Abstract
Proteases (PRs) catalyze the cleavage of peptide bonds by hydrolysis in proteins and peptides playing crucial functions in organisms all over the phylogenetic tree. These enzymes are present in all types of bacteria and are involved in critical processes such as acquisition of nutrients for growth and proliferation, facilitation of dissemination, colonization and evasion of host immune defenses or tissue damage during infection. Bacterial pathogens use their PRs to acquire or activate the function of host PRs to help them in their growth or progression of disease. Research into bacterial PRs and their substrates will allow the development of novel PRs inhibiting compounds that could potentially be used to limit host virulence or to block cell growth. The emergence resistances to traditional antibiotics have created clinical difficulties for nosocomial treatment on a global scale. Thus the pharmacological development of new PRs inhibitors that target essential proteins in the bacterial pathogen is of great interest, and it is the focus of this review.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Proteolytic enzymes also known as proteases (PRs), peptidases, or proteinases are one of the largest functional groups of proteins, with more than 4000 members actually described (Rawlings et al. 2012). PRs catalyze the cleavage of peptide bonds by their hydrolysis in proteins and peptides playing crucial functions in organisms all over the phylogenetic tree, starting from viruses, bacteria, protozoa, metazoa, fungi, and ending with plants and animals. PRs enzymes are extensively applied in several sectors of industry and biotechnology (Rao et al. 1998; Kirk et al. 2002; Rani et al. 2012; Ray 2012). Therefore, several research applications require the use of PRs, including peptide synthesis, digestion of unwanted proteins during nucleic acid purification and the production of Klenow fragments, use of proteases in cell culture experiments and in tissue dissociation, preparation of recombinant antibody fragments for research, peptide sequencing, proteolytic digestion of proteins in proteomics diagnostics and therapy, exploration of the structure–function relationships by structural studies, and removal of affinity tags from fusion proteins in recombinant protein techniques.
PRs are present in all types of bacteria and are involved in critical processes such as acquisition of nutrients for growth and proliferation, facilitation of dissemination, colonization and evasion of host immune defenses or tissue damage during infection (Miyoshi and Shinoda 1997; Travis and Potempa 2000). Bacteria produce a variety of PRs and their roles in virulence factors of the various bacteria genera are well known. Bacterial PRs have the ability to destroy important host defense proteins, such as those present in the complement system, which is the main part of a host’s innate immunity (Popadiak et al. 2007), as well as degrade the functional and structural proteins present in the host organism (Maeda 1996). Besides, it should be noted the ability of bacteria to use the PRs it produces to pass the proteinaceous barriers present inside a host organism, which is a major part of bacterial virulence. Bacterial pathogens use its PRs to acquire or activate the function of host PRs to help in its growth or progression of disease (Lantz 1997).
Bacterial pathogens rely on proteolysis for variety of purposes during the infection process and they are involved in the interruption of the cascade activation pathways, disruption of cytokine network, excision of cell surface receptors, and inactivation of host protease inhibitors (Maeda 1996; Miyoshi and Shinoda 1997; Rice et al. 1999; Travis and Potempa 2000). Thus, the proteolysis phenomena have been adopted by bacterial pathogens at multiple levels to ensure their success in contact with the host. It goes without saying that bacterial PRs may represent very attractive targets for the development of novel types of antibiotics (Travis and Potempa 2000).
Bacterial PRs inhibition would presumably lead to the death of the invading pathogen. Moreover, consider the specific role that bacterial PRs play in such critical steps for the successful invasion of the host. Taking into account the constant emergence of antibiotic resistance (Rolain et al. 2012; Bond 2015), it would be extremely beneficial to develop bacterial PRs inhibitors as a second antibiotic generation. Research into bacterial PRs and their substrates will allow the development of novel PRs inhibiting compounds that could potentially be used to limit the action of destructive PRs produced during the bacterial infection process (Zindel et al. 2013; Drag and Salvesen 2010). Though targeting PRs for creating PRs inhibitors has proven to be a useful idea for therapeutic intervention against pathogenic bacteria, utilizing these PRs or taking advantage of their functions to help in drug delivery techniques has not been widely tried.
In a general way, the inhibitor binds to the enzyme and decreases its activity; the classification of inhibitors is based on where and how such binding occurs, as well as, the type of binding (Fersht 1985; Otto and Schirmeister 1997). PRs inhibitors as enzymatic inhibitors bind either into the active site as active site-directed inhibitors or at another site as allosteric inhibitors. An active site-directed inhibitor can either bind covalently or noncovalently. Covalent inhibitors are electrophiles that are attacked by the active site to form a covalent bond which prevents further enzymatic reactions. Noncovalent inhibitors must depend on other interactions in order to remain in the binding site. Electrostatic interactions such as hydrogen bonds, salt bridges, and pi–pi interactions between aryl groups are the most important for protein recognition and binding with high specificity. Noncovalently bound inhibitors are reversible and the enzyme can be reactivated. Covalently bound inhibitors are either irreversible or reversible. Finally, the irreversible inhibitor reacts with the active site and blocks further modifications (Fersht 1985; Gohlke and Klebe 2002).
According to the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology, PRs belong to the hydrolase class of enzymes (EC 3) and are grouped into the subclass of the peptide hydrolases or peptidases (EC 3.4) (Table 1). Depending on the reaction they catalyze, proteases are divided into exopeptidases which specifically cleavage protein substrates from the carboxyl- or amino-termini and endopeptidases which preferentially hydrolyse peptide bonds in the inner regions of peptide chains. Aminopeptidases (Table 1) can liberate single amino acids (EC 3.4.11), dipeptides (dipeptidyl peptidases, EC 3.4.14), or tripeptides (tripeptidyl peptidases EC 3.4.14) from the N-terminal end of their substrates. Single amino acids can be released from dipeptide substrates by dipeptidases (EC 3.4.13) or from polypeptides by carboxypeptidases (EC 3.4.16, EC 2.4.17, EC 3.4.18) (Table 1), while peptidyl dipeptidases (EC 3.4.15) liberate dipeptides from the C-terminal end of a polypeptide chain. Finally, as mentioned above, endopeptidases (Table 1) cleave peptide bonds within and distant from the ends of a polypeptide chain [serine endopeptidases (EC 3.4.21), cysteineendopeptidases (EC 3.4.22), asparticendopeptidases (EC 3.4.23), metalloendopeptidases (EC 3.4.24), threonineendopeptidases (EC 3.4.25) and endopeptidases of unknown catalytic mechanism (EC 3.4.99)]. Seventh catalytic types of PRs have been recognized according to the nature of the catalytic residues [MEROPS—the peptidase database (http://merops.sanger.ac.uk/) (Rawlings et al. 2010)]: serine—(first described in 1993), cysteine—(1993), aspartic—(1993), metallo—(1993), threonine—(1997), glutamic acid—(2004), and asparagine-protease—(2010) (Kohei 2012).
2 Bacterial PRs Functions and Their Inhibitors
2.1 Bacterial PRs for Growth and Proliferation
It is well known that during the process of bacterial cell division and growth, they utilize several different PRs for the assembly and disassembly of the bacterial cell wall (Humann and Lenz 2009). Peptidoglycan (PGN) synthesis and its hydrolysis are processes required by almost all bacteria for growth; nevertheless, the involved pathways for cell division and growth are function of the shape and Gram type of the bacterium (Humann and Lenz 2009; Margolin 2009).
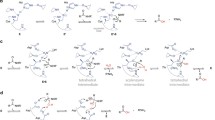
The structure of PGN in both Gram-positive and Gram-negative bacteria comprises repeating disaccharide backbones of N-acetylglucosamine (NAG) and β-(1-4)-N-acetylmuramic acid (NAM) that are cross-linked by peptide stem chains attached to the NAM residues (Fig. 1) (Bourhis and Werts 2007). PGN is concealed by an outer membrane in Gram-negative bacteria, or by layers of proteins and glycopolymers in Gram-positive bacteria (Fig. 2). During turnover of the bacterial cell wall for bacterial growth and division, bacteria employ several classes of PGN-hydrolyzing enzymes that participate in the assembly and disassembly of this cell wall. Bacterial PGN-hydrolyzing enzymes degrade the PGN of the producing organism or degrade intact bacterial cells.
PGN biosynthesis and mode of action of β-lactam antibiotics. As illustrated, in absence of β–lactam antibiotics, PBPs work in PGN biosynthesis creating a rigid cell wall. Nevertheless, in the presence of β-lactam antibiotics, they bind to the transpeptidase site of PBP inhibiting cell wall synthesis, resulting in a weakened cell wall which ends with bacterial death
Scheme of bacterial cell wall composition in Gram-positive and Gram-negative bacteria (Based on: Santos-Beneit et al. 2014)
These bacterial PRs are defined by their catalytic specificities and are classified as autolysins. Two classes of enzymes are involved in the PGN glycan backbone digestion: N-acetylmuramidases and N-acetylglucosaminidases. While the N-acetylmuramidases cleave PGN between the N-acetylglucosamine (NAG) and β-(1-4)-N-acetylmuramic acid (NAM) bond upstream of NAM; N-acetylglucosaminidases cleave the NAM–NAG bond.
Another group of autolysins, N-acetylmuramyl-l-alanine amidases, separates the PGN sugar backbones from the stem peptide chain concretely these enzymes cleave between NAM and the first alanine of the peptide chain (Fournier and Philpott 2005; Scheurwater et al. 2007). Finally, the lytic transglycosylases cleave between the N-acetylmuramic acid and N-acetylglucosamine sugar chains to facilitate cell growth among other functions (Zahrl et al. 2005; Scheurwater et al. 2008; Cloud-Hansen et al. 2008).
Bacilluis subtilis is the most studied microorganism in autolysin-related research, since it is a nonpathogenic model system for investigating the roles of autolysins in the cell wall metabolism (Smith et al. 2000). This knowledge, which was obtained through the study of PGN fine structure, the analysis of multiply inactivated mutants by sequencing of the entire B. subtilis genome, has become possible to define the roles played by individual autolysins in a number of important cellular processes (Rogers et al. 1984; Kunst et al. 1997; Smith et al. 1996). Cell separation in B. subtilis relies on the activity of the autolysins LytF, LytE, and CwlS.
All autolysins are cysteine proteases inside of the NlpC/p60 endopeptidase family (Smith et al. 2000; Yamamoto et al. 2008). LytN is another autolysin produced by Staphylococcus aureus that works as amidase and endopeptidase during the synthesis of PGN (Sugai et al. 1998). These bacterial PRs contribute to the release of the staphylococcal protein A (SpA) by removing amino sugars [i.e., NAM–NAG] from attached PGN (Becker et al. 2014). In a general way, autolysins and in particular B. subtilis and S. aureus are essential for the synthesis of PGN because its deletion in these microorganisms results in cell growth defects and altered growth morphology (Hashimoto et al. 2012; Frankel and Schneewind 2012).
Penicillin-binding proteins (PBPs) are a distinct group of autolysins that play an important role in cell morphology and viability. PBPs process d -amino acid bonds during PGN synthesis (Wise and Park 1965) by the degradation of the d -alanine– d -alanine bonds (Fig. 1). In addition, PPBs catalyze the terminal stages of PGN synthesis by creating crossbridges between the stem peptides (Frère and Page 2014). Taking into account that not all of the PBPs produced by bacteria are essential, as it was checked in an Escherichia coli mutant (Denome et al. 1999), they are validated targets for the β-lactam antibiotics in the antibacterial therapy.
2.2 β-Lactam Antibiotics as Tools for Bacterial Growth Inhibition
β-lactam antibiotics (cephalosporins, cephamycins, cephabacims, olivanic acids, thienamycins, epithienamycins, monolactams (nocardicins and monobactams), and clavams) (Fig. 3) are the most frequently prescribed antibiotics worldwide used to treat bacterial infections (Fernández-Aguado et al. 2014).
The general antimicrobial mechanism of action of β-lactam antibiotics consists of the inhibition of PGN biosynthesis, which weakens the bacterial cell wall during the cellular division, leading to cytolysis and death. These antibiotics covalently bind to the active site of PBPs (Fig. 1, detail), which catalyze the linking of PGN molecules in bacteria in the last step of the bacterial cell wall biosynthesis. It is due to the structural similarity between β-lactam antibiotics and the last two amino acids (acyl-d-alanine-d-alanine) of the pentapeptide that links the PGN molecule.
PBP enzymes, including transglycosylases (PBP1 complex), transpeptidases (PBP3), and carboxypeptidases (PBP4, PBP5, and PBP6) are irreversibly inhibited by β-lactam antibiotics, no longer catalyzing the linking reaction. These covalent complexes block the normal transpeptidation reaction inhibiting cell wall biosynthesis. In addition, these antibiotics lead to cell stress responses by the activation of bacterial cell wall hydrolases and autolysins, which result in bacterial cell lysis (Tomasz 1979).
The activity of β-lactam antibiotics is initially higher against Gram-positive bacteria (Fig. 2). In these microorganisms, PBPs are located on the cytoplasmic membrane exposed to the environment, unlike Gram-negative bacteria, where PBPs are present in the periplasmic space protected by the external outer membrane, which acts as a barrier for different molecules (Fig. 2). However, the incorporation of new molecules to the penam and cephem nuclei has given rise to the synthesis of semisynthetic antibiotics with a higher activity against Gram-negative microorganisms. Currently, many used β-lactams have very broad spectrum activity against most aerobic and anaerobic Gram-positive and Gram-negative bacteria, as well as, low-toxicity profiles making them popular first line antibiotics. Nevertheless, bacteria have acquired resistance to β-lactams mainly through three different strategies: active expulsion of β-lactams via efflux pumps, production of a specific β-lactamases, and presence of low-affinity PBPs (Poole 2004; Worthington and Melander 2013).
The first β-lactam antibiotic to be discovered was penicillin in 1932 (Clutterbuck et al. 1932) though it was not introduced to the clinic until the early 1940s. This antibiotic discovery started by an accidental experiment performed by Sir Alexander Fleming in his lab in 1928. Fleming evaluated the antimicrobial activity generated by a fungus culture, later identified as Penicillium notatum that contaminated a Petri dish with Staphylococcus sp. (Fleming 1929).
Penicillin (Fig. 3) and more specifically benzylpenicillin (penicillin G) is obtained by submerged fermentation of industrial strains of the filamentous fungus Penicillium chrysogenum, recently renamed as Penicillium rubens (Fernández-Aguado et al. 2013, 2014). Nowadays, several reports indicated the presence of a penicillin-resistant Staphylococcus aureus that produces enzymes that inactivate the β-lactam antibiotic by hydrolyzing the β-lactam core (Jovetic et al. 2010). These enzymes are known as β-lactamases. Later in 1959, the isolation of 6-aminopenicillanic acid (6-APA) from the P. chrysogenum fermentation end product [penicillin G or penicillin V (phenoxymethylpenicillin)] allowed the development of numerous semisynthetic penicillins that are resistant to the staphylococcal β-lactamases as a result of steric protection of the β-lactam ring. Methicillin-susceptible S. aureus produces five kinds of PBPs: PBP1, PBP2, PBP2B, PBP3, and PBP4, for which the genes have been cloned and sequenced (Boyle-Vavra et al. 2003). Nevertheless, Staphylococci have developed another mechanism for resistance to β-lactam antibiotics by the production of a new penicillin-binding protein 2a (PBP2a). These bacterial strains have been designated as methicillin-resistant S. aureus (MRSA), from 1960s to present. β-lactam resistance in MRSA is primarily due to the expression of mecA gene which encodes the low-affinity penicillin-binding protein PBP2 (Wu et al. 2001; Smith 2007). Bioinformatic analysis of PBP2a protein revealed a clear similarity to those of the shape-determining protein (PBP2) and septum forming (PBP3) of E. coli (Song et al. 1987).
These beta-lactam resistance gene mecA of S. aureus is carried on a large mobile genetic element known as SCCmec, which integrates it into the chromosome of MRSA strains (Katayama et al. 2000; Ito et al. 2001; Jansen et al. 2006). The mechanism of resistance of PBP2a producers is the outcome of the limited accessibility of the antibiotics to the active site which results in a reduced rate constant for acylation (3–4 orders of magnitude) as compared to other PBPs, and an increased dissociation constant for the preacylation complex. In contrast to the low accessibility of the PBP2a active site to β-lactam antibiotics, the native PGN substrate is still able to access the active site. It is believed as a consequence of conformational changes brought about by allosteric binding of PGN to the enzyme, resulting in effective PGN cross-linking and subsequent cell wall viability (Fuda et al. 2004, 2005; Worthington and Melander 2013).
It could be considered that PBP2a is an essential element for the prevalence of MRSA. Likewise in Streptococcus peneumoniae resistance to b-lactams is commonly caused by expression of a variety of low-affinity PBPs (Walsh 2003). These β-lactam resistances have proliferated dramatically and have created clinical difficulties for nosocomial treatment on a global scale. Strains of S. aureus exhibiting either PBP2a production or β-lactamase (or both) have established a considerable ecological niche among human pathogens which constitutes a serious health problem.
Inside the β-lactam antibiotics cephalosporins (Fig. 3) are widely and successfully used in medicine in the treatment of different bacterial infections. The history of cephalosporins started in 1945, when the filamentous fungus Cephalosporium acremonium (renamed as Acremonium chrysogenum) was isolated by Giuseppe Brotzu from the bay water at Cagliary (Italy) (Brotzu 1948). A. chrysogenum was found to produce at least three types of antimicrobial compounds, which were isolated and identified (Hamilton-Miller 2000). The first compounds isolated in 1949 were members of the cephalosporin P complex, tetracyclic triterpenes chemically related to helvolic acid (fumigacin). Later the same year, a second compound active against Gram-negative and Gram-positive bacteria, which was penicillin with a d -α-aminoadipic side chain was isolated and named penicillin N. Finally, the third compound isolated and identified in 1953 was cephalosporin C. This antibiotic was active against Gram-negative and Gram-positive bacteria and it was not hydrolyzed by penicillinase, a feature especially relevant due to the appearance of penicillin-resistant bacteria. Cephalosporin C is produced by Acremonium chrysogenum and a few other filamentous fungi (Ullán et al. 2002, 2007, 2010).
Early generations of semisynthetic cephalosporins improved their cellular penetration to increase their spectrum of activity against Gram-negative pathogens and to enhance their pharmacokinetics. Next generations have become increasingly focused on combating β-lactam resistance also by chemical side chain modifications on the cephem nucleus (Fig. 3). Actually, in their fifth generation, an extensive search for cephalosporins with efficacy against MRSA bacteria has been made. As a result of this search, new cephalosporin derivate drugs have been found like ceftobiprole that inhibits MRSA at 1–2 mg/L under standard conditions (Livermore 2006; Zhanel et al. 2008).
Following this line, ceftaroline as cephalosporin has the ability to bind to PBP2a to inhibit the bacterial growth of MRSA strains (Zhanel et al. 2009). Another novel class of anti-MRSA semisynthetic-cephalosporins are propenylamide and propenylsulfonamide cephalosporins that act against MRSA strains with minimum inhibitory concentrations as low as 1 μg/mL (Pohlmann et al. 2010). However, nowadays new resistances to ceftaroline and ceftobiprole cephalosporins have been reported (Kelley et al. 2015; Chan et al. 2015; Schaumburg et al. 2015).
In addition to the classical β-lactam compounds, many nonconventional β-lactam structures of scientific and industrial interest have been discovered and characterized since 1970. As shown in Fig. 3, these nonconventional β-lactams contain a β-lactam ring and they usually have a distinct bicyclic structure. The second ring in the molecule of clavulanic acid and other clavams is an oxazolidinic ring that includes oxygen instead of the sulfur atom occurring in classical β-lactams. The members of carbapenem family have a carbapenem ring containing a carbon atom instead of sulfur. Carbapenem members attack a wide range of PBPs, have low toxicity, and they are much more resistant to β-lactamases than the penicillins or cephalosporins. Monolactams (nocardicins and monobactams) contain a monocyclic structure (the β-lactam ring) and different side chains. Nonconventional β-lactams are also inhibitors of PGN biosynthesis (monolactams), others are potent β-lactamase inhibitors with weak antibiotic activity (clavulanic acid), or have antifungal activity (some clavams) (Brakhage et al. 2005; Coulthurst et al. 2005). β-lactamase inhibitors bind irreversibly to β-lactamases but do not have good activity against PBPs. These β-lactam compounds are combined with β-lactams in clinical treatments.
Besides fungi, some Gram-positive (actinomycetes such as Streptomyces clavuligerus or Amycolatopsis lactamdurans) and Gram-negative bacteria (e.g., Lysobacter lactamgenus) also synthesize hydrophilic β-lactam structures including cephalosporins (mainly as intermediates of biosynthetic pathways), cephamycins, cephabacins, clavams, carbapenems, and monolactams (Liras 1999). Cephamycins are 7-methoxy-cephalosporins active against penicillin-resistant bacteria. Cephabacins contain a formylamino group at C-7 and an oligopeptide side chain at C-3. The C-7 formylamino substituent of cephabacins and the C-7 metoxyl group of cephamycins confer to these antibiotics their characteristic β-lactamase resistance.
2.3 Inhibition of PBPs by Non-β-Lactam Antibiotics
An alternative to β-lactam antibiotics, to impede bacterial growth by inhibition of PBPs, are the noncovalent inhibitors. This way, Toney et al. (1998) developed a methodology for the identification of several non-β-lactam inhibitors, which exhibit IC50 values between 10 and 30 mM. Besides, the noncovalent inhibitors arylalkylidene rhodanines and arylalkylidene iminothiazolidin-4-ones can interfere with bacterial growth across the inhibition of PBPs (Zervosen et al. 2004). Recently, Turk et al. (2011) found new noncovalent inhibitors of PBPs after carrying out a screening of more than 250 compounds. As a result, among others, these authors identified a compound which inhibited PBP2a with a promising IC50 of 97 mM.
Other identified potential inhibitors of PBPs are the 4-quinolones (Shilabin et al. 2012) and the penicilloic acids (van Berkel et al. 2013). Inside this research line, a new option that has emerged recently to treat systemic MRSA infection are the biodegradable antimicrobial polycarbonates (Pascual et al. 2015; Cheng et al. 2015). Its antimicrobial mechanism is disrupting the bacterial membrane.
Studies involving FtsZ (Filamenting temperature sensitive mutant Z) indicated that it is an essential protein required for cell division in prokaryotes. (Addinall and Holland 2002). Tan et al. (2012) described that the combination of the FtsZ-specific inhibitor PC190723 and β-lactam antibiotics restored MRSA susceptibility to β-lactams. These kinds of FtsZ inhibitors act synergistically with β-lactams. Therefore, Merck has developed a patent for the combination of FtsZ inhibitors with β-lactam antibiotics to target methicillin-resistant Staphylococcus epidermidis and MRSA stains [Merck Sharp & Dohme Corp. WO2011112435 (2011)]. Nevertheless in general, vancomycin is the drug of choice to treat MRSA.
2.4 Role of PRs in Bacterial Virulence—PRs Inhibitors
In pathogenic bacteria, many secreted bacterial PRs are virulence factors that aid in bacterial invasion into host cells by the degradation of host-associated proteins (Table 2). During the infective process bacterial PRs involved in growth and proliferation contribute to bacterial virulence indirectly since they are necessary to the bacterium survival within the host environment.
There are several examples of targets of bacterial PRs such as coagulation factors, fibrinogen, and fibrin. The degradation of these proteins that are involved in blood clot formation in the host, may lead to disease states causing, among others, disseminated intravascular coagulation and the formation of small blood clots in the blood vessel (Imamura et al. 1997, 2001; Komori et al. 2001; Massimi et al. 2002). The role of bacterial PRs in disseminated intravascular coagulation is based on the experimental evidences obtained by Komori et al. (2001) who observed that Pseudomonas aeruginosa LasA (elastase A) PRs together with its fibrinogenolytic activity affects endothelial cells and destroys the basement membrane of blood vessels to cause hemorrhage in mice.
Therefore, LasA enhances the virulence activity of the metalloendopeptidase LasB (elastase) in the establishment of a P. aeruginosa infection. LasA (Table 2) cleaves a wider range of glycine-containing proteins, including, glycine-rich synthetic peptides, specific sequences present in elastin and tropoelastin-derived pentapeptides. This metalloprotein endopeptidase known as LasA also enhances the activity of several other host elastolytic proteases, including, human neutrophil elastase, human leukocyte elastase, and other proteases (Hoge et al. 2010). During the infective process the combination of both P. aeruginosa PRs LasA and LasB result in the degradation of elastin that is a component of the connective tissue of vertebrates that give them elastic properties (Morihara 1964; Kessler et al. 1997; Cowell et al. 2003). The key role of LasB in pseudomonal virulence makes it a potential target for the development of antimicrobial agents to attenuate virulence processes without bactericidal action and to avoid the emergence of resistant strains. This way, Cathcart et al. (2011) described the ability of N-mercaptoacetyl-Phe-Tyr-amide (K(i) = 41 nM) to block the pseudomonal virulence processes across the inhibition of LasB. Besides, diethylene triamine pentaacetic acid (pentetic acid) suppresses elastase production by P. aeruginosa (Gi et al. 2014). The advantage of this compound resides in the fact that nowadays, it is clinically used as a contrast agent for diagnostic imaging. Nevertheless, identification of new antimicrobial agents may not always result in clinical application due to concerns associated with safety for use in the human body.
2.4.1 Lethal Factor Inhibitors
Bacillus anthracis is a spore-forming bacterium that secretes, among others, the metalloendopectidase known as lethal factor (LF) (Table 2). This toxin is lethal to the host through disruption of signaling pathways, cell destruction, and circulatory shock. Treatment against B. anthracis must be given early after infection, delay of treatment reduces survival of infected patients because antibiotics can eliminate an anthrax infection but not combat the LF-mediated toxemia that persists even after the bacteria have been eliminated by antibiotics. The discovery of efficient LF inhibitors can increase the probability of host survival by blocking late stage effects of LF in post-exposure anthrax cases (Forino et al. 2005). Thus Shoop et al. (2005) described a hydroxamate LF inhibitor that can be administrated with conventional antibiotics to treat the anthrax infectious disease. Hydroxamate chelates the Zn++ from the LF active site resulting in reduced protease activity of LF in the mitogen-activated protein kinase (MAPK) cleavage assay (Li et al. 2012). Other LF inhibitors are the catecholate siderophores enterobactin and the cationic polyamine from salmon sperm, protamine. However, protamine, exhibits the best results in the LF inhibition assays (Thomas and Castignetti 2009). In the same research line, Dell’Aica et al. (2004) described that the green tea polyphenols epigallocatechin-3-gallate and catechin gallate also inhibit the LF and show reduced MAPK kinase cleavage activity. Benzylamine-derived inhibitors, PT8420 and PT8541 are also LF inhibitors, provide protection against B. anthracis in combination with other treatments (Moayeri et al. 2013).
Another target of LF inhibitors is the LF exocites which contribute to the catalysis process and participate in the efficient binding of substrate at other points of the active site. Their inhibition action results in an LF conformational alteration and, therefore, in a reduction of LF activity. As result, Bannwarth et al. (2012) screened a library of LF exosites inhibitors, which allowed the identification and evaluation of the depsidone stictic acid as a possible inhibitor. Lethal toxin (LT) is the combination of LF and protective antigen, it has been reported that the organogold compound auranofin inhibited LT-mediated toxicity in mouse macrophages. Auranofin, due to its anti-inflammatory activity, acts downstream of MAPKK cleavage by inhibiting the caspase-1 activation (Newman et al. 2011).
2.4.2 Periodontium Infective Process, Gingipain Inhibitors
Another target of bacterial PRs virulence factors are the proteins of the host connective tissue. A clear example is the periodontium infective process by Porphyromonas gingivalis, where the proteolytic activity of the secreted gingipains results in periodontal connective tissue destruction as a result of host protein degradation (Potempa et al. 2000). The gingipain family of P. gingivalis comprises three related cysteineendopeptidases that hydrolyze peptide bonds at the carbonyl groups of lysine (Lys-Xaa) and arginine (Arg-Xaa) residues. The Lys-specific gingipain, Lys-gingipain (Kgp), is encoded by the kgp gene; whereas the homologous arginine-specific gingipains, RgpA and RgpB, are products of two related genes rgpA and rgpB (Fitzpatrick et al. 2009). Proteolytic activities of these PRs play a fundamental role in the attachment and colonization, acquisition of nutrients, evasion of host defense and tissue invasion, and dissemination of P. gingivalis (Guo et al. 2010).
Discovery and characterization of gingipain inhibitors could prevent or slow down the progression of this dental disease. In a review article of Olsen and Potempa (2014) described a battery of gingipain inhibitors that include gingipain N-terminal prodomains, synthetic compounds, inhibitors from natural sources, antibiotics, antiseptics, antibodies, and bacteria. The use of nonpathogenic bacteria as antagonists of pathogenic bacteria to combat tooth decay, deserves special mention. Thus, Tenorio et al. (2011) isolated and identified 19 bacterial strains from the subgingival plaque from 20 patients who were diagnosed as having periodontitis. Some of the identified isolates decreased the gingipain activity and interfering with the growth of P. gingivalis in vitro. Besides, Camelo-Castillo et al. (2014) isolated and identified several bacterial strains from the supragingival dental plaque from adult individuals who had never suffered dental caries. Two of these isolates [Streptococcus dentisani Str. 7746 (CECT83135, DSM 27089) and Streptococcus dentisani Str. 7747T (CECT 8312T, DSM 27088)] are considered for these authors as potential probiotics for human oral health.
2.4.3 Immune System PRs Inhibitors
Another virulence factor of some pathogenic bacterial PRs is its role in the “escape response” to several host defense mechanisms (Potempa and Pike 2009). Pathogens cause immunosuppression in their infected host that shows depressed immune responses to antigens in general, including those of the infecting pathogen. Thus, the contact system (also known as the intrinsic pathway of coagulation or the kallikrein/kinin system) of host delivers antimicrobial peptides (AMPs) derived from kininogen and traps bacteria in the thrombus. Furthermore, the bradykinin peptide causes spreading of macrophages and induces an influx of neutrophils into the surrounding host tissue (Frick et al. 2007).
Immunoglobulins (Igs) play a key role in the immune defense system of humans. Igs recognize, in a specific way, the invading microorganisms and mediating their killing by professional phagocytes or the complement system, or both. Igs consist of antigen-recognizing Fab (fragment antigen-binding) regions, which are linked through a flexible hinge region with the constant Fc (fragment crystallizable) effector region (von Pawel-Rammingen and Björck 2003).
Streptopain [also called streptococcal pyogenic exotoxin B (speB), SCP] is a cysteine protease encoded by the speB gene in the Gram-positive bacterial pathogen Streptococcus pyogenes (Elliott 1945). The enzyme is secreted as a zymogen and autocatalyzes cleavage into the mature SpeB. It is involved in host–pathogen interactions and increases the invasive action of S. pyogenes by hydrolysis of human fibronectin and vitronectin (Kapur et al. 1993). SpeB prevents normal functions of the human immune system by cleavage of M proteins and C5a peptidases. SpeB cleaves the human antibody IgG between residues Gly236 and Gly237 and also other antibodies, such as IgA, IgD, IgE, and IgM (von Pawel-Rammingen and Björck 2003). S. pyogenes also secretes the cysteine protease IdeS (also called IgG-degrading enzyme of S. pyogenes or Mac-1), distinct from streptopain, which cleaves human IgG in the hinge region with a high degree of specificity (von Pawel-Rammingen et al. 2002). IdeS is secreted by S. pyogenes as a mature enzyme of 339 residues and cleaves IgG antibodies bound to bacterial surface structures, thereby inhibiting the killing of pathogen by phagocytic cells. Degradation of IgG by IdeS and SpeB could be complementary under certain conditions (Fig. 4).
Schematic picture showing the action of IdeS and SpeB secreted by S. pyogenes against human IgG antibodies (adapted from von Pawel-Rammingen and Björck 2003) (1) Y-shaped IgG binds to bacterial surface proteins and M surface proteins. Fab (fragment antigen–binding) fragments bound to specific bacterial surface proteins and Fc (fragment crystallizable) fragments bound by M proteins. (2) S. pyogenes secretes IdeS. (3) IdeS cleavages IgG in the hinge region (indicated by red arrows). (4) Unbound Fab and Fc fragments are released. (5) Secretion of SpeB. (6) SpeB degrades bacterial/M surface proteins (indicated by blue arrows). (7) Complexes of bacterial/M surface proteins and Fc and Fab fragments of IgG are released
Already nowadays IdeS has potential medical applications to block arthritis development induced by IgG due to its PRs activity against this immunoglobulin (Nandakumar et al. 2007). Studies in rabbits revealed that IdeS due to its highly selective degradation of IgG, may be used as an immunosuppressant drug used to prevent rejection after renal transplantation. In addition, autoimmune conditions where IgG labels endogenous compounds as pathogenic may be treated by IdeS injected into the circulatory system (Johansson et al. 2008).
Other examples of Igs degradation capacity are, among others, the pathogenic bacteria Neisseria spp., Streptococcus pneumoniae, Ureaplasma urealyticum, and Haemophilus influenza that secrete IgA1 proteases (degrade the heavy chain of human IgA1 in the hinge region) to avoid the host defense mechanisms at sites of infection and to promote disease. These bacterial PRs interfere with the protective functions of the principal mediator of specific immunity on mucosal surfaces, and especially in the upper respiratory tract (Plaut 1983; Poulsen et al. 1998). Gingipains (mentioned above) and staphopains (A and B) are cysteineendopeptidases produced by P. gingivalis and S. aureus, respectively (Table 2). Both bacterial PRs degrade kininogen to produce kinin which induces vascular permeability and promote an influx of plasma-containing nutrients into the site of infection (Imamura et al. 2005). In addition, it is well known that invasion of the systemic circulation by P. aeruginosa across the pseudomonal extracellular PRs production facilitates septicemia as well as toxemia through activation of the bradykinin-generating cascade (Sakata et al. 1996).
S. aureus produces the staphylococcal PRs inhibitors staphostatin A and B, respectively. Similar to the cystatins, the staphostatins interact specifically with their target PRs forming tight and stable noncovalent complexes, staphostatin A with staphopain A and staphostatin B with staphopain B. (Rzychon et al. 2003). The squamous cell carcinoma antigen (SCCA) 1 is an epithelial-derived serpin that inhibit the staphopains in an efficient way (Kantyka et al. 2011). Kantyka and Potempa (2011) indicated that the high association rate constant (k(ass)) for inhibitory complex formation (1.9 × 104 and 5.8 × 104 M−1 s−1 for staphopain A and staphopain B interaction with SCCA1, respectively) argues that SCCA1 can restrain staphopain activity in vivo at epithelial sites colonized by S. aureus.
Production of AMPs and proteins is an important means of host defense in eukaryotes against bacterial pathogens. In fact they are an important part of the innate immune system of all complex organisms as well as some microbes, with antibacterial, antifungal, antiparasitic, and antiviral activity (Jenssen et al. 2006; Guilhelmelli et al. 2013). For example, the antimicrobial activity of the human AMP LL-37 (member of the cathelicidin family of AMPs of the innate immune system) is mediated by disruption of the target cell lipid bilayer via toroidal pore formation, leading to osmotic lysis and cell death (Henzler-Wildman et al. 2004). Nevertheless, several bacterial PRs can inactivate these AMPs by their degradation. In this way, the human pathogens Proteus mirabilis, Group A Streptococci and Enterococcus faecalis have been demonstrated to target and inactivate LL-37 thought PRs secretion (Kindrachuk et al. 2010). Likewise, gingipain R of P. gingivalis and LasB of P. aeruginosa are able to degrade the AMP LL-37 (Schmidtchen et al. 2002; Carlisle et al. 2009). Furthermore, S. aureus can cleave and inactivate LL-37 in a time- and concentration-dependent manner by the production of the metalloproteinase aureolysin (Sieprawska-Lupa et al. 2004).
Other targets of these bacterial PRs are cytokines and chemokines that play an essential role in regulating both immunity adaptive and innate systems. The interaction between bacterial PRs and these small proteins made by host cells in the immune system disturb the communication network in the host which affects the bacterial pathogenicity (Mikolajczyk-Pawlinska et al. 1998; Baba et al. 2002; Leidal et al. 2003; Matheson et al. 2006; Sheets et al. 2008). For example, during the infective process of the human host by P. aeruginosa in lung disease, LasB PRs degrade RANTES and MCP-1 resulting in a loss of chemotactic activity, which suggests that this pathogen may alter the relative amounts of critical immunomodulatory cytokines in the airway (Leidal et al. 2003).
Nevertheless, further research into interaction between bacterial PRs and these host immune communication signals is necessary to extrapolate the in vitro experimental results to the situation in infected tissue. In summary, for the pathogenic bacteria the secretion of bacterial PRs at the site of infection is essential to progress in the infective process of the host, giving them an evolutionary advantage over non-protease-secreting bacteria.
3 Plants as Source of Bacterial PRs Inhibitors
Relying on the fact that PRs play an important role in the protection of plant tissues from pest and pathogen attack; Rakashanda et al. (2012) isolated the serine PRs inhibitor LC-pi I from the seeds of the plant Lavatera cashmeriana. This PRs inhibitor showed strong antibacterial activity against the pathogenic bacteria Klebsiella pneumoniae and P. aeruginosa. These kinds of bacteria can cause urinary tract infection, pneumonia, and septicemia in humans. Besides, the Soap Nut Trypsin inhibitor isolated from Sapindus trifoliatus exhibited antibacterial activity against B. subtilis, S. aureus, E. coli, and Proteus vulgaris (Rachel et al. 2013).
As mentioned above B. anthracis produces de metalloendopeptidase LF during the invective process. The plant derivate curcumin (diferuloylmethane) and its chemically modified 4-phenylaminocarbonylbis-demethoxycurcumin inhibit LF by both increasing its substrate affinity and decreasing its catalytic capacity (Antonelli et al. 2014). Curcumin is the yellow pigment associated with the curry spice of the curcuminoid family that was originally isolated from the Indian spice turmeric (Curcumin longa) being an important component in the daily diet of several Asian countries.
Another source of plant PRs inhibitors is Coccinia grandis, which have antibacterial activity by killing or inhibiting the growth of B. subtillis [minimal bactericidal concentration (MBC) of 1.25 mg⁄mL; minimal inhibitory concentration (MIC) of 1 mg⁄mL], S. aureus (MBC of 1.2 mg⁄mL; MIC of 1 mg⁄mL), E. coli (MBC of 1 mg⁄mL; MIC of 0.63 mg⁄mL), P. vulgaris (MBC of 0.5 mg⁄mL; MIC of 0.2 mg⁄mL), and K. pneumoniae (MBC of 0.5 mg⁄mL; MIC of 0.01 mg⁄mL) (Satheesh and Murugan 2011). The authors purified and isolated the PRs inhibitor and proposed that the mechanisms of action of this natural compound would be the formation of a channel in the bacterial cell membrane that will originate the cell death as a result of the out of the cellular contents.
Finally, in the Brassica chinensis seed there is a napin-like polypeptide with trypsin inhibitor activity. This compound inhibits the bacterial growth of Pseudomonas fluorescens [concentration that inhibits bacterial growth by 50 % (IC50) of 66 μM], Bacillus megaterium (IC50 of 215 μM), Mycobacterium phlei (IC50 of 146 μM), Bacillus cereus (IC50 of 222 μM), and B. subtilis (IC50 of 236 μM) (Ngai and Ng 2004). It becomes clear that further research with these natural PRs inhibitors is necessary to elucidate both its mechanism of action and its possible application from a pharmacological point of view.
4 Conclusions and Future Perspectives
It is well known that animals and plants have been developed defense mechanisms against pathogenic microorganisms (fungi, bacteria, and viruses) by biochemical compounds production. Between these biochemical compounds there is a production of proteins and other metabolites, which inhibit bacterial pathogen-secreted proteins by different mechanisms. They are known as bacterial PRs inhibitors that in some cases could limit host virulence or, in others, should block cell growth.
Targeting virulence proteins with PRs inhibitors to prevent colonization of the host and enable removal of bacteria by the host immune system represents an amazing opportunity to develop new antimicrobial compounds with novel mechanisms of action. Unlike classical antibiotics, it is expected a reduction of the risk of development of resistant strains in a clinical setting. The inhibition of virulence pathways will apply only a mild evolutionary pressure since these pathways are not essential for normal bacterial growth (Rasko and Sperandio 2010).
In fact, MRSA strains, as mentioned previously, or even the totally drug-resistant Mycobacterium tuberculosis (Velayati et al. 2009) make necessary the discovery, that is, a development of new antibiotics to combat these resistances. Nowadays, in the market there are several PRs inhibitors for the treatment of a range of diseases. For example, telaprevir (Incivek; Vertex Pharmaceuticals) and boceprevir (Victrelis; Merck) are PRs inhibitors that target the hepatitis C virus (HCV) NS3-4A protease2,3 (Vermehren and Sarrazin 2011). Another example of PRs inhibitor is the oral anticoagulant rivaroxaban (marketed as Xarelto by Bayer) that inhibits the factor Xa in the coagulation cascade (Eriksson et al. 2008). Nevertheless, this class of enzymes has not yet been exploited in the treatment of bacterial infections notwithstanding the pharmaceutical successes with the above-mentioned PRs modulators.
References
Addinall SG, Holland B (2002) The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J Mol Biol 318:219–236
Antonelli AC, Zhang Y, Golub LM, Johnson F, Simon SR (2014) Inhibition of anthrax lethal factor by curcumin and chemically modified curcumin derivatives. J Enzyme Inhib Med Chem 29:663–669
Baba A, Kadowaki T, Asao T, Yamamoto K (2002) Roles for Arg and Lys-gingipains in the disruption of cytokine responses and loss of viability of human endothelial cells by Porphyromonas gingivalis infection. Biol Chem 383:1223–1230
Bannwarth L, Goldberg AB, Chen C, Turk BE (2012) Identification of exosite–targeting inhibitors of anthrax lethal factor by high-throughput screening. Chem Biol 19:875–882
Becker S, Frankel MB, Schneewind O, Missiakas D (2014) Release of protein A from the cell wall of Staphylococcus aureus. Proc Natl Acad Sci USA 111:1574–1579
Bond C (2015) Antibiotic resistance: turning back the tide. Int J Pharm Pract 23:307–308
Bourhis LL, Werts C (2007) Role of nods in bacterial infection. Microbes Infect 9:629–636
Boyle-Vavra S, Yin S, Challapalli M, Daum RS (2003) Transcriptional induction of the penicillin–binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob Agents Chemother 47:1028–1036
Brakhage AA, Al-Abdallah Q, Tüncher A, Spröte P (2005) Evolution of beta–lactam biosynthesis genes and recruitment of trans-acting factors. Phytochemistry 66:1200–1210
Brotzu G (1948) Ricerche su di un nuovo antibiotico, Lavoratorio dell ́Istituto di Igiene di Cagliari 1–11
Camelo-Castillo A, Benítez-Páez A, Belda-Ferre P, Cabrera-Rubio R, Mira A (2014) Streptococcus dentisani sp. nov., a novel member of the mitis group. Int J Syst Evol Microbiol 64:60–65
Carlisle MD, Srikantha RN, Brogden KA (2009) Degradation of human alpha- and beta-defensins by culture supernatants of Porphyromonas gingivalis strain 381. J Innate Immun 1:118–122
Cathcart GR, Quinn D, Greer B, Harriott P, Lynas JF, Gilmore BF, Walker B (2011) Novel inhibitors of the Pseudomonas aeruginosa virulence factor LasB: a potential therapeutic approach for the attenuation of virulence mechanisms in pseudomonal infection. Antimicrob Agents Chemother 55:2670–2678
Chan LC, Basuino L, Diep B, Hamilton S, Chatterjee SS, Chambers HF (2015) Ceftobiprole– and ceftaroline–resistant methicillin–resistant Staphylococcus aureus. Antimicrob Agents Chemother 59:2960–2963
Cheng J, Chin W, Dong H, Xu L, Zhong G, Huang Y, Li L, Xu K, Wu M, Hedrick JL, Yang YY, Fan W (2015) Biodegradable Antimicrobial polycarbonates with in vivo efficacy against multidrug-resistant MRSA systemic infection. Adv Healthc Mater 4:2128–2136
Cloud-Hansen KA, Hackett KT, Garcia DL, Dillard JP (2008) Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J Bacteriol 190:5989–5994
Clutterbuck PW, Lovell R, Raistrick H (1932) Studies in the biochemistry of the microorganisms. XXVI. The formation from glucose by members of the Penicillium chrysogenum species of a pigment, an alkali soluble protein and penicillin. The antibacterial substance of Fleming. Biochem J 26:1907–1918
Coulthurst SJ, Barnard AML, Salmond GPC (2005) Regulation and biosyn–thesis of carbapenem antibiotics in bacteria. Nat Rev Microbiol 3:295–306
Cowell BA, Twining SS, Hobden JA, Kwong MS, Fleiszig SM (2003) Mutation of lasA and lasB reduces Pseudomonas aeruginosa invasion of epithelial cells. Microbiology 149:2291–2299
Dell’Aica I, Donà M, Tonello F, Piris A, Mock M, Montecucco C, Garbisa S (2004) Potent inhibitors of anthrax lethal factor from green tea. EMBO Rep 5:418–422
Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD (1999) Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181:3981–3993
Drag M, Salvesen GS (2010) Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 9:690–701
Elliott SD (1945) A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type–specific M antigen. J Exp Med 81:573–592
Eriksson BI, Smith H, Yasothan U, Kirkpatrick P (2008) Dabigatran etexilate. Nat Rev Drug Discov 7:557–558
Fernández-Aguado M, Teijeira F, Martín JF, Ullán RV (2013) A vacuolar membrane protein affects drastically the biosynthesis of the ACV tripeptide and the beta–lactam pathway of Penicillium chrysogenum. Appl Microbiol Biotechnol 97:795–808
Fernández-Aguado M, Martín JF, Rodríguez-Castro R, García-Estrada C, Albillos SM, Teijeira F, Ullán RV (2014) New insights into the isopenicillin N transport in Penicillium chrysogenum. Metab Eng 22:89–103
Fersht A (1985) Enzyme structure and mechanism, 2nd edn. W. H. Freeman and Company, New York
Fitzpatrick RE, Wijeyewickrema LC, Pike RN (2009) The gingipains: scissors and glue of the periodontal pathogen, Porphyromonas gingivalis. Future Microbiol 4:471–487
Fleming A (1929) On the antibacterial action of cultures of a penicillium with special reference to their use in the isolation of B. influenza. Br J Exp Pathol 10:226–236
Forino M, Johnson S, Wong TY, Rozanov DV, Savinov AY, Li W, Fattorusso R, Becattini B, Orry AJ, Jung D, Abagyan RA, Smith JW, Alibek K, Liddington RC, Strongin AY, Pellecchia M (2005) Efficient synthetic inhibitors of anthrax lethal factor. Proc Natl Acad Sci USA 102:9499–9504
Fournier B, Philpott DJ (2005) Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev 18:521–540
Frankel MB, Schneewind O (2012) Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J Biol Chem 287:10460–10471
Frère JM, Page MG (2014) Penicillin–binding proteins: evergreen drug targets. Curr Opin Pharmacol 18:112–119
Frick IM, Björck L, Herwald H (2007) The dual role of the contact system in bacterial infectious disease. Thromb Haemost 98:497–502
Fuda C, Suvorov M, Vakulen SB, Mobashery S (2004) The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J Biol Chem 279:40802–40806
Fuda CC, Fisher JF, Mobashery S (2005) Beta-Lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cell Mol Life Sci 62:2617–2633
Gi M, Jeong J, Lee K, Lee KM, Toyofuku M, Yong DE, Yoon SS, Choi JY (2014) A drug-repositioning screening identifies pentetic acid as a potential therapeutic agent for suppressing the elastase-mediated virulence of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:7205–7214
Gohlke H, Klebe G (2002) Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew Chem Int Ed 41:2645–2676
Guilhelmelli F, Vilela N, Albuquerque P, Derengowski Lda S, Silva-Pereira I, Kyaw CM (2013) Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front Microbiol 4:353
Guo Y, Nguyen K-A, Potempa J (2010) Dichotomy of gingipains action as virulence factors: from cleaning substrates with the precision of a surgeon’s knife to a meat chopper–like brutal degradation of proteins. Periodontol 2000 54:15–44
Hamilton-Miller JMT (2000) Sir Edward Abraham’s contribution to the development of the cephalosporins: a reassessment. Int J Antimicrob Agents 15:179–184
Hashimoto M, Ooiwa S, Sekiguchi J (2012) Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D, L-endopeptidase activity at the lateral cell wall. J Bacteriol 194:796–803
Henzler-Wildman KA, Martinez GV, Brown MF, Ramamoorthy A (2004) Perturbation of the hydrophobic core of lipid bilayers by the human antimicrobial peptide LL-37. Biochemistry 43:8459–8469
Hoge R, Pelzer A, Rosenau F, Wilhelm S (2010) Weapons of a pathogen: proteases and their role in virulence of Pseudomonas aeruginosa. In: Mendez–Vilas A (ed) Current research, technology and education topics in applied microbiology and microbial biotechnology, vol 2, pp 383–395. Microbiology book series, Formatex Research Center
Humann J, Lenz LL (2009) Bacterial peptidoglycan degrading enzymes and their impact on host muropeptide detection. J Innate Immun 1:88–97
Imamura T, Potempa J, Tanase S, Travis J (1997) Activation of blood coagulation factor X by arginine–specific cysteine proteinases (gingipain–Rs) from Porphyromonas gingivalis. J Biol Chem 272:16062–16067
Imamura T, Tanase S, Hamamoto T, Potempa J, Travis J (2001) Activation of blood coagulation factor IX by gingipains R, arginine–specific cysteine proteinases from Porphyromonas gingivalis. Biochem J 353:325–331
Imamura T, Tanase S, Szmyd G, Kozik A, Travis J, Potempa J (2005) Induction of vascular leakage through release of bradykinin and a novel kinin by cysteine proteinases from Staphylococcus aureus. J Exp Med 201:1669–1676
Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K (2001) Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin–resistant Staphylococcus aureus. Antimicrob Agents Chemother 45:1323–1336
Jansen WT, Beitsma MM, Koeman CJ, van Wamel WJ, Verhoef J, Fluit AC (2006) Novel mobile variants of staphylococcal cassette chromosome mec in Staphylococcus aureus. Antimicrob Agents Chemother 50:2072–2208
Jenssen H, Hamill P, Hancock RE (2006) Peptide antimicrobial agents. Clin Microbiol Rev 19:491–511
Johansson BP, Shannon O, Björck L (2008) Ides: a bacterial proteolytic enzyme with therapeutic potential. PLoS ONE 3:e1692
Jovetic S, Zhu Y, Marcone GL, Marinelli F, Tramper J (2010) β–Lactam and glycopeptide antibiotics: first and last line of defense? Trends Biotechnol 28:596–604
Kantyka T, Potempa J (2011) Human SCCA serpins inhibit staphylococcal cysteine proteases by forming classic “serpin–like” covalent complexes. Methods Enzymol 499:331–345
Kantyka T, Plaza K, Koziel J, Florczyk D, Stennicke HR, Thogersen IB, Enghild JJ, Silverman GA, Pak SC, Potempa J (2011) Inhibition of Staphylococcus aureus cysteine proteases by human serpin potentially limits staphylococcal virulence. Biol Chem 392:483–489
Kapur V, Topouzis S, Majesky MW, Li LL, Hamrick MR, Hamill RJ, Patti JM, Musser JM (1993) A conserved streptococcus-pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog 15:327–346
Katayama Y, Ito T, Hiramatsu K (2000) A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44:1549–1555
Kelley WL, Jousselin A, Barras C, Lelong E, Renzoni A (2015) Missense mutations in PBP2A Affecting ceftaroline susceptibility detected in epidemic hospital–acquired methicillin-resistant Staphylococcus aureus clonotypes ST228 and ST247 in Western Switzerland archived since 1998. Antimicrob Agents Chemother 59:1922–1930
Kessler E, Safrin M, AbramsWR Rosenbloom J, Ohman DE (1997) Inhibitors and specificity of Pseudomonas aeruginosa LasA. J Biol Chem 272:9884–9889
Kindrachuk J, Nijnik A, Hancock REW (2010) Host defense peptides: bridging antimicrobial and immunomodulatory activities. In: Mander L, Lui H-W (eds) Comprehensive natural products II chemistry and biology. Elsevier, Oxford, pp 175–216
Kirk O, Borchert TV, Fuglsang CC (2002) Industrial enzyme applications. Curr Opin Biotechnol 13:345–351
Komori Y, Nonogaki T, Nikai T (2001) Hemorrhagic activity and muscle damaging effect of Pseudomonas aeruginosa metalloproteinase (elastase). Toxicon 39:1327–1332
Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, Haiech J, Harwood CR, Hènaut A, Hilbert H, Holsappel S, Hosono S, Hullo MF, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee SM, Levine A, Liu H, Masuda S, Mauël C, Médigue C, Medina N, Mellado RP, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park SH, Parro V, Pohl TM, Portelle D, Porwollik S, Prescott AM, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror SJ, Serror P, Shin BS, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Togoni A, Tosato V, Uchiyama S, Vandebol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa HF, Zumstein E, Yoshikawa H, Danchin A (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256
Lantz MS (1997) Are bacterial proteases important virulence factors? J Periodontal Res 32:126–132
Leidal KG, Munson KL, Johnson MC, Denning GM (2003) Metalloproteases from Pseudomonas aeruginosa degrade human RANTES, MCP–1, and ENA–78. J Interferon Cytokine Res 23:307–318
Li F, Chvyrkova I, Terzyan S, Wakeham N, Turner R, Ghosh AK, Zhang XC, Tang J (2012) Inhibition of anthrax lethal factor: lability of hydroxamate as a chelating group. Appl Microbiol Biotechnol 94:1041–1049
Liras P (1999) Biosynthesis and molecular genetics of cephamycins. Antonie Van Leeuwenhoek 75:109–124
Livermore DM (2006) Can beta-lactams be re–engineered to beat MRSA? Clin Microbiol Infect 12(Suppl 2):11–16
Maeda H (1996) Role of microbial proteases in pathogenesis. Microbiol Immunol 40:685–699
Margolin W (2009) Sculpting the bacterial cell. Curr Biol 19:R812–R822
Massimi I, Park E, Rice K, Muller-Esterl W, Sauder D, McGavin MJ (2002) Identification of a novel maturation mechanism and restricted substrate specificity for the SspB cysteine protease of Staphylococcus aureus. J Biol Chem 277:41770–41777
Matheson NR, Potempa J, Travis J (2006) Interaction of a novel form of Pseudomonas aeruginosa alkaline protease (aeruginolysin) with interleukin-6 and interleukin-8. Biol Chem 387:911–915
Mikolajczyk-Pawlinska J, Travis J, Potempa J (1998) Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett 440:282–286
Miyoshi SI, Shinoda S (1997) Bacterial metalloproteases as the toxic factor in infection. J Toxicol Toxin Rev 16:177–194
Moayeri M, Crown D, Jiao GS, Kim S, Johnson A, Leysath C, Leppla SH (2013) Small–molecule inhibitors of lethal factor protease activity protect against anthrax infection. Antimicrob Agents Chemother 57:4139–4145
Morihara K (1964) Production of elastase and proteinase by Pseudomonas aeruginosa. J Bacteriol 88:745–757
Nandakumar KS, Johansson BP, Björck L, Holmdahl R (2007) Blocking of experimental arthritis by cleavage of igg antibodies in vivo. Arthritis Rheum 56:3253–3260
Newman ZL, Sirianni N, Mawhinney C, Lee MS, Leppla SH, Moayeri M, Johansen LM (2011) Auranofin protects against anthrax lethal toxin–induced activation of the Nlrp1b inflammasome. Antimicrob Agents Chemother 55:1028–1035
Ngai PHK, Ng TB (2004) A napin-like polypeptide from dwarf Chinese white cabbage seeds with translation-inhibitory, trypsininhibitory, and antibacterial activities. Peptides 25:171–176
Oda Kohei (2012) New families of carboxyl peptidases: serine-carboxyl peptidases and glutamic peptidases. J Biochem 151:13–25
Olsen I, Potempa J (2014) Strategies for the inhibition of gingipains for the potential treatment of periodontitis and associated systemic diseases. J Oral Microbiol 18:6
Otto HH, Schirmeister T (1997) Cysteine proteases and their inhibitors. Chem Rev 97:133–171
Pascual A, Tan JP, Yuen A, Chan JM, Coady DJ, Mecerreyes D, Hedrick JL, Yang YY, Sardon H (2015) Broad-spectrum antimicrobial polycarbonate hydrogels with fast degradability. Biomacromolecules 16:1169–1178
Plaut AG (1983) The IgA1 proteases of pathogenic bacteria. Annu Rev Microbiol 37:603–622
Pohlmann J, Vasilevich NI, Glushkov AI, Kellenberger L, Shapiro S, Caspers P, Page MG, Danel F (2010) Propenylamide and propenylsulfonamide cephalosporins as a novel class of anti–MRSA beta–lactams. Bioorg Med Chem Lett 20:4635–4638
Poole K (2004) Resistance to beta-lactam antibiotics. Cell Mol Life Sci 61:2200–2223
Popadiak K, Potempa J, Riesbeck K, Blom AM (2007) Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol 178:7242–7250
Potempa J, Pike RN (2009) Corruption of innate immunity by bacterial proteases. J Innate Immun 1:70–87
Potempa J, Banbula A, Travis J (2000) Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontology 24:153–192
Poulsen K, Reinholdt J, Jespersgaard C, Boye K, Brown TA, Hauhe MA (1998) comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect Immun 66:181–190
Rachel KV, Vimala Y, Apta Chaitanya D (2013) A trypsin inhibitor–SNTI with antidandruff activity from Sapindus trifoliatus. Indian J Appl Res 3:3–5
Rakashanda S, Ishaq M, Masood A, Amin S (2012) Antibacterial activity of a Trypsin–chymotrypsin–elastase inhibitor isolated from Lavatera cashmeriana camb. seeds. J Anim Plant Sci 22:983–986
Rani K, Rana R, Datt S (2012) Review on latest overview of proteases. Int J Curr Life Sci 2:12–18
Rao MB, Tanksale AM, Ghatge MS, Deshpande VV (1998) Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 62:597–635
Rasko DA, Sperandio V (2010) Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 9:117–128
Rawlings ND, Barrett AJ, Bateman A (2010) MEROPS: the peptidase database. Nucleic Acids Res 38 (Database issue):D227–33
Rawlings ND, Barrett AJ, Bateman A (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40:D343–D350
Ray A (2012) Protease enzyme-potential industrial scope. Int J Technol 2:1–4
Rice SA, Givskov M, Steinberg P, Kjelleberg S (1999) Bacterial signals and antagonists: the interaction between bacteria and higher organisms. J Mol Microbiol Biotechnol 1:23–31
Rogers HJ, Taylor C, Rayter S, Ward JB (1984) Purification and properties of an autolytic endo-β-N-glucosaminidase and the N–acetylmuramyl-L-alanine amidase from Bacillus subtilis strain 168. J Gen Microbiol 130:2395–2402
Rolain JM, Canton R, Cornaglia G (2012) Emergence of antibiotic resistance: need for a new paradigm. Clin Microbiol Infect 18:615–616
Rzychon M, Sabat A, Kosowska K, Potempa J, Dubin A (2003) Staphostatins: an expanding new group of proteinase inhibitors with a unique specificity for the regulation of staphopains, Staphylococcus spp. cysteine proteinases. Mol Microbiol 49:1051–1066
Sakata Y, Akaike T, Suga M, Ijiri S, Ando M, Maeda H (1996) Bradykinin generation triggered by Pseudomonas proteases facilitates invasion of the systemic circulation by Pseudomonas aeruginosa. Microbiol Immunol 40:415–423
Santos-Beneit F, Martín JF, Barreiro C (2014) Glycopeptides and Bacterial Cell Walls. In: Villa TG, Veiga-Crespo P (eds) Antimicrobial compounds. Springer, Berlin, pp 285–311
Satheesh LP, Murugan K (2011) Antimicrobial activity of protease inhibitors from leaves of Coccinia grandis (L.) Voigt. Indian J Exp Biol 49:366–374
Schaumburg F, Peters G, Alabi A, Becker K, Idelevich EA (2015) Missense mutations of PBP2a are associated with reduced susceptibility to ceftaroline and ceftobiprole in African MRSA. J Antimicrob Chemother. doi:10.1093/jac/dkv325 (first published online October 5)
Scheurwater EM, Pfeffer JM, Clarke AJ (2007) Production and purification of the bacterial autolysin N-acetylmuramoyl-L-alanine amidase B from Pseudomonas aeruginosa. Protein Expr Purif 56:128–137
Scheurwater E, Reid CW, Clarke AJ (2008) Lytic transglycosylases: bacterial space–making autolysins. Int J Biochem Cell Biol 40:586–591
Schmidtchen A, Frick IM, Andersson E, Tapper H, Björck L (2002) Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL–37. Mol Microbiol 46:157–168
Sheets SM, Robles-Price AG, McKenzie RM, Casiano CA, Fletcher HM (2008) Gingipain–dependent interactions with the host are important for survival of Porphyromonas gingivalis. Front Biosci 13:3215–3238
Shilabin AG, Dzhekieva L, Misra P, Jayaram B, Pratt RF (2012) 4–quinolones as noncovalent inhibitors of high molecular mass penicillin-binding proteins. ACS Med Chem Lett 3:592–595
Shoop WL, Xiong Y, Wiltsie J, Woods A, Guo J, Pivnichny JV, Felcetto T, Michael BF, Bansal A, Cummings RT, Cunningham BR, Friedlander AM, Douglas CM, Patel SB, Wisniewski D, Scapin G, Salowe SP, Zaller DM, Chapman KT, Scolnick EM, Schmatz DM, Bartizal K, MacCoss M, Hermes JD (2005) Anthrax lethal factor inhibition. Proc Natl Acad Sci USA 102:7958–7963
Sieprawska-Lupa M, Mydel P, Krawczyk K, Wójcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, McAleese F, Foster T, Travis J, Potempa J (2004) Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother 48:4673–4679
Smith A (2007) Bacterial resistance to antibiotics. In: Denyer SP, Hodges NA, Gorman SP (eds) Hugo and Russell’s pharmaceutical microbiology. Blackwell Science Ltd., Oxford
Smith TJ, Blackman SA, Foster SJ (1996) Peptidoglycan hydrolases of Bacillus subtilis 168. Microb Drug Resist 2:113–118
Smith TJ, Blackman SA, Foster SJ (2000) Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249–262
Song MD, Wachi M, Doi M, Ishino F, Matsuhashi M (1987) Evolution of an inducible penicillin–target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett 221:167–171
Sugai M, Fujiwara T, Komatsuzawa H, Suginaka H (1998) Identification and molecular characterization of a gene homologous to epr (endopeptidase resistance gene) in Staphylococcus aureus. Gene 224:67–75
Tan CM, Therien AG, Lu J, Lee SH, Caron A, Gill CJ, Lebeau-Jacob C, Benton-Perdomo L, Monteiro JM, Pereira PM, Elsen NL, Wu J, Deschamps K, Petcu M, Wong S, Daigneault E, Kramer S, Liang L, Maxwell E, Claveau D, Vaillancourt J, Skorey K, Tam J, Wang H, Meredith TC, Sillaots S, Wang-Jarantow L, Ramtohul Y, Langlois E, Landry F, Reid JC, Parthasarathy G, Sharma S, Baryshnikova A, Lumb KJ, Pinho MG, Soisson SM, Roemer T (2012) Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci Transl Med 4:126ra35
Tenorio EL, Klein BA, Cheung WS, Hu LT (2011) Identification of interspecies interactions affecting Porphyromonas gingivalis virulence phenotypes. J Oral Microbiol 3:8396
Thomas M, Castignetti D (2009) Examination of anthrax lethal factor inhibition by siderophores, small hydroxamates and protamine. J Microbiol Immunol Infect 42:284–289
Tomasz A (1979) The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annu Rev Microbiol 33:113–137
Toney JH, Hammond GG, Leiting B, Pryor KD, Wu JK, Cuca GC, Pompliano DL (1998) Soluble penicillin–binding protein 2a: beta-lactam binding and inhibition by non-betalactams using a 96-well format. Anal Biochem 255:113–119
Travis J, Potempa J (2000) Bacterial proteinases as targets for the development of second generation antibiotics. Biochim Biophys Acta 1477:35–50
Turk S, Verlaine O, Gerards T, Zivec M, Humljan J, Sosič I, Amoroso A, Zervosen A, Luxen A, Joris B, Gobec S (2011) New noncovalent inhibitors of penicillin–binding proteins from penicillin-resistant bacteria. PLoS ONE 6(5):e19418
Ullan RV, Casqueiro J, Banuelos O, Fernandez FJ, Gutierrez S, Martin JF (2002) A novel epimerization system in fungal secondary metabolism involved in the conversion of isopenicillin N into penicillin N in Acremonium chrysogenum. J Biol Chem 277:46216–46225
Ullán RV, Campoy S, Casqueiro J, Fernández FJ, Martín JF (2007) Deacetylcephalosporin C production in Penicillium chrysogenum by expression of the isopenicillin N epimerization, ring expansion, and acetylation genes. Chem Biol 14:329–339
Ullán RV, Teijeira F, Guerra SM, Vaca I, Martín JF (2010) Characterization of a novel peroxisome membrane protein essential for conversion of isopenicillin N into cephalosporin C. Biochem J 432:227–236
van Berkel SS, Nettleship JE, Leung IK, Brem J, Choi H, Stuart DI, Claridge TD, McDonough MA, Owens RJ, Ren J, Schofield CJ (2013) Binding of (5S)-penicilloic acid to penicillin binding protein 3. ACS Chem Biol 8:2112–2116
Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, Ziazarifi AH, Hoffner SE (2009) Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug–resistant tuberculosis or totally drug-resistant strains in iran. Chest 136:420–435
Vermehren J, Sarrazin C (2011) New HCV therapies on the horizon. Clin Microbiol Infect 17:122–134
von Pawel-Rammingen U, Björck L (2003) IdeS and SpeB: immunoglobulin–degrading cysteine proteinases of Streptococcus pyogenes. Curr Opin Microbiol 6:50–55
von Pawel-Rammingen U, Johansson BP, Tapper H, Björck L (2002) IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J 21:1607–1615
Walsh C (2003) Antibiotics: actions, origins, resistance. ASM Press, Washington DC
Wise EM Jr, Park JT (1965) Penicillin: its basic site of action as an inhibitor of a peptide cross-linking reaction in cell wall mucopeptide synthesis. Proc Natl Acad Sci USA 54:75–81
Worthington RJ, Melander C (2013) Overcoming resistance to β–lactam antibiotics. J Org Chem 78:4207–4213
Wu SW, de Lencastre H, Tomasz A (2001) Recruitment of the mecA Gene Homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J Bacteriol 183:2417–2424
Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, Sekiguchi J (2008) The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL–endopeptidase LytF in Bacillus subtilis. Mol Microbiol 70:297–310
Zahrl D, Wagner M, Bischof K, Bayer M, Zavecz B, Beranek A, Ruckenstuhl C, Zarfel GE, Koraimann G (2005) Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455–3467
Zervosen A, Lu WP, Chen Z, White RE, Demuth TP Jr, Frère JM (2004) Interactions between penicillin-binding proteins (PBPs) and two novel classes of PBP inhibitors, arylalkylidene rhodanines and arylalkylidene iminothiazolidin-4-ones. Antimicrob Agents Chemother 48:961–969
Zhanel GG, Lam A, Schweizer F, Thomson K, Walkty A, Rubinstein E, Gin AS, Hoban DJ, Noreddin AM, Karlowsky JA (2008) Ceftobiprole: a review of a broad–spectrum and anti-MRSA cephalosporin. Am J Clin Dermatol 9:245–254
Zhanel GG, Sniezek G, Schweizer F, Zelenitsky S, Lagacé-Wiens PR, Rubinstein E, Gin AS, Hoban DJ, Karlowsky JA (2009) Ceftaroline: a novel broad-spectrum cephalosporin with activity against meticillin-resistant Staphylococcus aureus. Drugs 69:809–831
Zindel S, Kaman WE, Fröls S, Pfeifer F, Peters A, Hays JP, Fuchsbauer HL (2013) The papain inhibitor (SPI) of Streptomyces mobaraensis inhibits bacterial cysteine proteases and is an antagonist of bacterial growth. Antimicrob Agents Chemother 57:3388–3391
Acknowledgments
The authors want to thank the European Union (FP7-ENVIRONMENT 2012-two-stage (BIOCORIN, project reference: 28288) and the Spanish Ministry of Economy and Competitively (Madrid, Spain) [Subprogramme for Non-Guided Fundamental Research Projects 2012 (FAES, project reference: CTM2012-320269)].
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Ullán, R.V., Barreiro, C. (2016). Bacterial Proteases as Targets to Control Bacterial Growth. In: Villa, T., Vinas, M. (eds) New Weapons to Control Bacterial Growth. Springer, Cham. https://doi.org/10.1007/978-3-319-28368-5_7
Download citation
DOI: https://doi.org/10.1007/978-3-319-28368-5_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-28366-1
Online ISBN: 978-3-319-28368-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)