Abstract
The identification and segmentation of focal hyperintense lesions on magnetic resonance images (MRI) are essential steps in the assessment of disease burden in multiple sclerosis (MS) patients. Manual lesion segmentation is considered to be the gold standard, although it is time-consuming and has poor intra- and inter-operator reproducibility. Here, we present a segmentation method based on dual-echo MR images initialized by manual identification of lesions and a priori information. The classification technique is based on a region growing approach with a final segmentation refinement step. The results have revealed high similarity between the segmentation performed with this method and that performed manually by an expert operator, as well as a low misclassification of lesions. Moreover, the time required for segmentation is drastically reduced.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The analysis of disease burden on magnetic resonance images (MRI) from patients with multiple sclerosis (MS), both for research and clinical trials, requires the quantification of the volume of hyperintense lesions on a T2-weighted MRI sequence [1].
While many automatic methods for MS lesion segmentation have been proposed in the last 15 years, manual segmentation is still considered the gold standard although it is time-consuming and introduces inter and intra-observer variability [2].
The situation on available automatic methods for lesion segmentation is somewhat confused and fragmentary, complicating the difficult task of selecting one of the methods. Methods for fully-automated MS lesion segmentation are usually validated on a restricted dataset of cases and without a common framework, using different evaluation metrics, making the results difficult to compare.
Moreover, those methods are usually not trained or validated using dual-echo (DE) PD/T2-weighted MRI scans that have historically been used for the quantification of hyperintense MS lesions. The FLAIR sequence is now more commonly used because of the better contrast between focal lesions and the surrounding tissue [3, 7]; however, large dual-echo datasets are in existence, and these represent a great resource for research, so there is a need to implement new methods to speed up lesion segmentation on those datasets.
The correct segmentation of all lesions is an important issue of the fully-automatic methods, since they often identify false positives and false negatives [9].
With these considerations in mind, we chose to implement a new method, based on DE MR images, that could guarantee the correct identification of all lesions by having an expert physician manually perform this task, but then automating the lesion segmentation phase which is the most time-consuming part, contributes most to variability.
This paper presents a semi-automatic method for MS lesion segmentation based on manual identification of lesions on DE MR images, using a priori information. It gave high similarity with the ground truth and it also provides a considerable reduction in the time required for whole task of lesion segmentation.
2 Materials and Methods
2.1 Patients
The dataset consisted of 10 MS patients used for training the algorithm, and 20 MS patients with a range of lesion loads [0.3 – 9 ml] used for the validation. For each patient, a brain DE turbo spin-echo MRI sequence was obtained using a 3.0 T scanner (Achieva Philips Medical Systems, Best, The Netherlands), (TR/TE = 2910/16,80 ms, ETL=6; flip angle=90\(^\circ \), matrix size=256\(\,\times \,\)256, FOV=240\(\,\times \,\)240 mm\(^{2}\), 50 axial 3 mm-thick slices).
Manual identification of lesions by an expert physician was used to initialize the algorithm, whereas manual segmentation, performed by the same expert, was used for validation purposes. Both steps were performed using software for medical image analysis (Jim Version 6, Xinapse Systems, Colchester, UK).
Approval was received from the ethical standards committee on human experimentation of San Raffaele Scientific Institute. Written informed consent was obtained from all subjects prior to study enrollment.
2.2 Methods
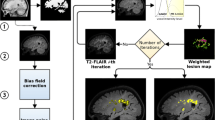
The following are the operational phases of the method.
Image Standardization. One difficulty with non-quantitative MRI techniques is that image intensities are arbitrary, even within the same protocol, for the same scanner and the same subject. This is a problem if a threshold value is to be used for a region growing approach, as described in the next section. Thus, proton density weighted (PD-w) image intensity values were standardized to correct for the arbitrary intensity scaling for different acquisitions [5]. The method used requires a training step, to be performed only once for a given MRI protocol on a cohort of patients, in which three intensity parameters are estimated from each histogram: the brightest peak position (\(\mu \)) that corresponds to the grey matter (GM) peak, and the first and last percentiles (\(p_1\) and \(p_2\) respectively) set at 1 % and 98 %.
The intensity range of values [\(s_1\),\(s_2\)] for the standard histogram in which to project the first and last percentiles intensity values of each input image, is selected according to a theorem, stated in [6], that guarantees in its formulation that each intensity value of the original image corresponds unequivocally a new intensity value on the standard image, so that no image compression is performed during the transformation. Thus, if standardization is done respecting these conditions, then there is no loss of information and the original image can be obtained by inverting the standardized image. The \(s_1\) value is fixed to 1, while \(s_2\) is extracted as follows, according to the cited theorem, where the index i identified each volume V of the training set:
The intensity value for the standard GM peak is calculated as the mean of the GM peak intensities of the training dataset.
During the transformation phase, the intensity value of the GM peak (brightest peak) of each input volume was fixed to the standard GM peak intensity value, and a linear intensity transformation that passes through this point and minimizes the distance from the two percentiles to the standard intensity range was applied. In this way the intensity histogram of each given image is rescaled into the standard one.
Figure 1 shows three PD-w MRI histograms after the standardization process.
Region Growing Algorithm. The core of the algorithm is the pixel-based region growing segmentation method. This approach to segmentation examines neighbouring pixels of initial “seed points” and determines whether the pixel neighbours should be added to the region according to similarity constraints [4]. The process is iterated as a clustering algorithm and stops when the similarity condition is violated.
The main constraint used for the growth of the segmented region is the intensity similarity, based on a threshold that varies according to a relationship determined by a training process described below.
Training. The region growing segmentation approach is applied to the training dataset where lesions were manually identified using a marker point and outlined by an expert physician. Region growing starts in each lesion from the markers, and lesion outlines are used as reference results to find the optimal threshold that pushes/stops the growth of the segmented region as close as possible to the manual segmentation. In this way, the optimal values of the threshold associated with each seed point are extracted and collected, as shown in Fig. 2. The threshold values extracted represent the difference between the seed point intensity value and the minimum intensity value inside the segmented region which stop the segmentation of the lesion. Due to the heterogeneity of MS lesions, threshold values are very noisy, as shown in Fig. 2. A straight line is fitted to those data to obtain a function for the validation dataset that unequivocally associates a threshold with each marker point on the PD-w image.
Segmentation. Lesions are first manually identified by an expert physician who places markers on the PD-w images while also having the T2-w image visible as a reference.
Starting from each marker (seed point), expansion of the segmented region continues to the adjacent pixels constrained according to a threshold value. This value (\(T_i\)) is different for each lesion and it is extracted by the threshold function computed during the training phase:
where \(m_f\) and \(q_f\) are respectively the slope and the intercept of the threshold function; \(seed_i\) is the intensity value of the \(i - th\) lesion marker point.
To avoid the segmentation going outside lesions, the region growing approach is combined with edge detection of lesions. For this purpose a half-way contrast image is obtained by averaging the non-standardized PD-w and T2-w images. The PD-w image has better contrast between white matter (WM) and cerebrospinal fluid (CSF) than the T2-w image, while the latter shows better contrast between WM and GM than the PD-w image. The “mean” image is created to take advantage of both images tissue contrasts, as shown in Fig. 3.
This image is filtered using a high-pass unsharp filter to create an image in which the high-frequency components (edges) are amplified [8]. The edge-enhanced image is subtracted from the original image, to obtain an image in which lesion edges are zero-crossing points between negative and positive values, representing respectively the internal and the external side of the lesion.
A new image S is obtained, as shown in Fig. 4:
where I is the original image and filt(I) is the filtered image.
This result is finally employed to restrict the growth of lesion segmentation when a lesion edge is reached.
Since the two constraints did not perform satisfactorily if used alone, because of noise or artefacts on the images, the intensity threshold is combined with the detection of lesion edges to obtain the stop condition of the region growing algorithm:
where \(I_s\) is the intensity of the seed point, \(I_{pi}\) is the intensity of the \(i-th\) adjacent pixel to classify in the standardized PD-w image and T is the threshold value previously extracted, just once for each lesion before the start of the region growing algorithm.
To stop the growth of the segmented region both conditions need to be satisfied.
Threshold Refinement Step. The threshold curve is used only to initialize the growth of the segmented region, and after an initial segmentation a more robust intensity threshold is estimated. For each segmented lesion, the distribution of intensity values is extracted after the first step of segmentation and the refined threshold of this distribution is used as a new intensity threshold to restart the region growing.
The refined threshold is selected according to the dimensions of the lesion: if a lesion is small (less than 10 pixels) the intensity distribution extracted is unreliable due to the low number of samples, so that the twentieth percentile of the distribution is selected as the refined threshold to avoid the inclusion of outliers. On the other hand, if a lesion is large (more than 10 pixels), the fifth percentile of the distribution is selected as the new threshold, since the intensity distribution is more reliable.
According to the new threshold values, the region growing is restarted from the previous segmentation. A final refined segmentation is obtained using the same stopping condition but with a new threshold T (Fig. 5).
The method is implemented in MatLab\(\circledR \) and the output of the algorithm is the mask of the segmented lesions and the lesion load in \(mm^3\).
3 Validation
Manual segmentation by an expert operator was used as the gold standard.
The metrics used for the validation are computed considering each lesion separately and then overall lesions.
-
1.
Dice Similarity Coefficient (DSC), to assess the similarity between the segmentation performed manually and that performed with the proposed method for each lesion:
$$\begin{aligned} DSC = \frac{2 |A_v \cap M_v|}{|A_v|+|M_v|}; \end{aligned}$$(8)where \(| A_v \cap M_v |\) is the number of voxels classified as lesion by both this method and the expert operator. \(| A_v |\) is the number of voxels classified as lesion by this method and \(| M_v |\) is the number of voxels classified as lesion by the expert operator.
-
2.
Root Mean Square Error of lesion load (RMSE) in ml:
$$\begin{aligned} RMSE = \sqrt{\frac{1}{n} \displaystyle \sum _{i=1}^{n} (M_i - A_i)^2} \end{aligned}$$(9)where n is the number of lesions; \(M_i\) is the \(i-th\) manually detected lesion load and \(A_i\) is the \(i-th\) automatically detected lesion load.
-
3.
True Positive Fraction (TPF); False Positive Fraction (FPF); False Negative Fraction (FNF):
$$\begin{aligned} TPF = \frac{| A_v \cap M_v |}{| M_v |}; \end{aligned}$$(10)$$\begin{aligned} FPF = \frac{| A_v \cap \lnot M_v |}{| M_v |}; \end{aligned}$$(11)$$\begin{aligned} FNF = \frac{| \lnot A_v \cap M_v |}{| M_v |}; \end{aligned}$$(12)where \(| A_v \cap \lnot M_v |\) is the number of voxels classified as lesion only by the new method and not by the expert operator, while \(| \lnot A_v \cap M_v |\) is the number of voxels classified as lesions only by the expert operator and not by this method.
4 Results
Fig. 6 shows example lesion segmentations. The manually segmented lesion mask can be visually compared to the output lesion mask of the new method. The validation metrics were extracted for each lesion load of each patient. Lesions are labelled in 3-D to compute these metrics.
In Fig. 7, the metrics evaluated over all lesions for each patient are graphically reported. Averaging the metrics over all patients the following values were obtained: DSC = 0.78; RMSE = 0.17 ml; TPF = 0.81; FPF = 0.14; FNF = 0.20.
In the top left graph a scatter plot is shown to compare manually estimated lesion load to that estimated by the new method for each patient. In the top right graph the mean DSC values for each patient are reported. In the bottom graph, the mean TPF (blue squares), FPF (red crosses) and FNF (black circles) values for each patient are shown (Color figure online).
5 Discussion
In this paper, a semi-automatic method is presented for segmenting MS lesions on DE MRI, based on the manual identification of lesions and a trained region-growing algorithm with prior intensity standardization.
Lesion segmentation obtained using the new method was very similar to the ground truth, with a high degree of overlap (DSC = 0.78 and TPF = 0.81). The lesion load obtained with this segmentation method is comparable with that obtained with the manual segmentation (RMSE = 0.17 ml). FPF and FNF values indicated that there was low misclassification of lesion voxels.
Moreover, the operator time required to process the images was drastically reduced: for the images evaluated here, the average time for manual lesion segmentation was about 40 min, while for the method proposed the average time was about 50 s, regardless manually marking the seed points.
The comparison of this method with other proposed automatic or semi-automatic MS lesion segmentation methods is very challenging. The difficulty would be to find an available method that can be used with our own data (PD/T2-w scans). On the other hand, the full re-implementation of a published, but not freely-accessible method might introduce some small differences or errors in the code which could mean that the method performs badly. We therefore chose to compare the method with expert manual segmentation, which is still considered to be the gold standard. Moreover, it was difficult to find a MS lesion challenge with a shared PD-T2w MRI dataset for an easy comparison of the results with other lesion segmentation methods.
Due to the heterogeneous nature of MS lesions, the method sometimes encountered difficulties in segmenting those lesions with blurred and poorly-defined borders, which are also difficult for a human observer to delineate. Those lesions have poor contrast on PD/T2-w scans, thus confounding the constraints for the region growing approach, and the segmentation exceeded the external borders of the lesion. This might be improved by introducing further information about the spatial location of lesions, perhaps using co-registered T1-w images.
The method has been validated on data from a single center, and from a single type of MRI scanner. Further validation is required by testing the method on a multi-center dataset with different scanners and scanner operators. Another additional validation would be to test the sensitivity of the method with respect to the location of the seed points.
While accuracy is certainly important, it is essential that we assess the reproducibility in future. If a technique is inaccurate or has a bias, as long as this bias is consistent it should still be possible to measure changes over time. However, if the reproducibility is poor, real changes in longitudinal studies can be masked by random variations due to poor measurement.
In future, it may be also possible to fully automate the method, by removing the need to manually identify lesions by employing FLAIR or double inversion recovery (DIR) sequences.
References
Filippi, M., Rocca, M.A., De Stefano, N., Enzinger, C., Fisher, E., Horsfield, M.A., Inglese, M., Pelletier, D., Comi, G.: Magnetic resonance techniques in multiple sclerosis: the present and the future. Arch. Neurol. 68(12), 1514–1520 (2011). doi:10.1001/archneurol.2011.914.Review
Garcia-Lorenzo, D., Francis, S., Narayanan, S., Arnold, D.L., Collins, D.L.: Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med. Image Anal. 17, 1–18 (2013)
Garcia-Lorenzo, D., Prima, S., Arnold, D.L., Collins, D.L., Barillot, C.: Trimmed-likelihood estimation for focal lesions and tissue segmentation in multisequence MRI for multiple sclerosis. IEEE Trans. Med. Imaging 30, 1455–1467 (2011)
Kamdi, S., Krishna, R.K.: Image segmentation and region growing algorithm. Int. J. Comput. Technol. Electron. Eng. 2, 103–107 (2012)
Nyul, L.G., Udupa, J.K.: On standardizing the MR image intensity scale. Magn. Reson. Med. 42, 1072–1081 (1999)
Nyul, L.G. and Udupa, J.K.: On standardizing the MR image intensity scale. Technical Report MIPG.250, Medical Image Processing Group, Department of Radiology, University of Pennsylvania (1999)
Schmidt, P., Gaser, C., Arsic, M., Buck, D., Forschler, A., Erthele, A., Hoshi, M., Ilg, R., Schmid, V.J., Zimmer, C., Hemmer, B., Muhlau, M.: An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 59, 3774–3783 (2012)
Luft, T., Colditz, C., Deussen, O.: Image enhancement by unsharp masking the depth buffer. Association for Computing Machinery (2006)
Van Leemput, K., Maes, F., Vandermeulen, D., Colchester, A., Suetens, P.: Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Trans. Med. Imaging 20(8), 677–688 (2001)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Storelli, L., Pagani, E., Rocca, M.A., Horsfield, M.A., Filippi, M. (2016). A Semi-automatic Method for Segmentation of Multiple Sclerosis Lesions on Dual-Echo Magnetic Resonance Images. In: Crimi, A., Menze, B., Maier, O., Reyes, M., Handels, H. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2015. Lecture Notes in Computer Science(), vol 9556. Springer, Cham. https://doi.org/10.1007/978-3-319-30858-6_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-30858-6_8
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-30857-9
Online ISBN: 978-3-319-30858-6
eBook Packages: Computer ScienceComputer Science (R0)