GPR56 has been strongly linked with development. HSPCs in the mouse embryo and adult bone marrow abundantly express GPR56, but levels substantially decrease as cells differentiate. Recent studies have explored the role of GPR56 in HSPCs. Solaimani-Kartalaei et al. found a critical role of GPR56 in the formation of hematopoietic clusters during endothelial to hematopoietic cell transition [93]. In contrast, GPR56 deficiency does not impair HSPC maintenance or function during steady-state or myeloablative stress-induced hematopoiesis, and GPR56-deficient cells respond normally to physiological and pharmacological mobilization signals, despite the reported role of this aGPCR as a regulator of cell adhesion and migration in neuronal cells [94]. Thus, GPR56 expression is required for generating the first hematopoietic stem cells, but largely dispensable for steady-state and regenerative hematopoiesis.
Flow cytometric analysis with newly generated mAbs detected the presence of GPR56 in human cytotoxic NK and T lymphocytes, including CD8+, CD4+, and γδ T cells [95, 96]. Primary infection with cytomegalovirus, which generates a vast population of CD8+ T cells with an effector phenotype, induced a strong increase in GPR56 expression in virus-specific CD8+ T cells that remained detectable during latency. In NK-92 cells, ectopic expression of GPR56 inhibits spontaneous and SDF-1-stimulated cell migration [96]. Investigation of NK cells from polymicrogyria patients with a null mutation in the GPR56 gene and NK-92 cells overexpressing GPR56 revealed that GPR56 suppresses the production of inflammatory cytokines and cytolytic proteins, degranulation, and target cell killing [97]. GPR56 pursues this activity by associating with the tetraspanin CD81. Expression of GPR56 was triggered by Hobit, a homolog of the transcription factor Blimp-1, and declined upon cell activation. Thus, GPR56 controls natural cytotoxicity displayed by NK cells in order to protect the body against harmful viruses and neoplasms (Fig. 2d). Circulating T and NK cells in the mouse do not express GPR56, which may be due to the different way they acquire cytotoxic capacity.
4.2 GPR97 (PB99, ADGRG3) and GPR114 (ADGRG5)
A microarray study of mature human leukocyte populations detected GPR97 transcripts in neutrophils and eosinophils [96]. Moreover, GPR97 gene expression that was found in murine pre-B cells and thymocytes, but not in mature B and T cells, suggested a role in early lymphoid development. Investigation of independently raised knockout mice revealed no necessity of GPR97 for B- or T-cell development [98], but implied an essential role in follicular versus marginal zone B-lymphocyte fate decision [99]. In spite of its expression in eosinophilic granulocytes, GPR97 was found to be dispensable for inflammation in ovalbumin-induced asthmatic mice [100]. GPR114 is another aGPCR transcribed in lymphoid and myeloid cells [96]; yet, as for GPR97, expression needs to be confirmed at the protein level.
5 Biological and Translational Implications
aGPCRs have been implicated in the development and function of HSPCs, monocytes, Mφs, DCs, granulocytes, lymphocytes, and, thus, most lineages of hematopoietic cells. Nevertheless, it has not been clear for quite some time whether these noncanonical GPCRs regulate cellular homeostasis and activation or whether they also control specific immune functions. Studies on the role of BAI1 in Mφs and GPR56 in cytotoxic lymphocytes imply that aGPCRs indeed regulate innate, and possibly also adaptive, effector functions. Thereby, these studies paved the way for further exploring the biological role of these receptors using novel genetic models and pharmacological tools. As a prime example, BAI1 facilitates clearance of apoptotic cells by Mφs and intestinal epithelial cells and modulates the inflammatory status within tissue.
EGF-TM7 subfamily aGPCRs figure prominently in the biology of polymorphonuclear granulocytes, an aspect that has been overlooked in the beginning, due to the prominent presence of the EMR1 ortholog F4/80 on Mφs in the mouse and the upregulation of CD97 during lymphocyte activation. Legrand et al. recently showed that an afucosylated mAb directed against EMR1 efficiently eradicates eosinophils through Ab-dependent cell-mediated cytotoxicity, resulting in long-term in vivo depletion of eosinophils in monkeys and, thus, providing a treatment option for eosinophilic disorders [19]. Similarly, mAbs to mouse CD97 eliminate neutrophils (and possible also eosinophils) in an Fc receptor-dependent manner, thereby ameliorating various inflammatory conditions [64]. These studies imply that biologicals targeting aGPCRs may be useful in treating immune cell-related diseases.
Association of aGPCRs with hematopoietic malignancies has not been investigated systematically so far, despite their role in solid tumors (see [101]). A recent study reported the association of CD97 expression with internal tandem duplications within the juxtamembrane region of the FMS-like tyrosine kinase receptor FLT3 (FLT3-ITD) in patients with acute myeloid leukemia (AML ), which is associated with an aggressive clinical phenotype [102]. CD97 knockdown resulted in reduced cell adhesion and trans-well migration in vitro. Moreover, GPR56 expression identifies primary human AML cells with high repopulating potential in vivo [103]. Elevated expression of GPR56 was found in AML cells with high ecotropic viral integration site-1 expression, a refractory type of the disease with a poor prognosis. Knockdown of GPR56 expression decreased the cellular adhesion ability through inactivation of RhoA signaling, resulting in a reduction of cellular growth rates and enhanced apoptosis [104].
Despite an increasing number of studies that have linked aGPCRs with immune function(s), main uncertainties persist. For example, though many of the protein folds found in the NTF of aGPCRs are involved in cell–cell interactions, evidence is scarce that aGPCRs contribute to the communication between immune cells. Further, it is not clear to what extent aGPCR autoproteolysis and signaling facilitate immune functions. Moreover, functional redundancy between members of the EGF-TM7 subfamily and the GPR56/GPR97/GPR114 cluster seems possible and warrants further investigation. Related to this, the aGPCR signature of immune cells need to be unraveled comprehensively. Exploring these issues eventually will improve our understanding of the biology of aGPCRs and, thereby, may widen the canon of GPCR activities in the immune system.
Abbreviations
- 7TM:
-
Seven transmembrane
- Ab:
-
Antibody
- ABCA:
-
ATP-binding cassette transporter
- ACAID:
-
Anterior chamber-associated immune deviation
- Ag:
-
Antigen
- AML:
-
Acute myeloid leukemia
- BAI:
-
Brain-specific angiogenesis inhibitor
- BDCA:
-
Blood dendritic cell antigen
- CTF:
-
C-terminal fragment
- DC:
-
Dendritic cell
- EGF:
-
Epidermal growth factor
- ELMO:
-
Engulfment and cell motility
- EMR:
-
EGF-like module-containing mucin-like hormone receptor-like
- GAIN:
-
GPCR autoproteolysis inducing
- GEF:
-
Guanine nucleotide exchange factor
- GPCR:
-
G protein-coupled receptor
- GPS:
-
GPCR proteolysis site
- HDL:
-
High-density lipoprotein
- HRM:
-
Hormone receptor motif
- HSPC:
-
Hematopoietic stem and progenitor cell
- IFN:
-
Interferon
- IL:
-
Interleukin
- LDL:
-
Low-density lipoprotein
- LPS:
-
Lipopolysaccharide
- mAb:
-
Monoclonal antibody
- Mφ:
-
Macrophages
- NK:
-
Natural killer
- NTF:
-
N-terminal fragment
- PtdSer:
-
Phosphatidylserine
- ROS:
-
Reactive oxygen species
- SIRS:
-
Systemic inflammatory response syndrome
- TGF:
-
Tumor growth factor
- TNF:
-
Tumor necrosis factor
- Treg:
-
Regulatory T
- TSR:
-
Thrombospondin type 1 repeat
References
Rot A, von Andrian UH (2004) Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol 22:891–928
Austyn JM, Gordon S (1981) F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol 11:805–815
Eichler W, Aust G, Hamann D (1994) Characterization of an early activation-dependent antigen on lymphocytes defined by the monoclonal antibody BL-Ac(F2). Scand J Immunol 39:111–115
McKnight AJ, Gordon S (1996) EGF-TM7: a novel subfamily of seven-transmembrane-region leukocyte cell-surface molecules. Immunol Today 17:283–287
McKnight AJ, Gordon S (1998) The EGF-TM7 family: unusual structures at the leukocyte surface. J Leukoc Biol 63:271–280
Kwakkenbos MJ, Kop EN, Stacey M, Matmati M, Gordon S, Lin H-H et al (2004) The EGF-TM7 family: a postgenomic view. Immunogenetics 55:655–666
Lin H-H, Stacey M, Stein-Streilein J, Gordon S (2010) F4/80: the macrophage-specific adhesion-GPCR and its role in immunoregulation. Adv Exp Med Biol 706:149–156
Gordon S, Hamann J, Lin H-H, Stacey M (2011) F4/80 and the related adhesion-GPCRs. Eur J Immunol 41:2472–2476
McGarry MP, Stewart CC (1991) Murine eosinophil granulocytes bind the murine macrophage-monocyte specific monoclonal antibody F4/80. J Leukoc Biol 50:471–478
Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S (2005) Macrophage receptors and immune recognition. Annu Rev Immunol 23:901–944
Hume DA, Gordon S (1983) Mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Identification of resident macrophages in renal medullary and cortical interstitium and the juxtaglomerular complex. J Exp Med 157:1704–1709
Hume DA, Robinson AP, MacPherson GG, Gordon S (1983) The mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Relationship between macrophages, Langerhans cells, reticular cells, and dendritic cells in lymphoid and hematopoietic organs. J Exp Med 158:1522–1536
Morris L, Graham CF, Gordon S (1991) Macrophages in haemopoietic and other tissues of the developing mouse detected by the monoclonal antibody F4/80. Development 112:517–526
Qian B-Z, Pollard JW (2010) Macrophage diversity enhances tumor progression and metastasis. Cell 141:39–51
Baud V, Chissoe SL, Viegas-Péquignot E, Diriong S, N’Guyen VC, Roe BA et al (1995) EMR1, an unusual member in the family of hormone receptors with seven transmembrane segments. Genomics 26:334–344
Lin HH, Stubbs LJ, Mucenski ML (1997) Identification and characterization of a seven transmembrane hormone receptor using differential display. Genomics 41:301–308
McKnight AJ, Macfarlane AJ, Dri P, Turley L, Willis AC, Gordon S (1996) Molecular cloning of F4/80, a murine macrophage-restricted cell surface glycoprotein with homology to the G-protein-linked transmembrane 7 hormone receptor family. J Biol Chem 271:486–489
Hamann J, Koning N, Pouwels W, Ulfman LH, van Eijk M, Stacey M et al (2007) EMR1, the human homolog of F4/80, is an eosinophil-specific receptor. Eur J Immunol 37:2797–2802
Legrand F, Tomasevic N, Simakova O, Lee C-CR, Wang Z, Raffeld M et al (2014) The eosinophil surface receptor epidermal growth factor-like module containing mucin-like hormone receptor 1 (EMR1): a novel therapeutic target for eosinophilic disorders. J Allergy Clin Immunol 133:1439–1447 1447.e1–8
Warschkau H, Kiderlen AF (1999) A monoclonal antibody directed against the murine macrophage surface molecule F4/80 modulates natural immune response to Listeria monocytogenes. J Immunol 163:3409–3416
Lin H-H, Faunce DE, Stacey M, Terajewicz A, Nakamura T, Zhang-Hoover J et al (2005) The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J Exp Med 201:1615–1625
Schaller E, Macfarlane AJ, Rupec RA, Gordon S, McKnight AJ, Pfeffer K (2002) Inactivation of the F4/80 glycoprotein in the mouse germ line. Mol Cell Biol 22:8035–8043
van den Berg TK, Kraal G (2005) A function for the macrophage F4/80 molecule in tolerance induction. Trends Immunol 26:506–509
Kwakkenbos MJ, Matmati M, Madsen O, Pouwels W, Wang Y, Bontrop RE et al (2006) An unusual mode of concerted evolution of the EGF-TM7 receptor chimera EMR2. FASEB J 20:2582–2584
Lin HH, Stacey M, Hamann J, Gordon S, McKnight AJ (2000) Human EMR2, a novel EGF-TM7 molecule on chromosome 19p13.1, is closely related to CD97. Genomics 67:188–200
Stacey M, Lin HH, Hilyard KL, Gordon S, McKnight AJ (2001) Human epidermal growth factor (EGF) module-containing mucin-like hormone receptor 3 is a new member of the EGF-TM7 family that recognizes a ligand on human macrophages and activated neutrophils. J Biol Chem 276:18863–18870
Kwakkenbos MJ, Chang G-W, Lin H-H, Pouwels W, de Jong EC, van Lier RAW et al (2002) The human EGF-TM7 family member EMR2 is a heterodimeric receptor expressed on myeloid cells. J Leukoc Biol 71:854–862
Chang G-W, Davies JQ, Stacey M, Yona S, Bowdish DME, Hamann J et al (2007) CD312, the human adhesion-GPCR EMR2, is differentially expressed during differentiation, maturation, and activation of myeloid cells. Biochem Biophys Res Commun 353:133–138
van Eijk M, Aust G, Brouwer MSM, van Meurs M, Voerman JSA, Dijke IE et al (2010) Differential expression of the EGF-TM7 family members CD97 and EMR2 in lipid-laden macrophages in atherosclerosis, multiple sclerosis and Gaucher disease. Immunol Lett 129:64–71
Lin HH, Stacey M, Saxby C, Knott V, Chaudhry Y, Evans D et al (2001) Molecular analysis of the epidermal growth factor-like short consensus repeat domain-mediated protein-protein interactions: dissection of the CD97-CD55 complex. J Biol Chem 276:24160–24169
Stacey M, Chang G-W, Davies JQ, Kwakkenbos MJ, Sanderson RD, Hamann J et al (2003) The epidermal growth factor-like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood 102:2916–2924
Yona S, Lin H-H, Dri P, Davies JQ, Hayhoe RPG, Lewis SM et al (2008) Ligation of the adhesion-GPCR EMR2 regulates human neutrophil function. FASEB J 22:741–751
Lewis SM, Treacher DF, Edgeworth J, Mahalingam G, Brown CS, Mare TA et al (2015) Expression of CD11c and EMR2 on neutrophils: potential diagnostic biomarkers for sepsis and systemic inflammation. Clin Exp Immunol 182:184–194
Huang Y-S, Chiang N-Y, Hu C-H, Hsiao C-C, Cheng K-F, Tsai W-P et al (2012) Activation of myeloid cell-specific adhesion class G protein-coupled receptor EMR2 via ligation-induced translocation and interaction of receptor subunits in lipid raft microdomains. Mol Cell Biol 32:1408–1420
Boyden SE, Desai A, Cruse G, Young ML, Bolan HC, Scott LM et al (2016) Vibratory urticaria associated with a missense variant in ADGRE2. N Engl J Med 374:656–663
Nieberler M, Kittel RJ, Petrenko AG, Lin H-H, Langenhan T (2016) Control of adhesion GPCR function through proteolytic processing. In: Langenhan T, Schöneberg T (eds) Adhesion G protein-coupled receptors: molecular, physiological and pharmacological principles in health and disease. Springer, Heidelberg
Scholz N, Monk KR, Kittel RJ, Langenhan T (2016) Adhesion GPCRs as a putative class of metabotropic mechanosensors. In: Langenhan T, Schöneberg T (eds) Adhesion G protein-coupled receptors: molecular, physiological and pharmacological principles in health and disease. Springer, Heidelberg
Liebscher I, Schöneberg T (2016) Tethered agonism: a common activation mechanism of adhesion GPCRs. In: Langenhan T, Schöneberg T (eds) Adhesion G protein-coupled receptors: molecular, physiological and pharmacological principles in health and disease. Springer, Heidelberg
Kishore A, Hall RA (2016) Versatile signaling activity of adhesion GPCRs. In: Langenhan T, Schöneberg T (eds) Adhesion G protein-coupled receptors: molecular, physiological and pharmacological principles in health and disease. Springer, Heidelberg
Matmati M, Pouwels W, van Bruggen R, Jansen M, Hoek RM, Verhoeven AJ et al (2007) The human EGF-TM7 receptor EMR3 is a marker for mature granulocytes. J Leukoc Biol 81:440–448
Drewniak A, van Raam BJ, Geissler J, Tool ATJ, Mook ORF, van den Berg TK et al (2009) Changes in gene expression of granulocytes during in vivo granulocyte colony-stimulating factor/dexamethasone mobilization for transfusion purposes. Blood 113:5979–5998
Caminschi I, Lucas KM, O’Keeffe MA, Hochrein H, Laâbi Y, Köntgen F et al (2001) Molecular cloning of F4/80-like-receptor, a seven-span membrane protein expressed differentially by dendritic cell and monocyte-macrophage subpopulations. J Immunol 167:3570–3576
Stacey M, Chang G-W, Sanos SL, Chittenden LR, Stubbs L, Gordon S et al (2002) EMR4, a novel epidermal growth factor (EGF)-TM7 molecule up-regulated in activated mouse macrophages, binds to a putative cellular ligand on B lymphoma cell line A20. J Biol Chem 277:29283–29293
Hamann J, Kwakkenbos MJ, de Jong EC, Heus H, Olsen AS, van Lier RAW (2003) Inactivation of the EGF-TM7 receptor EMR4 after the Pan-Homo divergence. Eur J Immunol 33:1365–1371
Caminschi I, Vandenabeele S, Sofi M, McKnight AJ, Ward N, Brodnicki TC et al (2006) Gene structure and transcript analysis of the human and mouse EGF-TM7 molecule, FIRE. DNA Seq 17:8–14
Hamann J, Eichler W, Hamann D, Kerstens HM, Poddighe PJ, Hoovers JM et al (1995) Expression cloning and chromosomal mapping of the leukocyte activation antigen CD97, a new seven-span transmembrane molecule of the secretion receptor superfamily with an unusual extracellular domain. J Immunol 155:1942–1950
van Pel M, Hagoort H, Hamann J, Fibbe WE (2008) CD97 is differentially expressed on murine hematopoietic stem-and progenitor-cells. Haematologica 93:1137–1144
Kop EN, Matmati M, Pouwels W, Leclercq G, Tak PP, Hamann J (2009) Differential expression of CD97 on human lymphocyte subsets and limited effect of CD97 antibodies on allogeneic T-cell stimulation. Immunol Lett 123:160–168
Jaspars LH, Vos W, Aust G, van Lier RA, Hamann J (2001) Tissue distribution of the human CD97 EGF-TM7 receptor. Tissue Antigens 57:325–331
Veninga H, Becker S, Hoek RM, Wobus M, Wandel E, van der Kaa J et al (2008) Analysis of CD97 expression and manipulation: antibody treatment but not gene targeting curtails granulocyte migration. J Immunol 181:6574–6583
Gray JX, Haino M, Roth MJ, Maguire JE, Jensen PN, Yarme A et al (1996) CD97 is a processed, seven-transmembrane, heterodimeric receptor associated with inflammation. J Immunol 157:5438–5447
Qian YM, Haino M, Kelly K, Song WC (1999) Structural characterization of mouse CD97 and study of its specific interaction with the murine decay-accelerating factor (DAF, CD55). Immunology 98:303–311
Hamann J, van Zeventer C, Bijl A, Molenaar C, Tesselaar K, van Lier RA (2000) Molecular cloning and characterization of mouse CD97. Int Immunol 12:439–448
Hamann J, Vogel B, van Schijndel GM, van Lier RA (1996) The seven-span transmembrane receptor CD97 has a cellular ligand (CD55, DAF). J Exp Med 184:1185–1189
Hamann J, Stortelers C, Kiss-Toth E, Vogel B, Eichler W, van Lier RA (1998) Characterization of the CD55 (DAF)-binding site on the seven-span transmembrane receptor CD97. Eur J Immunol 28:1701–1707
Karpus ON, Kiener HP, Niederreiter B, Yilmaz-Elis AS, van der Kaa J, Ramaglia V et al (2015) CD55 deposited on synovial collagen fibers protects from immune complex-mediated arthritis. Arthritis Res Ther 17:6
Kwakkenbos MJ, Pouwels W, Matmati M, Stacey M, Lin H-H, Gordon S et al (2005) Expression of the largest CD97 and EMR2 isoforms on leukocytes facilitates a specific interaction with chondroitin sulfate on B cells. J Leukoc Biol 77:112–119
Wang T, Ward Y, Tian L, Lake R, Guedez L, Stetler-Stevenson WG et al (2005) CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood 105:2836–2844
Wandel E, Saalbach A, Sittig D, Gebhardt C, Aust G (2012) Thy-1 (CD90) is an interacting partner for CD97 on activated endothelial cells. J Immunol 188:1442–1450
Leemans JC, te Velde AA, Florquin S, Bennink RJ, de Bruin K, van Lier RAW et al (2004) The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. J Immunol 172:1125–1131
Kop EN, Adriaansen J, Smeets TJM, Vervoordeldonk MJ, van Lier RAW, Hamann J et al (2006) CD97 neutralisation increases resistance to collagen-induced arthritis in mice. Arthritis Res Ther 8:R155
de Groot DM, Vogel G, Dulos J, Teeuwen L, Stebbins K, Hamann J et al (2009) Therapeutic antibody targeting of CD97 in experimental arthritis: the role of antigen expression, shedding, and internalization on the pharmacokinetics of anti-CD97 monoclonal antibody 1B2. J Immunol 183:4127–4134
Hamann J, Veninga H, de Groot DM, Visser L, Hofstra CL, Tak PP et al (2010) CD97 in leukocyte trafficking. Adv Exp Med Biol 706:128–137
Veninga H, de Groot DM, McCloskey N, Owens BM, Dessing MC, Verbeek JS et al (2011) CD97 antibody depletes granulocytes in mice under conditions of acute inflammation via a Fc receptor-dependent mechanism. J Leukoc Biol 89:413–421
Wang T, Tian L, Haino M, Gao J-L, Lake R, Ward Y et al (2007) Improved antibacterial host defense and altered peripheral granulocyte homeostasis in mice lacking the adhesion class G protein receptor CD97. Infect Immun 75:1144–1153
Veninga H, Hoek RM, de Vos AF, de Bruin AM, An F-Q, van der Poll T et al (2011) A novel role for CD55 in granulocyte homeostasis and anti-bacterial host defense. PLoS One 6:e24431
Hoek RM, de Launay D, Kop EN, Yilmaz-Elis AS, Lin F, Reedquist KA et al (2010) Deletion of either CD55 or CD97 ameliorates arthritis in mouse models. Arthritis Rheum 62:1036–1042
Capasso M, Durrant LG, Stacey M, Gordon S, Ramage J, Spendlove I (2006) Costimulation via CD55 on human CD4+ T cells mediated by CD97. J Immunol 177:1070–1077
Karpus ON, Veninga H, Hoek RM, Flierman D, van Buul JD, Vandenakker CC et al (2013) Shear stress-dependent downregulation of the adhesion-G protein-coupled receptor CD97 on circulating leukocytes upon contact with its ligand CD55. J Immunol 190:3740–3748
Cork SM, Van Meir EG (2011) Emerging roles for the BAI1 protein family in the regulation of phagocytosis, synaptogenesis, neurovasculature, and tumor development. J Mol Med 89:743–752
Park D, Ravichandran KS (2010) Emerging roles of brain-specific angiogenesis inhibitor 1. Adv Exp Med Biol 706:167–178
Stephenson JR, Purcell RH, Hall RA (2014) The BAI subfamily of adhesion GPCRs: synaptic regulation and beyond. Trends Pharmacol Sci 35:208–215
Nishimori H, Shiratsuchi T, Urano T, Kimura Y, Kiyono K, Tatsumi K et al (1997) A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene 15:2145–2150
Kaur B, Brat DJ, Calkins CC, Van Meir EG (2003) Brain angiogenesis inhibitor 1 is differentially expressed in normal brain and glioblastoma independently of p53 expression. Am J Pathol 162:19–27
Park D, Tosello-Trampont A-C, Elliott MR, Lu M, Haney LB, Ma Z et al (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450:430–434
Das S, Owen KA, Ly KT, Park D, Black SG, Wilson JM et al (2011) Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc Natl Acad Sci U S A 108:2136–2141
Koh JT, Kook H, Kee HJ, Seo Y-W, Jeong BC, Lee JH et al (2004) Extracellular fragment of brain-specific angiogenesis inhibitor 1 suppresses endothelial cell proliferation by blocking alphavbeta5 integrin. Exp Cell Res 294:172–184
Kaur B, Brat DJ, Devi NS, Van Meir EG (2005) Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene 24:3632–3642
Kaur B, Cork SM, Sandberg EM, Devi NS, Zhang Z, Klenotic PA et al (2009) Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res 69:1212–1220
Cork SM, Kaur B, Devi NS, Cooper L, Saltz JH, Sandberg EM et al (2012) A proprotein convertase/MMP-14 proteolytic cascade releases a novel 40 kDa vasculostatin from tumor suppressor BAI1. Oncogene 31:5144–5152
Mazaheri F, Breus O, Durdu S, Haas P, Wittbrodt J, Gilmour D et al (2014) Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat Commun 5:4046
Fond AM, Lee CS, Schulman IG, Kiss RS, Ravichandran KS (2015) Apoptotic cells trigger a membrane-initiated pathway to increase ABCA1. J Clin Invest 125:2748–2758
Lee CS, Penberthy KK, Wheeler KM, Juncadella IJ, Vandenabeele P, Lysiak JJ et al (2016) Boosting apoptotic cell clearance by colonic epithelial cells attenuates inflammation in vivo. Immunity 44(4):807–820
Billings EA, Lee CS, Owen KA, D’Souza RS, Ravichandran KS, Casanova JE (2016) The adhesion GPCR BAI1 mediates macrophage ROS production and microbicidal activity against Gram-negative bacteria. Sci Signal 9:ra14
Hochreiter-Hufford AE, Lee CS, Kinchen JM, Sokolowski JD, Arandjelovic S, Call JA et al (2013) Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497:263–267
Duman JG, Tzeng CP, Tu Y-K, Munjal T, Schwechter B, Ho TS-Y et al (2013) The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. J Neurosci 33:6964–6978
Zhu D, Li C, Swanson AM, Villalba RM, Guo J, Zhang Z et al (2015) BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. J Clin Invest 125:1497–1508
Shiratsuchi T, Nishimori H, Ichise H, Nakamura Y, Tokino T (1997) Cloning and characterization of BAI2 and BAI3, novel genes homologous to brain-specific angiogenesis inhibitor 1 (BAI1). Cytogenet Cell Genet 79:103–108
Lanoue V, Usardi A, Sigoillot SM, Talleur M, Iyer K, Mariani J et al (2013) The adhesion-GPCR BAI3, a gene linked to psychiatric disorders, regulates dendrite morphogenesis in neurons. Mol Psychiatry 18:943–950
Hamoud N, Tran V, Croteau L-P, Kania A, Côté J-F (2014) G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc Natl Acad Sci U S A 111:3745–3750
Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R et al (2004) G protein-coupled receptor-dependent development of human frontal cortex. Science 303:2033–2036
Hamann J, Aust G, Araç D, Engel FB, Formstone C, Fredriksson R et al (2015) International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol Rev 67:338–367
Solaimani Kartalaei P, Yamada-Inagawa T, Vink CS, de Pater E, van der Linden R, Marks-Bluth J et al (2015) Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J Exp Med 212:93–106
Rao TN, Marks-Bluth J, Sullivan J, Gupta MK, Chandrakanthan V, Fitch SR et al (2015) High-level Gpr56 expression is dispensable for the maintenance and function of hematopoietic stem and progenitor cells in mice. Stem Cell Res 14:307–322
Chiesa Della M, Falco M, Parolini S, Bellora F, Petretto A, Romeo E et al (2010) GPR56 as a novel marker identifying the CD56dull CD16+ NK cell subset both in blood stream and in inflamed peripheral tissues. Int Immunol 22:91–100
Peng Y-M, van de Garde MDB, Cheng K-F, Baars PA, Remmerswaal EBM, van Lier RAW et al (2011) Specific expression of GPR56 by human cytotoxic lymphocytes. J Leukoc Biol 90:735–740
Chang G-W, Hsiao C-C, Peng Y-M, Vieira Braga FA, Kragten NAM, Remmerswaal EBM et al (2016) The adhesion G protein-coupled receptor GPR56/ADGRG1 is an inhibitory receptor on human NK cells. Cell Rep 15:1757–1770
Sleckman BP, Khan WN, Xu W, Bassing CH, Malynn BA, Copeland NG et al (2000) Cloning and functional characterization of the early-lymphocyte-specific Pb99 gene. Mol Cell Biol 20:4405–4410
Wang J-J, Zhang L-L, Zhang H-X, Shen C-L, Lu S-Y, Kuang Y et al (2013) Gpr97 is essential for the follicular versus marginal zone B-lymphocyte fate decision. Cell Death Dis 4:e853
Shi J-P, Li X-N, Zhang X-Y, Du B, Jiang W-Z, Liu M-Y et al (2015) Gpr97 is dispensable for inflammation in OVA-induced asthmatic mice. PLoS One 10:e0131461
Aust G, Zhu D, Van Meir EG, Xu L (2016) Adhesion GPCRs in tumorigenesis. In: Langenhan T, Schöneberg T (eds) Adhesion G protein-coupled receptors: molecular, physiological and pharmacological principles in health and disease. Springer, Heidelberg
Wobus M, Bornhäuser M, Jacobi A, Kräter M, Otto O, Ortlepp C et al (2015) Association of the EGF-TM7 receptor CD97 expression with FLT3-ITD in acute myeloid leukemia. Oncotarget 6:38804–38815
Pabst C, Bergeron A, Lavallée V-P, Yeh J, Gendron P, Norddahl GL et al (2016) GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 127:2018–2027
Saito Y, Kaneda K, Suekane A, Ichihara E, Nakahata S, Yamakawa N et al (2013) Maintenance of the hematopoietic stem cell pool in bone marrow niches by EVI1-regulated GPR56. Leukemia 27:1637–1649
Acknowledgments
We thank the members of our laboratories for generating a large part of the data discussed in this chapter. This work was supported by grants to J.H. from the Deutsche Forschungsgemeinschaft (Research Unit 2149) and the Thyssen Foundation (2015-00387), to K.S.R. from the National Institutes of Health, USA (GM064709, HD074981, and MH096484), and to H.H.L. from the Ministry of Science and Technology, Taiwan (MOST-104-2320-B-182-035-MY3) and the Chang Gung Memorial Hospital (CMRPD1C0633, CMRPD1D0072-3, and CMRPD1D0392).
Author Contributions J.H., C.C.H., H.H.L., C.S.L., and K.S.R. wrote the manuscript.
Competing Financial Interests The authors declare no competing financial interests.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this chapter
Cite this chapter
Hamann, J., Hsiao, CC., Lee, C.S., Ravichandran, K.S., Lin, HH. (2016). Adhesion GPCRs as Modulators of Immune Cell Function. In: Langenhan, T., Schöneberg, T. (eds) Adhesion G Protein-coupled Receptors. Handbook of Experimental Pharmacology, vol 234. Springer, Cham. https://doi.org/10.1007/978-3-319-41523-9_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-41523-9_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-41521-5
Online ISBN: 978-3-319-41523-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)