Abstract
MicroRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs) are two groups of small non-coding RNAs with different functional roles. miRNAs are post-transcriptional regulators of gene expression in a plethora of critical processes in multicellular eukaryotes. Therefore, it comes as no surprise that viral pathogens have evolved ways to subvert the miRNA network. It is increasingly evident that miRNAs have functional roles in viral replication as well as their potential employment by host cells to combat viral infection. A number of viruses are now known to encode for miRNAs, predominantly in DNA viruses such as herpesviruses. Although virus-encoded miRNAs have been reported in retroviruses such as HIV-1 , their functional significance is under debate. This controversy also extends to RNA viruses and their ability to express miRNAs. Identification of target genes for some of these viral miRNAs suggests they may function in the regulation of lytic and latent viral replication and in restricting antiviral responses. Viruses have also evolved the ability to downregulate or upregulate the expression of specific cellular miRNAs to enhance their replication. I will also briefly review evidence that demonstrate the role of piRNAs in silencing transposable elements to maintain germline genome integrity . This chapter provides an overview of our current understanding of the complex relationship between viruses and cellular miRNA and piRNA machineries.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 miRNA and piRNA Biogenesis and Function
1.1 miRNAs
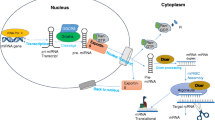
miRNAs are noncoding RNAs ~21–23 nucleotide (nt) in length that post-transcriptionally regulate the expression of a plethora of eukaryotic genes. miRNAs are first transcribed as primary transcripts (pri-miRNAs) by RNA polymerase II (RNAP II) (Fig. 1.1). Many pri-miRNA are capped and polyadenylated, and all contain a stem-loop secondary structure. At the nucleus, this secondary structure is recognized by the RNAse type-III Drosha , in association with its co-factor DGCR8, which cleaves the pri-miRNA into its intermediate form (pre-miRNA) of about 70 nt in length with a 2 bp overhang at its 3′ end. The excised hairpin loop is then recognized by the nuclear export factor exportin 5, which facilitates the transport of the pre-miRNA from the nucleus to the cytoplasm where it is processed by another RNAse III Dicer , and its co-factor TRBP, into its mature duplex form. The “guide” strand of the mature miRNA (often the anti-sense strand) is subsequently recruited to the RNA-induced silencing complex (RISC) by its interaction with the Argonaute protein, whereas the “passenger” strand frequently gets degraded (Shukla et al. 2011; Van Wynsberghe et al. 2011; Erson-Bensan 2014). Although the mechanism in allocating the “guide” and “passenger” strand designation is not fully understood, it is thought that once a strand is selected and loaded onto RISC, the other (“star”) strand is destroyed. The proteins present in RISC vary between species, but the core proteins include Dicer and the Argonaute protein family. Argonaute proteins are phylogenetically categorized into two clades based on sequence similarity: the Argonaute (Ago) clade and the P-element induced wimpy testis (Piwi) clade (Carmell et al. 2002). The Ago clade is associated with miRNA and short interfering RNA (siRNA) activities whereas the Piwi proteins are associated with a different sncRNA pathway to be discussed in the next subsection. There are 4 Argonaute proteins in human cells but so far only Argonaute 2 (Ago2) has been reported to contain endonuclease activity (Meister 2013). The miRNA acts as a guide to direct the RISC complex to the target mRNA via base complementarity between the miRNA 5′ seed region (nucleotides at positions 2–8) and the mRNA 3′ untranslated region (3′-UTR ) (Bartel 2009; Agarwal et al. 2015). Perfect sequence complimentary between the miRNA and target mRNA may result in target cleavage by the endonucleolytic activity of RISC and significant mRNA destabilization . However, if sequence complimentarity is only partial then RISC binding induces translational inhibition. miRNA regulation of genes was initially known to occur mainly through translational repression , but subsequent observations revealed that miRNAs can also induce mRNA degradation as a result of deadenylation of target mRNAs (Bagga et al. 2005; Krutzfeldt et al. 2005; Valencia-Sanchez et al. 2006). A relatively recent study reported that miRNA mode of regulation may encompass both aspects with initial repression of target mRNAs followed by deadenylation and subsequent degradation (Djuranovic et al. 2012). Moreover, Bartel’s group showed that mRNA destabilization may be a major consequence of mRNA repression by miRNA (Eichhorn et al. 2014).
The canonical miRNA biogenesis pathway. Pri-miRNA are transcribed from viral or cellular genomic DNA by RNA polymerase II. The transcript is processed by Drosha and its cofactor DCGR8 to produce pre-miRNA, which gets exported to the cytoplasm by cellular exportin-5 protein. In the cytoplasm, pre-miRNA is further processed into a miRNA duplex by Dicer and its cofactor TRBP. The mature strand of the miRNA duplex (~21–23 nt) is loaded into the miRISC complex. Depending on the degree by which the miRNA “seed” sequence binds to the target mRNA, the resultant mRNA target can be translationally inhibited, deacetylated and degraded, or sequestered to P-bodies for storage
Despite significant advances in our understanding of miRNA activity, the molecular mechanism by which miRNAs suppresses protein production of targeted mRNAs is not completely understood. It has been proposed that miRNA translocation of the targeted mRNA into cytoplasmic processing bodies (P-bodies) leads to induction of translational inhibition, deadenylation , and degradation of the target (Leung and Sharp 2013). P-bodies lack ribosomes and are reported to regulate mRNA turnover and degradation (Leung and Sharp 2013). They also may participate in miRNA regulation of gene expression based on evidence that miRNA-mRNA complexes and components of the miRISC complex such as DGCR8 and Ago localize to these cytoplasmic foci (Leung and Sharp 2013; Baril et al. 2015; Chen and Shyu 2013; Jakymiw et al. 2007; Nishi et al. 2015). It has been proposed that the translocation of miRNA-bound target mRNA complexes to P-bodies promote their catalytic function or for temporal storage, and that this translocation may be mediated by GW182, a component of P-bodies, through its binding to the Argonaute protein in miRISC. Deadenylases that reside within P-bodies can deadenylate targeted mRNAs, which are then decapped and degraded. P-bodies can also function as a temporary storage compartment where targeted mRNAs are held in stasis, spatially removed from the translational machinery (Nilsen 2007).
1.2 piRNAs
PiRNAs were first identified as small RNAs that specifically interact with Piwi proteins in mouse and rat germ cells (Aravin et al. 2006; Girard et al. 2006; Lau et al. 2006). Subsequent studies revealed an extremely complex population of piRNAs that is highly enriched in the germline tissues of most metazoans examined to date (Lim and Kai 2015). Unlike miRNAs, piRNAs are transcribed by RNA polymerase II from intergenic loci called piRNA clusters as long continuous, single-stranded precursor transcripts which are processed by a Dicer-independent mechanism into ~24–31 nt with 2′–O–methyl modification sites at the 3′ end (Hirakata and Siomi 2015; Vagin et al. 2006) (Fig. 1.2), which may be targets for the murine methylase HENMT1 (Kirino and Mourelatos 2007). PiRNAs constitute the largest class of noncoding RNAs and have the greatest sequence diversity among known classes of cellular RNAs (Moazed 2009; Lim and Kai 2015). PiRNAs predominantly regulate transposon activities within the genome to preserve normal gametogenesis and reproduction because the expression and transposition of these transposable elements pose a high risk of destabilizing genome integrity . Piwi proteins and piRNAs are conserved in a broad range of metazoans. The absence of Piwi resulted in fertility defects in diverse animal species, indicating the Piwi/piRNA pathway has an important role in maintaining fertility (Carmell et al. 2007; Das et al. 2008; Houwing et al. 2007). Notably, piRNAs possess the ability to distinguish between “self” and “non-self” through a complex mechanism that effectively identifies non-self genes for regulation (Malone and Hannon 2009), the details of which won’t be discussed in this chapter.
piRNA biogenesis pathway. piRNA precursors are transcribed by RNA polymerase II from piRNA clusters . These precursors have a 5′ cap and 3′ poly-A tail. The precursor piRNAs are exported to the cytoplasm where they are further processed by an unidentified exonuclease at the 3′ end, which is methylated by the murine homolog HENMTI. The mature piRNA associates with Piwi proteins and gets recruited into the piRISC complex
After assembly, the piRISC complex gets imported into the nucleus where it directs histone 3 lysine 9 (H3K9me3) methylation of target transposon loci to induce a heterochromatin state that transcriptionally silences transposons (Lim and Kai 2015; Le Thomas et al. 2013). In Drosophila , the nuclear protein Asterix/DmGTSF1 is required for piRISC to mediate the addition of this silencing histone marker (Ohtani et al. 2013). The precise mechanisms for how piRISC directs the deposition of H3K9me3 at targeted transposon loci have yet to be elucidated.
2 Current Methods Used for Viral MiRNA Identification
The very nature and function of miRNAs make them an attractive strategy for viruses to use to manipulate their host environments. Due to the limited size of most viral genomes, the low coding capacity needed to encode the small size of miRNAs, coupled with their non-immunogenic characteristics, makes them an attractive tool to incorporate into a virus’ arsenal. Furthermore, a single miRNA has the potential to target numerous host and viral RNAs, which allows a virus to modulate the infection cycle with only limited virus-encoded factors. Viral miRNAs (vmiRNAs) are encoded by many viruses, but the large dsDNA herpesvirus family is the predominant group of viruses that have the most miRNAs characterized within their genomes. The biogenesis of vmiRNAs utilize the same cellular machinery involved in processing cellular miRNAs, and they undergo a similar cascade of steps from the transcription of pri-vmiRNAs transcribed in the nucleus to their subsequent maturation in the cytoplasm. VmiRNAs have been documented to modulate the host environment by targeting either viral or cellular mRNAs to facilitate different facets of the viral lifecycle such as latency .
The most commonly used method to identify vmiRNAs requires the isolation of total small RNAs from infected cells, reverse transcription into cDNA followed by sequencing. Computer algorithms such as TargetScan (Agarwal et al. 2015), miRanda (John et al. 2004; Betel et al. 2008), and RNAhybrid (Rehmsmeier et al. 2004), are also used to predict potential miRNA coding regions which are then verified by direct experimental assays (Bennasser et al. 2004; Pfeffer et al. 2005).
The identification of each miRNA target(s) is not a simple task because a single miRNA can potentially target multiple cellular mRNAs. Bioinformatics computation is used to query for miRNA-seed sequences in the 3′-UTR of potential target mRNAs (Kim and Nam 2006; Rajewsky 2006). Predicting miRNA targets is complicated by the variability in “seed” sequence complementarity between miRNA-mRNA, and that a single miRNA has the potential to regulate the expression of up to 100 discrete mRNAs (Brennecke et al. 2005). Currently, the use of bioinformatics platforms to identify the entire complement of potential mRNA targets (the ‘targetome’) of a given miRNA results in long lists that very likely contain many false positives. Nevertheless, successful identifications of miRNA targets have been reported using this method and advances are continually being made in this area.
Messenger RNA microarrays have also been employed to identify targets of a given miRNA by measuring the change in global gene expression in the presence or absence of the miRNA. Differential expression of specific mRNAs in the presence or absence of a given miRNA suggests it may be a potential target of the miRNA, and bioinformatics tools also help predict a target site for the miRNA in the identified mRNA, providing stronger support that this is likely a real target.
Alternatively, target mRNAs can be recovered and sequenced through methods such as mRNA-protein crosslinking followed by immunoprecipitation (CLIP) with a miRISC component such as AGO2 or from P bodies by immunopurification (Easow et al. 2007). CLIP is a powerful tool for the global recovery of miRISC target sites, but the accurate identification of the compliment miRNA responsible for mediating the recruitment of the mRNA to miRISC remains a challenge. A common assay used to confirm that a miRNA targets an identified mRNA is the use of reporter constructs that contain a chimeric transcription with the 3′-UTR from the target mRNA. When the miRNA is overexpressed, its ability to silence the reporter transcript with the target 3′-UTR would appease one criterion supportive of specific targeting.
3 Herpesviruses
Herpesviruses are a group of DNA viruses whose infectious lifecycle encompasses both lytic and latent cycles. During latency , viral gene expression is limited to a few specialized genes that maintain the latency state. Life-long persistence in hosts infected with herpesvirus is closely associated with the virus’ ability to evade immune detection and establish latency (Feldman and Tibbetts 2015; Frappier 2015). The first virally encoded miRNA identified arose from a cloning experiment in human B cells latently infected with the herpesvirus Epstein-Barr virus (EBV) . This initial discovery spurred the prospect that other herpesviruses or large DNA viruses in general might also encode for vmiRNAs (Pfeffer et al. 2004). Indeed, vmiRNAs were recovered from cells infected with herpes simplex 1 (HSV-1), human cytomegalovirus (HCMV), and Kaposi’s sarcoma herpesvirus (KSHV) (Feldman and Tibbetts 2015; Kincaid and Sullivan 2012; Pfeffer et al. 2005). Interestingly, most herpesvirus vmiRNAs identified to date are expressed during latency, and have been found to regulate both viral and cellular functions to allow the virus to evade immune detection and persist in the infected host. Nevertheless, certain herpesviruses such as HHV-6, HHV-7, and Varacella Zoster virus (VZV) do not appear to encode miRNAs. A group reported the inability to identify viral miRNAs in cells latently infected with VZV, but this does not rule out the possibility that there may be miRNAs produced during VZV lytic infection (Umbach et al. 2009). Nevertheless, this observation is particularly interesting given that Varicelloviruses such as Bovine Herpesvirus 1 and Suid Herpesvirus 1 do encode miRNAs (Anselmo et al. 2011; Glazov et al. 2010). This raises the question as to what is different between viruses that do and do not encode miRNAs, the answer of which will be informative in understanding virus miRNA function. Notably, most vmiRNAs encoded by different herpesviruses are not conserved with each other or with host miRNAs, which suggests herpesvirus vmiRNA genes may undergo rapid evolution . However, Poxviruses , which are DNA viruses that replicate in the cytoplasm, do not appear to encode for vmiRNAs (Skalsky and Cullen 2010).
3.1 Herpes Simplex Virus
The most studied virally encoded miRNAs among the herpesvirus family are encoded by HSV-1, the prototypical alpha herpesvirus (Table 1.1). miR-H1 is a late gene product initially identified from cells lytically infected with HSV-1 and reported to downregulate an ND10 component alpha-thalassemia /mental retardation syndrome X-linked (ATRX) (Jurak et al. 2012). During HSV-1 latent infections in the sensory ganglia, the latency associated transcript (LAT) is expressed. LAT is encoded antisense to the immediate early (IE) gene ICP0 in the long terminal repeat end of the unique long genome segment (Roizman and Whitley 2013). AlteRNAtive splicing gives rise to 3 isoforms of the LAT transcript, all of which show different expression patterns. LAT transcripts have not been observed to translate into any peptides, but studies have reported further processing of LAT in HSV-1 latently infected cells to produce 6 functional miRNAs designated as miR-H2, miR-H3, miR-4, miR-H5, miR-H7, and miR-H8 (Umbach et al. 2009). miR-H2 expression leads to a reduction of ICP0 protein level by translational inhibition, as ICP0 mRNA level is not affected (Umbach et al. 2008). ICP0 is an E3 ubiquitin ligase that allows for a lytic mode of replication at low multiplicity of infection (MOI) (Roizman and Whitley 2013). This protein also facilitates the remodeling of ND10 or PML, which are repressive bodies in the nucleoplasm (Roizman and Whitley 2013). Despite being transcribed antisense to ICP34.5 transcript, miR-H3 and miR-H4 do not appear to effect ICP34.5 levels (Umbach et al. 2008). On the other hand, miR-H6, another HSV-1 encoded miRNA, inhibits translation of the viral transactivator for early viral gene expression, ICP4, via imperfect binding to the ICP4 mRNA. The downregulation of ICP0 and ICP4 by miR-H2 and miR-H6 may inhibit entry into lytic replication and maintenance of an established latent state.
Although there is limited sequence homology between miRNAs expressed by the closely related herpes simplex virus 2 (HSV-2), its miRNAs, also expressed from LAT transcripts, also target ICP0 and ICP34.5 for downregulation, suggesting similar mechanisms for establishing and maintaining latency .
3.2 Human Cytomegalovirus
Human cytomegalovirus (HCMV) has the largest genome of the human herpesvirus at 230 kb and is the prototype of beta herpesviruses . To date, experimental evidence has uncovered 14 HCMV miRNAs from lytically infected primary cells (Table 1.1), 3 of which are transcribed from the antisense strand of known ORFs, 5 miRNAs are located in intergenic regions, and 1 is situated within an intron (Pfeffer et al. 2005).
HCMV miR-UL112-1 was reported to inhibit the transactivation of early gene expression during lytic infection by binding to the 3′ UTR of viral IE1 (Murphy et al. 2008). This suggests that miR-UL112-1 may contribute to the establishment and maintenance of latency, but expression of miR-UL112-1 during latent infection remains unknown (Murphy et al. 2008). miR-UL112-1 has recently been implicated in downregulating IL-32, which is critical for both innate and adaptive immune responses (Huang et al. 2013). miR-UL112-1 also has been shown to target the cellular major histocompatibility complex class-I-related chain B (MICB), a cell-surface protein recognized by natural killer (NK) cells, resulting in a decline in MICB protein levels (Stern-Ginossar et al. 2007). Additionally, miR-US25-2-3p was shown to downregulate tissue inhibitors of metalloprotease 3 (TIMP3) expression resulting in an increased shedding of soluble major histocompatibility complex class-I-related chain A (MICA) in patient serum (Esteso et al. 2014). The targeting of MICB by miR-UL112-1 and TIMP3 by miR-US25-2-3p most likely prevents NK cells from killing HCMV-infected cells. Another HCMV-encoded miRNA that targets NK cell activity is miR-UL148, which was determined to bind to the 3’ UTR of RANTES, a cellular protein that induces proliferation and activation of NK cells (Kim et al. 2012).
HCMV miR-US4-1 was also shown to indirectly inhibit cytotoxic T lymphocyte (CTL) response by targeting ERAP1 transcripts (Kim et al. 2012). ERAP1 is essential in antigenic peptide production in the ER and mediates the stability of MHC class I-β2-microglobulin-peptide heterotrimer. Thus, miR-US4-1 targetting of ERAP1 would have profound affects on antigen presentation and the resultant CTL response.
HCMV miRNA activity also affects other signalling cascades such as NF-κB signalling. miR-UL112-3p inhibits NF-κB signalling by targeting Toll-like receptor 2 (TLR2), a major pathogen recognition receptor (PRR) of NF-κB signalling (Landais et al. 2015). There is also miR-US25-1, which has been implicated in the downregulation of cell cycle control protein cyclin E2 (Grey et al. 2010). Thus, HCMV encodes for several miRNAs which have been shown to target both viral and cellular transcripts that affect a number of key signalling pathways.
3.3 Epstein–Barr Virus
Epstein-Barr virus (EBV) is a gamma herpesvirus that chronically persists in human B-lymphocytes after primary infection (Young and Rickinson 2004). In vitro studies have shown that EBV is capable of transforming normal human B-cells into malignant cells, and infection with EBV is associated with malignant diseases such as Hodgkin’s lymphoma, endemic Burkitt’s lymphoma, and nasopharyngeal carcinoma (Young and Rickinson 2004). During latency , EBV expresses latency-associated genes that include latency-associated membrane proteins (LMP) and Epstein–Barr virus -associated nuclear antigens (EBNAs) (Young and Rickinson 2004). Two regions within the EBV genome , the Bam HI fragment H rightward open reading frame 1 (BHRF1) gene and in the Bam HI-A-region rightward transcript (BART) gene, encode for viral miRNAs (Table 1.1). miR-BHRF1-3 activity may be associated with host immune evasion by downregulating a T-cell attractant CXCL-11 (Xia et al. 2008). miR-BART2 targets BALF5 transcripts resulting in a reduction of BALF5 viral DNA polymerase , which may serve to stabilize latency in EBV by suppressing lytic cycle viral replication (Barth et al. 2008). The late membrane protein (LMP1) modulates NF-κB signalling and contributes to EBV-mediated transformation. However, several BART miRNAs target the 3’ UTR of EBV LMP1 transcripts, resulting in a reduction of late membrane protein (LMP1) levels thereby affecting the protein’s activity in viral-mediated transformation and NF-kB signalling. miR-BART5 has been shown to target apoptosis signalling by downregulating pro-apoptotic protein PUMA (Choy et al. 2008). In contrast, a relatively recent study reported EBV miR-BART15-3p promotes apoptosis by targeting anti-apoptotic protein BRUCE (Choi et al. 2013). The targeting of PUMA implies that miR-BART5 inhibits apoptosis to promote infected cell survival and persistent viral progeny production. However, it remains to be seen what role miR-BART15-3p plays in EBV lifecycle by promoting apoptosis.
3.4 Kaposi’s Sarcoma-Associated Herpesvirus
The oncogenic Kaposi’s sarcoma-associated herpesvirus (KSHV or HHV8) is also a gamma herpesvirus associated with the development of several human malignancies including Kaposi’s sarcoma, primary effusion lymphoma (PEL), and Castleman’s disease (Ensser and Fleckenstein 2005). KSHV expresses 12 viral miRNAs in latently infected cells from a 5-kb latency-associated region of the viral genome (Table 1.1). Cellular mRNA BCLAF1 is believed to be involved in KSHV latency, since its downregulation by miR-K5, miR-K9, and mir-K10 resulted in significant reduction in progeny virus recovery after exit from latency (Ziegelbauer et al. 2009). Similarly, miR-K9 targets an important lytic switch protein RTA, which suggests this miRNA may have a role in regulating KSHV latency (Bellare and Ganem 2009). KSHV miR-K1, miR-K3-3p, and miR-K6-3p may be involved in host immune response evasion by targeting thrombospondin-1 (THBS1), which acts as a chemoattractant to recruit monocytes and T cells to sites of infection (Narizhneva et al. 2005). miR-K1 inhibits cell arrest by downregulating the cyclin-dependent kinase inhibitor p21 in B cells latently infected with KSHV (Gottwein and Cullen 2010).
4 Polyomavirus , Adenovirus, and Papillomavirus
Polyomaviruses are small dsDNA viruses that can establish persistent infections as well as immortalize infected cells (White et al. 2013). Current human diseases caused by polyomaviruses are limited to 4 virus strains: BK polyomavirus, which causes kidney and urinary tract diseases; JC polyomavirus , which causes progressive multifocal leukoencephalopathy in immunocompromised individuals; Merkel cell polyomavirus, which causes Merkel cell carcinoma ; and trichodysplasia spinulosa-associated polyomavirus, which causes a rare condition of its namesake (White et al. 2013). miRNAs encoded by polyomaviruses were first identified in SV40, a well-studied monkey polyomavirus (Alwine and Khoury 1980; Sullivan et al. 2005). Since then, many polyomaviruses are reported to encode for miRNAs including medically significant BK, JC and Merkel cell polyomaviruses (Seo et al. 2008, 2009). SV40 miRNA appears to be involved in immune evasion. SV40 miRNAs target the viral large T-antigen transcript, leading to cleavage of the transcription and a reduction in both transcript and protein levels (Sullivan et al. 2005). Since the large T-antigen is a target for CTL, miRNA-mediated reduction of large T-antigen levels may allow infected cells to escape immune detection.
Adenoviruses are also small dsDNA viruses that encode for 2 noncoding virus associated RNA (VAI and VAII). VAI confers resistance to cellular interferon-related defenses and contributes to viral replication . Both VAI and VAII have been reported to be processed by Dicer to yield miRNAs that are loaded onto miRISC (Aparicio et al. 2006, 2010; Xu et al. 2007; Sano et al. 2006). However, the functions of these miRNAs remain to be determined.
Initial pursuit to discover miRNAs in papillomaviruses (HPV-31), another DNA virus family, did not yield any positive identifications, suggesting this group of virus may not encode for any miRNAs (Cai et al. 2006). However, most recently, a study reported the identification of viral miRNAs encoded by HPV-16, HPV-38, and HPV-68 using advance sequencing technology SOLiD 4 (Qian et al. 2013). It remains to be seen how these miRNAs affect HPV infection and whether other groups may discover more miRNAs in other HPV genotypes.
5 RNA Viruses and Retroviruses
Contrary to DNA viruses, encryption of functional miRNAs in RNA virus genomes remains controversial. Functional miRNAs have not been detected from studies with RNA viruses such as hepatitis C virus (HCV) , yellow fever virus, and human immunodeficiency virus (HIV-1 ) (Swaminathan et al. 2013). The absence of miRNAs in RNA virus genomes may be due to the fact that most RNA viruses replicate in the cytoplasm, which seclude them from nuclear microprocessing machineries such as Drosha and TRBP. However, even with nuclear replicating RNA viruses such as influenza virus, no functional miRNAs have been identified so far (Umbach et al. 2010; Tycowski et al. 2015; Perez et al. 2010, 2012). It is quite possible that inclusion of non-coding miRNA regions within an RNA virus genome may lead to degradation of the entire viral genome by RISC-mediated mechanisms. Therefore, it has been generally accepted that RNA viruses do not encode vmiRNAs.
However, retroviruses, such as HIV-1, the causative agent of the acquired immunodeficiency syndrome (AIDS), are suspected to potentially encode for vmiRNAs due to the way they replicate within a host cell. HIV-1 viral RNA is reverse transcribed into double-stranded DNA that gets transported into the nucleus where it integrates into the host genome. Thus, HIV-1 should have access to RNAi machinery components present in both the nucleus and cytoplasm, similar to host miRNAs. As such, computational analysis predicts HIV-1 is capable of encoding several miRNA precursors (Bennasser et al. 2004, 2006). The vmiRNA Nef-U3-miR-N367, encoded within nef, was reported to target nef transcripts that resulted in its degradation and reduction in Nef protein levels, which led to a significant decline in viral replication through various mechanisms (Omoto et al. 2004). The inhibition of HIV-1 replication by Nef-U3-miR-N367 suggests this vmiRNA may play a crucial role in establishing persistent HIV-1 infection. Several other studies have reported the discovery of other vimiRNA encoded within the HIV-1 genome . Kaul et al. reported the identification of a pre-miRNA sequence in the 3′-end of the viral genome called hiv1-miR-H1. They observed this vmiRNA specifically targets a transcription factor that plays an important role negatively regulating cellular apoptosis (Kaul et al. 2009). The authors further noted hiv1-miR-H1 also suppresses other cellular proteins such as c-myc, Par-4, Bcl-2 and Dicer as well as downregulates cellular miR-149, which targets the HIV-1 Vpr protein (Kaul et al. 2009). The authors propose that this vmiRNA activates apoptosis in mononuclear cells by initiating an epigenomic pathway. Interestingly, the Env and Gag-Pol coding regions of different HIV-1 strains are reported to contain miRNA-like sequences with a surprisingly high similarity to human miR-196, miR-30d, miR-30e, miR-374a, and miR-424 (Holland et al. 2013). Additionally, the HIV-1 transactivation RNA (TAR) element has been found to undergo asymmetrical processing by Dicer to produce TAR-miR-5p and -3p (Ouellet et al. 2008). Recent studies using the extremely sensitive SOLiDTM 3 Plus System and a novel, sequence-targeted enrichment strategy identified several hundred small non-coding RNAs (sncRNAs) in infected T lymphocytes and macrophages ranging between 16 and 89 nt in length (Althaus et al. 2012; Schopman et al. 2012). However, controversy remains over whether HIV-1 truly encodes functional vmiRNAs given that some labs failed to detect the expression of any vmiRNA in HIV-1 infected cells (Lin and Cullen 2007; Whisnant et al. 2013) in addition to the lack of experimental demonstrations on the physiological relevance of these reported vmiRNAs.
6 Activities of Cellular MiRNAs and PiRNAs in Viral Infections
6.1 Influence of Cellular MiRNAs and PiRNAs on Virus Replication in Mammalian Host
The possibility that cellular miRNAs may modulate virus replication has been the subject of intense investigation. Relatively recent experimental evidence provides support that mammalian sncRNAs regulate virus replication but whether this is physiologically relevant in vivo is still debatable (Yeung et al. 2007). Nevertheless, the impact of cellular miRNA on viral replication was first hinted at when the knockdown of major miRNA-processing enzymes Drosha and Dicer led to more robust replication of viruses such as influenza, vesicular stomatitis virus (VSV), and HIV-1 as a result of reduced mature miRNAs levels in mammalian cells (Matskevich and Moelling 2007; Otsuka et al. 2007; Triboulet et al. 2007). Moreover, the overexpression of virus-encoded RNAi suppressors resulted in a 5- to 10 fold increase in virus replication (de Vries et al. 2008). Antagomirs, chemically modified antisense-oligoribonucleotides, are the current stable method used to inactivate individual cellular miRNAs to assess their functional roles in targeting viruses. (Bennasser et al. 2005; Krutzfeldt et al. 2005). These observations suggest mammalian miRNA/RNAi functionally regulates viral replication.
Studies on miR-122 in HCV replication provide the only current evidence for direct regulation of virus replication by cellular miRNA (Jopling et al. 2005). HCV establishes persistent infections in the liver that may lead to liver cirrhosis and hepatocellular carcinoma . This cellular miRNA is highly expressed in the liver where it has been shown to positively regulate HCV replication in Huh7 human liver cells (Jopling et al. 2005) by directly interacting with two adjacent sites in the 5′ UTR of the viral RNA (Jopling et al. 2005). The mechanism that underlies miR-122`s affect on HCV replication remains unknown but recent studies begin to shed light on this process. miR-122 binding to sites located upstream of the HCV IRES, which directs translation of the positive-sense viral RNA genome , resulted in moderate upregulation of viral protein translation (Henke et al. 2008). However, another study has shown that miR-122 may act at another, yet undefined, stage of viral replication (Jangra et al. 2010).
Many cellular miRNAs have been found to indirectly affect HIV-1 replication by targeting factors known to be important to HIV infection called HIV dependency factors (HDF). Evidence for the anti-viral nature of cellular miRNA in HIV-1 infection comes from a study carried out by Triboulet et al. where they knocked down Drosha and Dicer , two important miRNA processing proteins, and found viral replication was greatly enhanced in peripheral blood mononuclear cells (PBMCs) from HIV-1-infected patients and in latently infected cells (Triboulet et al. 2007). The same group further showed that viral mRNA co-localized with components of the miRISC complex. Interestingly, they reported virus reactivation in PBMCs isolated from patients undergoing cART when they knocked down cellular factors involved in miRNA-mediated silencing. These factors were also shown to inhibit viral gene expression by interfering with viral mRNA association with polysomes (Chable-Bessia et al. 2009). Also, a cluster of human miRNAs that include miR-28, miR-125b, miR-150, miR-223 and miR-382 was shown to target the 3′ end of HIV-1 mRNAs for repression (Huang et al. 2007). These observations highlight the important role miRNAs have in regulating HIV-1 infection.
Knockdown of Dicer greatly enhances influenza A virus (IAV) replication, suggesting cellular RNAi also has antiviral effects on influenza A virus replication (Matskevich and Moelling 2007). Song et al. used a 3′ UTR reporter construct to show that host miR-323, miR-491, and miR-654 inhibited influenza strain A/WSN/33 H1N1 replication through the binding of the PB1 transcript, resulting in degradation of the PB1 mRNA (Song et al. 2010). PB1 forms part of the IAV viral polymerase complex necessary for viral genome transcription and replication (Kobayashi et al. 1996). Several studies have reported modulation of host miRNA expression profiles upon viral infection, where infection of specific IAV subtypes resulted in different expression profiles of cellular miRNAs (Loveday et al. 2012; Tambyah et al. 2013; Li et al. 2010). This raises the question of whether the differing pathogenicities exhibited by different IAV subtypes may be associated with the differential expressions of specific cellular miRNAs. One recent study suggests that miR-24 may modulate the highly pathogenic H5N1 virus by downregulating the furin secretory pathway. This pathway is exploited by the virus to proteolytically cleave its haemagglutinin precursor to active forms that allow viral entry in host cells via viral-host membrane fusion (Loveday et al. 2015). Surprisingly, miR-24 was reported to have little effect on low-pathogenic 2009 pandemic H1N1 infection, suggesting its activity may be strain-specific.
The cellular RNAi arsenal is not limited to miRNAs but include short interfering RNAs (siRNAs) and a separate class of sncRNAs called piRNAs . In the nematode model organism Caenorhabditis elegans, RNAi has been demonstrated to be the main antiviral response to Flock House virus and the natural pathogen Orsay virus. This antiviral response involves the PIWI family of genes such as rde-1, which encodes an Argonaute protein, and rde-4, which encodes a dsRNA-binding protein (Felix et al. 2011). While there are no homologues of these proteins in mammalian cells, there are evidence that piRNAs or repeat-associated small interfering RNAs (rasiRNAs) functionally suppress mammalian endogenous retroviruses in germ and somatic cells (Carmell et al. 2007; Watanabe et al. 2006). It has also been shown that mouse ES cells, and possibly a subset of somatic stem cells, retain the ability to generate small RNA species from exogenous dsRNA and viral RNA, which are also implicated in repressing retrotransposon activities (Calabrese et al. 2007). However, the importance of these small RNA species as antiviral agents against human viral infections remains to be fully addressed.
6.2 Interferon-Mediated Antiviral Activity Via MiRNAs
The interferon (IFN) signalling pathway is an important part of cellular antiviral immunity. This pathway is activated upon the production of long dsRNAs within a cell. Interestingly, there is evidence to suggest the IFN pathway partially overlaps with the RNAi pathway. It has been noted that proteins such as PACT and TRBP, which function in the IFN antiviral network, also has a role in miRNA processing and function (Chendrimada et al. 2005; Haase et al. 2005; Kok et al. 2007; Lee et al. 2006). Additionally, RNA-binding proteins are capable of suppressing both the IFN effector protein kinase R (PKR) (Cai et al. 2000; Li et al. 2004; McMillan et al. 1995) and RNAi (Bennasser et al. 2005; Haasnoot et al. 2007). Most notably was the report that interferon beta (IFN-β) can regulate the expression of several cellular miRNAs, 8 of which are predicted to have binding sites within the HCV genomic RNA. In support of this, the introduction of synthetic miRNA-mimics resembling these IFN-β-induced miRNAs recapitulated the antiviral effects of IFN-β on HCV infection. Additionally, IFN-β’s antiviral effects against HCV were attenuated when antagomirs to these IFN-β-induced miRNAs were introduced into the infected cells (Pedersen et al. 2007). This evidence support the potential overlap of miRNAs and the IFN network in mammalian cells and possible IFN-dependent and IFN-independent defenses to viral infection. There also is evidence in a number of viruses that show vmiRNAs modulate the innate immune system by directly targeting the downregulation of immune factors, which affect processes such as the recruitment of effector cells of the immune system (Cullen 2013).
6.3 Viral Counter-Responses to Cellular RNAi Restriction
Viruses appear capable of countering the cell’s RNAi restriction by actively suppressing RNAi activation through viral dsRNA-binding proteins that sequester and neutralize the antiviral RNAi activities. An example of this may be influenza A virus’s NS1 protein which has been suggested to suppress host RNAi activity possibly through its dsRNA-binding capability (Haasnoot et al. 2007). HIV-1 counteracts this host defense by developing strategies to modulate cellular miRNA expression and interfere with the overall biogenesis of miRNAs. Viral protein Tat inhibits Dicer activity through its physical interaction with the helicase domain of Dicer and its binding to host mRNAs. However, the authors did not determine whether this was a direct protein-protein interaction (Chable-Bessia et al. 2009; Bennasser et al. 2005). Contradicting observations have been made in whether the SRS/RSS function of Tat alters miRNA expression profiles. Some have reported this function of Tat does result in altered miRNA expression in T cell lines and primary PBMCs, whereas others have shown that it does not affect miRNA expression in PBMCs (Mishra et al. 2012). AlteRNAtively, HCV evolved to adapt to miRNA-restriction in a way that benefitted the virus’ replication, for example the enhancement of HCV replication by liver-specific miR-122 (Jopling et al. 2005).
7 Concluding Remarks and Future Perspectives
Given the relatively recent discovery of sncRNAs , tremendous advances have been made in the discovery and characterization of these molecules and their effects on viral replication. However, the more we discover the clearer it becomes that viruses of diverse origin have evolved to exploit the host miRNA pathway in vast numbers of ways to effectively regulate infection and the host response. Nevertheless, several prominent issues remain to be addressed. The physiological importance of viral miRNAs in many groups of viruses aside from herpesviruses and polyomaviruses is still unclear. Likewise, it is very likely that the present list of targets identified for herpesvirus miRNAs represent only a partial number of potential targets; that many more targets remain to be discovered. Notably, the relationship between herpesvirus miRNA expression and viral latency and immune evasion remains uncertain. Although it is known that miRNAs from different herpesviruses target common pathways, it remains unclear whether these miRNAs also share a common function.
Another major area to expand future research is cellular miRNA regulation of viral infection. It appears that a broad spectrum of viruses employ complex mechanisms to regulate cellular miRNA expression to mold the host environment into something more permissible and advantageous for viral replication. The mechanistic details specific viruses use to modulate cellular miRNA expressions are largely unknown. Viral proteins may directly modulate host miRNA expression or indirectly regulate it due to a general host response to the infection. There have been several reports of direct cellular miRNA binding to viral RNA and negatively regulating the virus but most of these studies were performed in tissue culture or in organisms other than the natural host, which brings into question the physiological relevance of these interactions. It would be rational to imagine that evolutionary pressure would preclude viruses retaining cellular miRNA target sites that negatively regulate its replication, but if such sites were beneficial to the pathogen, then it seems possible the viruses might want to retain them in their genomes.
Although tremendous advances have been made to understand the relationship between miRNAs and viral replication, much remains to be discovered. This poses exciting potentials in terms of scientific discovery as well as prospects for novel therapeutic strategies to control viral infections.
References
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4. doi:10.7554/eLife.05005
Althaus CF, Vongrad V, Niederost B, Joos B, Di Giallonardo F, Rieder P, Pavlovic J, Trkola A, Gunthard HF, Metzner KJ, Fischer M (2012) Tailored enrichment strategy detects low abundant small noncoding RNAs in HIV-1 infected cells. Retrovirology 9:27. doi:10.1186/1742-4690-9-27
Alwine JC, Khoury G (1980) Simian virus 40-associated small RNA: mapping on the simian virus 40 genome and characterization of its synthesis. J Virol 36(3):701–708
Anselmo A, Flori L, Jaffrezic F, Rutigliano T, Cecere M, Cortes-Perez N, Lefevre F, Rogel-Gaillard C, Giuffra E (2011) Co-expression of host and viral microRNAs in porcine dendritic cells infected by the pseudorabies virus. PLoS ONE 6(3):e17374. doi:10.1371/journal.pone.0017374
Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P (2006) Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol 80(3):1376–1384. doi:10.1128/jvi.80.3.1376-1384.2006
Aparicio O, Carnero E, Abad X, Razquin N, Guruceaga E, Segura V, Fortes P (2010) Adenovirus VA RNA-derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Res 38(3):750–763. doi:10.1093/nar/gkp1028
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442(7099):203–207. doi:10.1038/nature04916
Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122(4):553–563. doi:10.1016/j.cell.2005.07.031
Baril P, Ezzine S, Pichon C (2015) Monitoring the spatiotemporal activities of miRNAs in small animal models using molecular imaging modalities. Int J Mol Sci 16(3):4947–4972. doi:10.3390/ijms16034947
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. doi:10.1016/j.cell.2009.01.002
Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA (2008) Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res 36(2):666–675. doi:10.1093/nar/gkm1080
Bellare P, Ganem D (2009) Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6(6):570–575. doi:10.1016/j.chom.2009.11.008
Bennasser Y, Le SY, Yeung ML, Jeang KT (2004) HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology 1:43. doi:10.1186/1742-4690-1-43
Bennasser Y, Le SY, Benkirane M, Jeang KT (2005) Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22(5):607–619. doi:10.1016/j.immuni.2005.03.010
Bennasser Y, Le SY, Yeung ML, Jeang KT (2006) MicroRNAs in human immunodeficiency virus-1 infection. Meth Mol Biol 342:241–253. doi:10.1385/1-59745-123-1:241
Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36 (Database issue):D149–153. doi:10.1093/nar/gkm995
Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3(3):e85. doi:10.1371/journal.pbio.0030085
Cai R, Carpick B, Chun RF, Jeang KT, Williams BR (2000) HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch Biochem Biophys 373(2):361–367. doi:10.1006/abbi.1999.1583
Cai X, Li G, Laimins LA, Cullen BR (2006) Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J Virol 80(21):10890–10893. doi:10.1128/jvi.01175-06
Calabrese JM, Seila AC, Yeo GW, Sharp PA (2007) RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci USA 104(46):18097–18102. doi:10.1073/pnas.0709193104
Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16(21):2733–2742. doi:10.1101/gad.1026102
Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12(4):503–514. doi:10.1016/j.devcel.2007.03.001
Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, Wagschal A, Jacquet JM, Reynes J, Levy Y, Saib A, Bennasser Y, Benkirane M (2009) Suppression of HIV-1 replication by microRNA effectors. Retrovirology 6:26. doi:10.1186/1742-4690-6-26
Chen CY, Shyu AB (2013) Deadenylation and P-bodies. Adv Exp Med Biol 768:183–195. doi:10.1007/978-1-4614-5107-5_11
Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436(7051):740–744. doi:10.1038/nature03868
Choi H, Lee H, Kim SR, Gho YS, Lee SK (2013) Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J Virol 87(14):8135–8144. doi:10.1128/jvi.03159-12
Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY (2008) An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 205(11):2551–2560. doi:10.1084/jem.20072581
Cullen BR (2013) MicroRNAs as mediators of viral evasion of the immune system. Nature Immunol 14(3):205–210. doi:10.1038/ni.2537
Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, Matthews N, Berezikov E, Ketting RF, Tavare S, Miska EA (2008) Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31(1):79–90. doi:10.1016/j.molcel.2008.06.003
de Vries W, Haasnoot J, van der Velden J, van Montfort T, Zorgdrager F, Paxton W, Cornelissen M, van Kuppeveld F, de Haan P, Berkhout B (2008) Increased virus replication in mammalian cells by blocking intracellular innate defense responses. Gene Therap 15(7):545–552. doi:10.1038/gt.2008.12
Djuranovic S, Nahvi A, Green R (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336(6078):237–240. doi:10.1126/science.1215691
Easow G, Teleman AA, Cohen SM (2007) Isolation of microRNA targets by miRNP immunopurification. RNA 13(8):1198–1204. doi:10.1261/rna.563707
Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villen J, Bartel DP (2014) mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell 56(1):104–115. doi:10.1016/j.molcel.2014.08.028
Ensser A, Fleckenstein B (2005) T-cell transformation and oncogenesis by gamma2-herpesviruses. Adv Cancer Res 93:91–128. doi:10.1016/s0065-230x(05)93003-0
Erson-Bensan AE (2014) Introduction to microRNAs in biological systems. Meth Mol Biol 1107:1–14. doi:10.1007/978-1-62703-748-8_1
Esteso G, Luzon E, Sarmiento E, Gomez-Caro R, Steinle A, Murphy G, Carbone J, Vales-Gomez M, Reyburn HT (2014) Altered microRNA expression after infection with human cytomegalovirus leads to TIMP3 downregulation and increased shedding of metalloprotease substrates, including MICA. J Immunol 193(3):1344–1352. doi:10.4049/jimmunol.1303441
Feldman ER, Tibbetts SA (2015) Emerging roles of herpesvirus microRNAs during in vivo infection and pathogenesis. Curr Pathobiol Rep 3(3):209–217. doi:10.1007/s40139-015-0085-z
Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, Sanroman M, Miska EA, Wang D (2011) Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol 9(1):e1000586. doi:10.1371/journal.pbio.1000586
Frappier L (2015) Regulation of herpesvirus reactivation by host microRNAs. J Virol 89(5):2456–2458. doi:10.1128/jvi.03413-14
Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442(7099):199–202. doi:10.1038/nature04917
Glazov EA, Horwood PF, Assavalapsakul W, Kongsuwan K, Mitchell RW, Mitter N, Mahony TJ (2010) Characterization of microRNAs encoded by the bovine herpesvirus 1 genome. J Gen Virol 91(Pt 1):32–41. doi:10.1099/vir.0.014290-0
Gottwein E, Cullen BR (2010) A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol 84(10):5229–5237. doi:10.1128/jvi.00202-10
Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA (2010) A viral microRNA down-regulates multiple cell cycle genes through mRNA 5’UTRs. PLoS Pathog 6(6):e1000967. doi:10.1371/journal.ppat.1000967
Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W (2005) TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6(10):961–967. doi:10.1038/sj.embor.7400509
Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B (2007) The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 3(6):e86. doi:10.1371/journal.ppat.0030086
Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M (2008) microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27(24):3300–3310. doi:10.1038/emboj.2008.244
Hirakata S, Siomi MC (2015) piRNA biogenesis in the germline: from transcription of piRNA genomic sources to piRNA maturation. Biochim Biophys Acta. doi:10.1016/j.bbagrm.2015.09.002
Holland B, Wong J, Li M, Rasheed S (2013) Identification of human microRNA-like sequences embedded within the protein-encoding genes of the human immunodeficiency virus. PLoS ONE 8(3):e58586. doi:10.1371/journal.pone.0058586
Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129(1):69–82. doi:10.1016/j.cell.2007.03.026
Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H (2007) Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+T lymphocytes. Nature Med 13(10):1241–1247. doi:10.1038/nm1639
Huang Y, Qi Y, Ma Y, He R, Ji Y, Sun Z, Ruan Q (2013) The expression of interleukin-32 is activated by human cytomegalovirus infection and down regulated by hcmv-miR-UL112-1. Virol J 10:51. doi:10.1186/1743-422x-10-51
Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, Satoh M, Fritzler MJ, Chan EK (2007) The role of GW/P-bodies in RNA processing and silencing. J Cell Sci 120(Pt 8):1317–1323. doi:10.1242/jcs.03429
Jangra RK, Yi M, Lemon SM (2010) Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol 84(13):6615–6625. doi:10.1128/jvi.00417-10
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human MicroRNA targets. PLoS Biol 2(11):e363. doi:10.1371/journal.pbio.0020363
Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309(5740):1577–1581. doi:10.1126/science.1113329
Jurak I, Silverstein LB, Sharma M, Coen DM (2012) Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity. J Virol 86(18):10093–10102. doi:10.1128/jvi.00930-12
Kaul D, Ahlawat A, Gupta SD (2009) HIV-1 genome-encoded hiv1-mir-H1 impairs cellular responses to infection. Mol Cell Biochem 323(1–2):143–148. doi:10.1007/s11010-008-9973-4
Kim VN, Nam JW (2006) Genomics of microRNA. Trends Genet: TIG 22(3):165–173. doi:10.1016/j.tig.2006.01.003
Kim Y, Lee S, Kim S, Kim D, Ahn JH, Ahn K (2012) Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog 8(3):e1002577. doi:10.1371/journal.ppat.1002577
Kincaid RP, Sullivan CS (2012) Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog 8(12):e1003018. doi:10.1371/journal.ppat.1003018
Kirino Y, Mourelatos Z (2007) The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13(9):1397–1401. doi:10.1261/rna.659307
Kobayashi M, Toyoda T, Ishihama A (1996) Influenza virus PB1 protein is the minimal and essential subunit of RNA polymerase. Arch Virol 141(3–4):525–539
Kok KH, Ng MH, Ching YP, Jin DY (2007) Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem 282(24):17649–17657. doi:10.1074/jbc.M611768200
Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438(7068):685–689. doi:10.1038/nature04303
Landais I, Pelton C, Streblow D, DeFilippis V, McWeeney S, Nelson JA (2015) Human cytomegalovirus miR-UL112-3p targets TLR2 and modulates the TLR2/IRAK1/NFkappaB signaling pathway. PLoS Pathog 11(5):e1004881. doi:10.1371/journal.ppat.1004881
Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE (2006) Characterization of the piRNA complex from rat testes. Science 313(5785):363–367. doi:10.1126/science.1130164
Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF (2013) Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27(4):390–399. doi:10.1101/gad.209841.112
Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN (2006) The role of PACT in the RNA silencing pathway. EMBO J 25(3):522–532. doi:10.1038/sj.emboj.7600942
Leung AK, Sharp PA (2013) Quantifying Argonaute proteins in and out of GW/P-bodies: implications in microRNA activities. Adv Exp Med Biol 768:165–182. doi:10.1007/978-1-4614-5107-5_10
Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding SW (2004) Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA 101(5):1350–1355. doi:10.1073/pnas.0308308100
Li Y, Chan EY, Li J, Ni C, Peng X, Rosenzweig E, Tumpey TM, Katze MG (2010) MicroRNA expression and virulence in pandemic influenza virus-infected mice. J Virol 84(6):3023–3032. doi:10.1128/jvi.02203-09
Lim RS, Kai T (2015) A piece of the pi(e): The diverse roles of animal piRNAs and their PIWI partners. Sem Cell Dev Biol. doi:10.1016/j.semcdb.2015.10.025
Lin J, Cullen BR (2007) Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol 81(22):12218–12226. doi:10.1128/jvi.01390-07
Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD (2007) Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA 104 (41):16164–16169. doi:10.1073/pnas.0702896104
Loveday EK, Svinti V, Diederich S, Pasick J, Jean F (2012) Temporal- and strain-specific host microRNA molecular signatures associated with swine-origin H1N1 and avian-origin H7N7 influenza A virus infection. J Virol 86(11):6109–6122. doi:10.1128/jvi.06892-11
Loveday EK, Diederich S, Pasick J, Jean F (2015) Human microRNA-24 modulates highly pathogenic avian-origin H5N1 influenza A virus infection in A549 cells by targeting secretory pathway furin. J Gen Virol 96(Pt 1):30–39. doi:10.1099/vir.0.068585-0
Malone CD, Hannon GJ (2009) Small RNAs as guardians of the genome. Cell 136(4):656–668. doi:10.1016/j.cell.2009.01.045
Matskevich AA, Moelling K (2007) Dicer is involved in protection against influenza A virus infection. J Gen Virol 88(Pt 10):2627–2635. doi:10.1099/vir.0.83103-0
McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE, Mak TW, Hovanessian AG, Jeang KT, Williams BR (1995) HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213(2):413–424. doi:10.1006/viro.1995.0014
Meister G (2013) Argonaute proteins: functional insights and emerging roles. Nature Rev Genet 14(7):447–459. doi:10.1038/nrg3462
Mishra R, Chhatbar C, Singh SK (2012) HIV-1 Tat C-mediated regulation of tumor necrosis factor receptor-associated factor-3 by microRNA 32 in human microglia. J Neuroinflam 9:131. doi:10.1186/1742-2094-9-131
Moazed D (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457(7228):413–420. doi:10.1038/nature07756
Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ (2008) Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci USA 105(14):5453–5458. doi:10.1073/pnas.0711910105
Narizhneva NV, Razorenova OV, Podrez EA, Chen J, Chandrasekharan UM, DiCorleto PE, Plow EF, Topol EJ, Byzova TV (2005) Thrombospondin-1 up-regulates expression of cell adhesion molecules and promotes monocyte binding to endothelium. FASEB J 19(9):1158–1160. doi:10.1096/fj.04-3310fje
Nilsen TW (2007) Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet 23(5):243–249. doi:10.1016/j.tig.2007.02.011
Nishi K, Takahashi T, Suzawa M, Miyakawa T, Nagasawa T, Ming Y, Tanokura M, Ui-Tei K (2015) Control of the localization and function of a miRNA silencing component TNRC6A by Argonaute protein. Nucleic Acids Res 43(20):9856–9873. doi:10.1093/nar/gkv1026
Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K (2013) DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev 27(15):1656–1661. doi:10.1101/gad.221515.113
Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR (2004) HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1:44. doi:10.1186/1742-4690-1-44
Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, Das SC, Pattnaik AK, Beutler B, Han J (2007) Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27(1):123–134. doi:10.1016/j.immuni.2007.05.014
Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P (2008) Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res 36(7):2353–2365. doi:10.1093/nar/gkn076
Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M (2007) Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449(7164):919–922. doi:10.1038/nature06205
Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, tenOever BR (2010) Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci USA 107(25):11525–11530. doi:10.1073/pnas.1001984107
Perez JT, Zlatev I, Aggarwal S, Subramanian S, Sachidanandam R, Kim B, Manoharan M, tenOever BR (2012) A small-RNA enhancer of viral polymerase activity. J Virol 86(24):13475–13485. doi:10.1128/jvi.02295-12
Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T (2004) Identification of virus-encoded microRNAs. Science 304(5671):734–736. doi:10.1126/science.1096781
Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T (2005) Identification of microRNAs of the herpesvirus family. Nat Methods 2(4):269–276. doi:10.1038/nmeth746
Qian K, Pietila T, Ronty M, Michon F, Frilander MJ, Ritari J, Tarkkanen J, Paulin L, Auvinen P, Auvinen E (2013) Identification and validation of human papillomavirus encoded microRNAs. PLoS ONE 8(7):e70202. doi:10.1371/journal.pone.0070202
Rajewsky N (2006) microRNA target predictions in animals. Nature Genet 38(Suppl):S8–13. doi:10.1038/ng1798
Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10(10):1507–1517. doi:10.1261/rna.5248604
Roizman B, Whitley RJ (2013) An inquiry into the molecular basis of HSV latency and reactivation. Ann Rev Microbiol 67:355–374. doi:10.1146/annurev-micro-092412-155654
Sano M, Kato Y, Taira K (2006) Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett 580(6):1553–1564. doi:10.1016/j.febslet.2006.01.085
Schopman NC, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, Berkhout B, Haasnoot J (2012) Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res 40(1):414–427. doi:10.1093/nar/gkr719
Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS (2008) Evolutionarily conserved function of a viral microRNA. J Virol 82(20):9823–9828. doi:10.1128/jvi.01144-08
Seo GJ, Chen CJ, Sullivan CS (2009) Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology 383(2):183–187. doi:10.1016/j.virol.2008.11.001
Shukla GC, Singh J, Barik S (2011) MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol 3(3):83–92
Skalsky RL, Cullen BR (2010) Viruses, microRNAs, and host interactions. Ann Rev Microbiol 64:123–141. doi:10.1146/annurev.micro.112408.134243
Song L, Liu H, Gao S, Jiang W, Huang W (2010) Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol 84(17):8849–8860. doi:10.1128/jvi.00456-10
Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O (2007) Host immune system gene targeting by a viral miRNA. Science 317(5836):376–381. doi:10.1126/science.1140956
Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D (2005) SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435(7042):682–686. doi:10.1038/nature03576
Swaminathan G, Martin-Garcia J, Navas-Martin S (2013) RNA viruses and microRNAs: challenging discoveries for the 21st century. Physiol Genom 45(22):1035–1048. doi:10.1152/physiolgenomics.00112.2013
Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR (2008) An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci USA 105(31):10931–10936. doi:10.1073/pnas.0801845105
Tang S, Patel A, Krause PR (2009) Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol 83(3):1433–1442. doi:10.1128/jvi.01723-08
Tambyah PA, Sepramaniam S, Mohamed Ali J, Chai SC, Swaminathan P, Armugam A, Jeyaseelan K (2013) MicroRNAs in circulation are altered in response to influenza A virus infection in humans. PLoS ONE 8(10):e76811. doi:10.1371/journal.pone.0076811
Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M (2007) Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315(5818):1579–1582. doi:10.1126/science.1136319
Tycowski KT, Guo YE, Lee N, Moss WN, Vallery TK, Xie M, Steitz JA (2015) Viral noncoding RNAs: more surprises. Genes Dev 29(6):567–584. doi:10.1101/gad.259077.115
Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR (2008) MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454(7205):780–783. doi:10.1038/nature07103
Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR (2009) Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol 83(20):10677–10683. doi:10.1128/jvi.01185-09
Umbach JL, Yen HL, Poon LL, Cullen BR (2010) Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio 1 (4). doi:10.1128/mBio.00204-10
Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313(5785):320–324. doi:10.1126/science.1129333
Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20(5):515–524. doi:10.1101/gad.1399806
Van Wynsberghe PM, Chan SP, Slack FJ, Pasquinelli AE (2011) Analysis of microRNA expression and function. Meth Cell Biol 106:219–252. doi:10.1016/b978-0-12-544172-8.00008-6
Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20(13):1732–1743. doi:10.1101/gad.1425706
Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH, Cullen BR (2013) In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio 4(2):e000193. doi:10.1128/mBio.00193-13
White MK, Gordon J, Khalili K (2013) The rapidly expanding family of human polyomaviruses: recent developments in understanding their life cycle and role in human pathology. PLoS Pathog 9(3):e1003206. doi:10.1371/journal.ppat.1003206
Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ Jr (2008) EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res 68(5):1436–1442. doi:10.1158/0008-5472.can-07-5126
Xu N, Segerman B, Zhou X, Akusjarvi G (2007) Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol 81(19):10540–10549. doi:10.1128/jvi.00885-07
Yeung ML, Benkirane M, Jeang KT (2007) Small non-coding RNAs, mammalian cells, and viruses: regulatory interactions? Retrovirology 4:74. doi:10.1186/1742-4690-4-74
Young LS, Rickinson AB (2004) Epstein-Barr virus: 40 years on. Nature Rev Cancer 4(10):757–768. doi:10.1038/nrc1452
Ziegelbauer JM, Sullivan CS, Ganem D (2009) Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nature Genet 41(1):130–134. doi:10.1038/ng.266
Acknowledgments
I would like to thank Dr. Brent Derry and Dr. Kevin Coombs for their reviews of the manuscript. ATT is supported by the Garron Family Cancer Centre and SickKids Research Training Centre Fellowship.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Tran, A.T. (2016). The Roles of MicroRNAs and PiRNAs in Virus-Host Interactions. In: Leitão, A., Enguita, F. (eds) Non-coding RNAs and Inter-kingdom Communication. Springer, Cham. https://doi.org/10.1007/978-3-319-39496-1_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-39496-1_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-39494-7
Online ISBN: 978-3-319-39496-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)