Abstract
This chapter deals with differences among fungi to solve specific metabolic problems. Although metabolic studies have mainly been performed on Ascomycetes, it is shown that even closely related species developed different solutions to cope with specific nutritional conditions. Especially, fungal morphology (yeast versus hyphae), the natural nutritional niches, gene loss and acquisition and organismal interaction led to a very large variety of metabolic pathways. To exemplify these variations, an emphasis is given on the degradation of the toxic metabolic intermediate propionyl-CoA, the difference in utilisation of gluconeogenic nutrient sources among yeasts and filamentous fungi, the ability of using histidine as a nitrogen source and the biosynthesis of the amino acid lysine that also provides the essential precursor alpha-aminoadipate for synthesis of penicillins and cephalosporins.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
I. Introduction
Sensing, uptake and metabolism of available nutrients is a prerequisite for all living organisms in order to proliferate, compete and maintain in the environment. Besides bacteria, fungi play a major role in carbon and nitrogen cycling by utilising plant-derived organic matter for growth. A large number of fungi are found as free-living saprophytes in the soil, and these species are generally able to synthesise all 20 proteinogenic amino acids de novo and do not depend on the external supply of vitamins. On the other hand, some fungi have specifically adapted to a pathogenic lifestyle infecting plants, insects or mammals. Although pathogenic adaptations may have led to the development of specific infection strategies that are unique to the pathogenic species, these adaptations can also lead to the loss of certain metabolic capacities that must be compensated by nutrient acquisition from the infected host. Extreme examples for such an adaptation are the Microsporidia, which are obligate intracellular pathogens that can infect a wide variety of animals from different phyla (Cuomo et al. 2012). Microsporidia seem to form the earliest branching clade within the sequenced fungi and have adapted to modulate the infected host cells to provide essential nutrients. These pathogens express specific nucleotide transporters to acquire these essential components from the host cells, because they have lost the capacity of de novo nucleotide synthesis. Additionally, they export hexokinases to the host cell, which most likely leads to activation of host storage sugars and increases the production of building blocks that are used by the pathogen for rapid proliferation (Cuomo et al. 2012). Another, but less dramatic form of adaptation that impacts cata- and anabolism is the synergism of fungi with phototrophic organisms such as algae or plants. In case of a synergism with algae - green algae and/or cyanobacteria - the resulting species are called lichens, which are able to colonise extreme environments (Stocker-Wörgötter 2008). However, in contrast to the Microsporidia, several fungal species identified from lichens can still be cultivated without their interacting partner (Crittenden et al. 1995). Specialised fungi that grow in synergistic association with plant roots form a so-called mycorrhiza. Depending on the mode of interaction with plant roots, mycorrhizae can be subdivided into endo- or arbuscular mycorrhizae and the ectomycorrhiza (Balestrini and Bonfante 2014). However, regardless of the detailed interaction, the common feature is that the fungus obtains sugars from plant photosynthesis and delivers inorganic substrates such as nitrogen and phosphorous to the plant cell (Nehls et al. 2007; Fellbaum et al. 2014). In this respect, especially those fungi forming an endomycorrhiza are frequently unable to proliferate without the direct interaction with their host or synergistic partner, which also results in adapted regulation of fungal metabolism that changes in contact with the host (Trepanier et al. 2005).
The way of nutrient acquisition and metabolic properties makes fungi also interesting for a wide variety of technologic applications. Small nutritive molecules such as sugars can easily diffuse through the fungal cell wall and penetrate the membrane by specific transporters. In contrast, large and insoluble polymers cannot be consumed by this uptake mechanism. Thus, fungi that degrade plant material need to secrete enzymes that decompose polymers like cellulose, pectin or lignin outside the cell (Hori et al. 2013). The resulting monomers from these hydrolyses are then taken up and metabolised. Therefore, fungi are interesting organisms for the production of cellulases, xylanase, pectinases, amylases and other hydrolysing enzymes (Chi et al. 2009; Abdeljalil et al. 2013; Tiwari et al. 2013; Tu et al. 2013). Furthermore, besides secreted enzymes some of the metabolic intermediates that are secreted by a variety of fungi are of major importance in our daily life. Yeasts, with the prominent example of baker’s yeast Saccharomyces cerevisiae, tend to rapidly metabolise glucose by fermentation accompanied by the secretion of ethanol, acetate and glycerol (Woo et al. 2014). In contrast to yeasts, filamentous fungi tend to prefer complete substrate oxidation to carbon dioxide. However, under certain conditions some filamentous fungi secrete large amounts of organic acids such as the citric acid producer Aspergillus niger (Papagianni 2007) or the itaconic acid producer Aspergillus terreus (Klement and Buchs 2013).
In conclusion, despite a sometimes very close phylogenetic relationship, the metabolic capacities of fungi show great diversity. Even more, some fungi have developed different metabolic pathways or strategies to make a given nutrient available. In this chapter selected metabolic pathways will be introduced that provide examples for metabolic diversity in fungi.
II. Propionyl-CoA, A Common Metabolic Intermediate
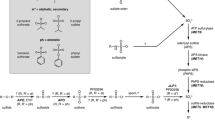
Propionyl-CoA is an activated short-chain fatty acid that can derive from the direct activation of propionate, the degradation of odd-chain fatty acids, the side chain of cholesterol and some amino acids such as isoleucine, valine and methionine (Maerker et al. 2005; Ballhausen et al. 2009). Interestingly, propionic acid, its potassium, calcium and sodium salts are used as preservatives (Coblentz et al. 2013). Especially feed of cattle is frequently enriched with propionate. This has two reasons. First, due to its ability to reduce growth of fungi, the addition of propionate prevents feed from getting mouldy and, thus, reduces the amount of toxins that is produced by several fungal species (Marin et al. 1999; Alam et al. 2010). Second, addition of propionate prevents or reduces metabolic ketoacidosis in cattle and sheep (Emmanuel and Kennelly 1984; Bigner et al. 1997). The reason for this is the ability of mammals to convert activated propionate via the methylmalonyl-CoA pathway into succinyl-CoA, an intermediate of the citric acid cycle with its end product oxaloacetate (Aschenbach et al. 2010). Oxaloacetate is an essential acceptor molecule for acetyl-CoA units that derive from β-oxidation of fatty acids. Additionally, oxaloacetate is required for gluconeogenesis to produce sugar molecules and is, thus, removed from the citric acid cycle and needs to be regenerated. Since mammals cannot produce oxaloacetate de novo from acetyl-CoA units, a lack of oxaloacetate can lead to severe ketoacidosis due to an arrest of fatty acid oxidation (Aschenbach et al. 2010). Thus, feeding of propionate can replenish the pool of oxaloacetate. In the methylmalonyl-CoA pathway, propionate is first carboxylated to yield S-methylmalonyl-CoA, which is subsequently converted into R-methylmalonyl-CoA via a specific racemase (Fig. 14.1). Then, a rearrangement of the carbon skeleton is required to convert the methylmalonyl-CoA into succinyl-CoA. This reaction is performed via a radical mechanism that is catalysed by the methylmalonyl-CoA mutase in the presence of the essential cofactor 5′deoxyadenosylcobalamin (coenzyme B12) (Takahashi-Iniguez et al. 2012). Interestingly, despite the importance of propionyl-CoA degradation pathways, mammals do not possess genes to produce the cofactor and depend on the uptake of vitamin B12 from the diet (Jiang et al. 2013). Similarly, fungi are unable to produce coenzyme B12, and a sufficient uptake from the environment is unlikely. Moreover, enzymatic activity determinations (Ledley et al. 1991) and genome analyses show that no gene for a methylmalonyl-CoA mutase is present (Otzen et al. 2014). Thus, since propionate is an abundant carbon source in the soil environment and propionyl-CoA is a common metabolic intermediate, alternative pathways for propionyl-CoA degradation need to exist.
Scheme of propionyl-CoA degradation pathways. Left: the methylmalonyl-CoA pathway leading to the formation of succinyl-CoA. This pathway is not present in fungi due to its dependence on coenzyme B12 as a cofactor in methylmalonyl-CoA mutase. 1 propionyl-CoA carboxylase, 2 methylmalonyl-CoA racemase, 3 methylmalonyl-CoA mutase. Centre: the methylcitrate cycle leading to the formation of pyruvate. This cycle is the main pathway for propionyl-CoA degradation in Ascomycetes and Basidiomycetes. 4 methylcitrate synthase, 5 methylcitrate dehydratase, 6 aconitase (methylisocitrate dehydratase), 7 methylisocitrate lyase. For details on the de- and rehydration reactions of (5) and (6), refer to the main text. Right: the 3-hydroxypropionate pathway leading to acetyl-CoA. This pathway is present in yeasts of the CUG clade. 8 acyl-CoA oxidase, 9 enoyl-CoA hydratase, 10 3-hydroxypropionyl-CoA hydrolase, 11 3-hydroxypropionate dehydrogenase, 12 malonate semialdehyde dehydrogenase
The first report for such an alternative pathway derived from investigations on the yeast Yarrowia lipolytica that was grown on n-paraffins (Tabuchi and Hara 1974). A mutant of this yeast accumulated methylisocitrate in the growth medium, and the so-called methylcitrate cycle was postulated. Although enzymatic activities of some of the enzymes required for a methylcitrate cycle had been identified (Tabuchi and Uchiyama 1975), the genome sequence of Y. lipolytica was not available, and several questions on this pathway remained unaddressed. However, since this pathway was also detected in some bacteria, subsequent analyses on fungi and bacteria led to a more complete picture on the degradation of propionyl-CoA via this cycle.
A. Degradation of Propionyl-CoA via the Methylcitrate Cycle
First, we will start with a short overview of the reactions of the methylcitrate cycle (Fig. 14.1). It is now well established that the methylcitrate cycle starts with the condensation of propionyl-CoA with oxaloacetate to yield methylcitrate (Brock et al. 2000; Domin et al. 2009; Chittori et al. 2010; Kobayashi et al. 2013). This first intermediate undergoes a two-step de- and rehydration reaction to form methylisocitrate (Horswill and Escalante-Semerena 2001; Brock et al. 2002), which is subsequently cleaved into succinate and pyruvate and, thus, resembles an α-oxidation of pyruvate (Brock et al. 2001).
In bacteria, genes of the methylcitrate cycle are organised in a so-called prp operon from which at least two different main types exist. The first type is found in bacteria such as Escherichia coli or Salmonella enterica (Textor et al. 1997; Tsang et al. 1998). It contains genes for the regulator PrpR, the methylisocitrate lyase PrpB, the methylcitrate synthase PrpC, the methylcitrate dehydratase PrpD and the propionyl-CoA synthetase PrpE. The second class of bacterial prp operons contains a gene substitution in which the prpD gene is replaced by two other genes called prpF and acnD. This type is present, for example, in Vibrio cholerae or Shewanella oneidensis (Grimek and Escalante-Semerena 2004).
This gene replacement is of interest, because it indicates a special problem in the dehydration reaction of methylcitrate. Methylcitrate synthase produces (2S, 3S)-2-methylcitrate (Brock et al. 2000) and the product of an PrpD-catalysed dehydration reaction is cis-2-methylaconitate (Brock et al. 2002). Thus, a dehydrating enzyme must perform a syn-elimination of water for the direct generation of cis-2-methylaconitate (Fig. 14.1). However, β-eliminations of water, in which a proton is removed adjacent to a carboxylate anion, generally follow an antistereochemical mechanism (Creighton and Murthy 1990; Brock et al. 2000). Therefore, the methylcitrate dehydratase might catalyse two reactions: (1) the elimination of water from the C3–C4 bond of (2S, 3S)-2-methylcitrate, leading to γ-methylaconitate, which is (2) subsequently isomerised via a Δ-isomerase activity into cis-2-methylaconitate. Although experimental evidence for such a mechanism is lacking, the replacement of the prpD gene by prpF and acnD in some bacterial prp-operons strongly points to a common dehydration/isomerisation mechanism. AcnD is an aconitase like enzyme that performs the dehydration of (2S, 3S)-2-methylcitrate leading to a methylaconitate molecule with yet unknown stereochemistry (Grimek and Escalante-Semerena 2004). However, it is unlikely that cis-2-methylaconitate is formed by this reaction, since AcnD alone cannot replace PrpD in S. enterica and does not generate a product that can be used by a citric acid cycle aconitase for rehydration. Since PrpF was shown to isomerise trans-aconitate into cis-aconitate, it can be assumed that PrpF acts as an isomerase on a trans-methylaconitate isomer that might be produced by AcnD to yield cis-2-methylaconitate (Garvey et al. 2007).
In fungi the methylcitrate cycle seems to follow the E. coli and S. enterica reaction mechanisms. A methylcitrate synthase (McsA) that is specifically induced in the presence of propionyl-CoA-generating nutrient sources initially forms methylcitrate from propionyl-CoA and oxaloacetate as shown for several fungi such as Aspergillus nidulans (Brock et al. 2000), Aspergillus fumigatus (Maerker et al. 2005), Fusarium species (Domin et al. 2009) and Saccharomyces cerevisiae (Graybill et al. 2007). The methylcitrate stereoisomer formed in this reaction is (2S, 3S)-2-methylcitrate as determined for the bacterial pathway. Since a deletion of the mcsA gene in Aspergillus spp. leads to accumulation of propionyl-CoA with severe disturbance of primary and secondary metabolism (Brock and Buckel 2004; Zhang et al. 2004), no alternative pathways for propionyl-CoA degradation seem to exist in these fungi. From an evolutionary point of view, methylcitrate synthases may have evolved from the citric acid cycle citrate synthase, because gene and protein alignments reveal >50 % sequence identity (Brock et al. 2000).
The formation of methylcitrate is followed by de- and rehydration reactions. Although hardly investigated in fungi, preliminary analyses indicate that the dehydration is performed by a methylcitrate dehydratase (McdA) that possesses 50–60 % sequence identity to PrpD from E. coli (unpublished). Interestingly, except for a few itaconate-producing fungi such as Aspergillus terreus and Aspergillus itaconicus that possess a cis-aconitate decarboxylase with sequence similarity to methylcitrate dehydratases (Kanamasa et al. 2008; Steiger et al. 2013), no other enzymes similar to McdA are encoded in fungal genomes. This indicates that fungal methylcitrate dehydratases were directly acquired from a bacterial source. The subsequent rehydration of cis-2-methylaconitate is most likely performed by a citric acid cycle aconitase, as shown for E. coli and S. enterica (Horswill and Escalante-Semerena 2001; Brock et al. 2002). This is possible, because cis-2-methylaconitate fits into the active site of aconitases when bound in the isocitrate mode resulting in (2R, 3S)-2-methylisocitrate (Lauble and Stout 1995).
The final pathway-specific reaction is the cleavage of (2R, 3S)-2-methylisocitrate into succinate and pyruvate by a methylisocitrate lyase. Succinate can be used to replenish the oxaloacetate pool, whereas pyruvate can be used for gluconeogenesis or energy metabolism. Phylogenetic analyses of fungal methylisocitrate lyases have shown that these enzymes likely evolved from the isocitrate lyase of the glyoxylate cycle (Müller et al. 2011) that cleaves isocitrate into succinate and glyoxylate (for further details of the glyoxylate cycle, see below and Fig. 14.2). However, the bacterial methylisocitrate lyases only share low sequence identity to isocitrate lyases and fungal methylisocitrate lyases, indicating an independent evolution of bacterial and fungal methylisocitrate lyases (Müller et al. 2011). Nevertheless, the structural fold of methylisocitrate lyases appears to be conserved (Liu et al. 2005), and a specific active site motif required to harbour the methyl group of methylisocitrate is conserved in these independently evolved enzymes (Müller et al. 2011). This shows that nature developed the same enzymatic solution for a specific metabolic problem.
Scheme of the glyoxylate cycle and its connection to gluconeogenesis. The glyoxylate cycle uses acetyl-CoA units for the anaplerosis of oxaloacetate. Acetyl-CoA condenses with oxaloacetate to form citrate, which is isomerised to isocitrate. The glyoxylate cycle-specific isocitrate lyase (ICL) cleaves isocitrate into succinate and glyoxylate. Succinate is regenerated by citric acid cycle enzymes to malate. Glyoxylate is condensed via malate synthase (MLS) to malate, which is oxidised to oxaloacetate and can enter gluconeogenesis (grey box). Green part in centre: yeasts decarboxylate cytosolic pyruvate via pyruvate decarboxylase (PDC) to acetyl-CoA. To enter gluconeogenesis, acetyl-CoA must be converted to oxaloacetate via the glyoxylate cycle. In filamentous fungi, pyruvate carboxylase (PC) carboxylates pyruvate to oxaloacetate, which can directly enter gluconeogenesis without the need for the glyoxylate cycle
B. Alternative Propionyl-CoA Degradation Pathways in Fungi
The methylcitrate cycle appears as the main metabolic pathway in fungi to cope with the metabolic intermediate propionyl-CoA. Key enzymes such as the methylcitrate synthase can be found encoded in genomes of Basidiomycetes and most Ascomycetes (Müller et al. 2011). However, by searching the currently available genomes of Zygomycetes, no homologous genes are detected. This indicates either that the fungal methylcitrate cycle has specifically evolved in a common ancestor of Basidiomycetes and Ascomycetes after the branch from the Zygomycetes or that the genes have been completely lost in the latter groups. Thus, it has remained unclear by which mechanisms Zygomycetes metabolise propionyl-CoA. However, even more surprisingly, the so-called CUG clade of the Saccharomycetales, among them the important human pathogen Candida albicans, is also lacking methylcitrate cycle genes, and no activities for methylcitrate cycle enzymes can be detected from cell-free extracts (Otzen et al. 2014).
Nevertheless, C. albicans is able to grow, though at low rates, on propionate and its growth is well supported on the odd-chain fatty acid valerate that is composed of five carbon units (Otzen et al. 2014). A valerate degradation via β-oxidation leads to equal amounts of acetyl- and propionyl-CoA. Since an intracellular accumulation of propionyl-CoA would interfere with other enzymatic reactions dealing with CoA-ester substrates such as the pyruvate dehydrogenase complex or the succinyl-CoA synthetase, an efficient removal of propionyl-CoA is also essentially required (Brock and Buckel 2004).
An alternative pathway for propionyl-CoA degradation had been suggested for some insects and plants that proceeds via the key intermediate 3-hydroxypropionate and finally ends in the formation of acetyl-CoA (Halarnkar and Blomquist 1989). However, experimental evidence for the existence of the respective enzymes was mostly lacking. This has recently changed by analysing the proteome and transcriptome of C. albicans cells grown on propionate or valerate medium (Otzen et al. 2014). As depicted in Fig. 14.1, it is assumed that propionyl-CoA initially enters the normal pathway of β-oxidation where a fatty acid oxidase generates acryloyl-CoA. In the next step acryloyl-CoA is hydrated by the bifunctional enoyl-CoA hydratase/dehydrogenase yielding 3-hydroxypropionyl-CoA. This intermediate is transported from peroxisomes to mitochondria, where the specific enzymes of the 3-hydroxypropionate pathway are located. A hydroxypropionyl-CoA hydrolase cleaves the CoA-ester, which leads to the intermediate 3-hydroxypropionate. This key intermediate is oxidised to malonate semialdehyde as shown by enzymatic characterisation of the essential 3-hydroxypropionate dehydrogenase. Additionally, a 3-hydroxypropionate dehydrogenase deletion mutant accumulates significant amounts of 3-hydroxypropionate when grown on acetate/propionate or valerate containing medium (Otzen et al. 2014). Finally, malonate semialdehyde is oxidatively decarboxylated to yield acetyl-CoA via an aldehyde dehydrogenase.
Although the 3-hydroxypropionate pathway is very elegantly used to degrade propionyl-CoA and to provide mitochondrial acetyl-CoA, it also causes a severe problem for C. albicans. While filamentous Ascomycetes are able to transport mitochondrial acetyl-CoA into the cytosol by using an export of citrate with subsequent cytosolic acetyl-CoA formation via ATP citrate lyase, this latter enzyme is not present in C. albicans (Hynes and Murray 2010). Therefore, acetyl-CoA is trapped within mitochondria and can only be used for energy metabolism but not for anabolic processes. This may explain the very low growth rate of C. albicans on propionate. On the other hand, growth on valerate is well supported.
Valerate first undergoes one round of β-oxidation, which leads to the formation of peroxisomal acetyl-CoA, whereas the resulting propionyl-CoA forms mitochondrial acetyl-CoA. The peroxisomal acetyl-CoA can now be used for biosynthetic purposes via conversion by the malate forming glyoxylate cycle, whereas the mitochondrial acetyl-CoA is available for energy metabolism. Thus, the modified β-oxidation with a branch into the 3-hydroxypropionate pathway can be efficiently used for the utilisation of propionyl-CoA from fatty acids, but appears less suitable for the utilisation of propionate. However, the yeast S. cerevisiae appears, from a phylogenetic and metabolic point of view, more closely related to C. albicans than to filamentous Ascomycetes but metabolises propionate via the methylcitrate cycle. Nevertheless, it should be mentioned that in contrast to filamentous fungi, S. cerevisiae can only metabolise propionate in co-metabolism with limited amounts of glucose and cannot use propionate as sole carbon source (Pronk et al. 1994). Thus, at least two questions remain open: (1) Is there a specific advantage of using either the methylcitrate cycle or the 3-hydroxypropionate pathway? (2) At which time did the fungi from the CUG clade lose the genes of a functional methylcitrate cycle? An answer to this question might derive from studies on Zygomycetes. Analyses of currently available genomes imply that genes for a methylcitrate cycle are lacking, but there is no experimental proof for the existence of a modified β-oxidation pathway via 3-hydroxypropionate. A survey on currently available zygomycete genomes is provided in Chap. 15. Therefore, Zygomycetes either developed a third fungal, yet unknown, pathway for propionyl-CoA degradation or they are using the C. albicans pathway. If so, the 3-hydroxypropionate pathway would be more ancient than the methylcitrate cycle. This means that modern Basidiomycetes and Ascomycetes lost this ancient pathway and developed the methylcitrate cycle, which might make them more independent of fatty acid β-oxidation and produces the more versatile intermediate pyruvate rather than acetyl-CoA.
III. Growth on Gluconeogenic Substrates
Sugars are of major importance for fungal growth and development. The fungal cell wall is mainly composed of a mixture of glucans (mainly β-1,3- and β-1,6-glucan), chitin, chitosan and glycosylated proteins (Free 2013). For more details on the fungal cell wall please refer to Chaps. 8 and 9. Additionally, sugars are required for nucleotide synthesis and for the provision of building blocks for amino acid synthesis. Therefore, when cultivated on sugar-free media, fungi require the de novo synthesis of glucose via gluconeogenesis. The common intermediate to initiate gluconeogenesis is pyruvate, which is carboxylated to the energy-rich intermediate oxaloacetate that is required to circumvent the irreversible pyruvate kinase reaction from glycolysis (Hers and Hue 1983).
As outlined above, propionyl-CoA degradation via the methylcitrate cycle generates pyruvate, which is directly suitable for gluconeogenesis. In contrast, the 3-hydroxypropionate pathway forms acetyl-CoA, which cannot serve as a direct gluconeogenic precursor. Similarly, degradation of fatty acids via β-oxidation or direct activation of acetate yields acetyl-CoA. The ability of using acetyl-CoA as a gluconeogenic precursor requires the action of a specific metabolic pathway, the so-called glyoxylate cycle (Fig. 14.2). In this cycle two acetyl units are condensed to yield malate, which is suitable for gluconeogenesis (Peraza-Reyes and Berteaux-Lecellier 2013). In humans genes for a functional glyoxylate cycle are lacking. Therefore, they are unable to survive for a longer period with fatty acids as a sole carbon and energy source although fatty acids are degraded to acetyl-CoA units that serve for energy metabolism. Thus, additional nutrients such as proteins are required to replenish the pool of biosynthetic precursors during fat-burning starvation periods frequently resulting in the degradation of muscle-derived proteins.
In contrast to humans, the glyoxylate cycle is common to most fungi, which allows them to live at the expense of fatty acids as sole carbon and energy source (Dunn et al. 2009). However, a prominent exception is the model yeast Schizosaccharomyces pombe. Although a gene for isocitrate lyase, which is one of the two key enzymes of the cycle, can be identified by BLAST analyses, its sequence seems to have accumulated several point mutations that might have resulted in a loss of function (Müller et al. 2011). In agreement, no glyoxylate cycle enzyme activity is detected in S. pombe and acetate is only utilised in the co-metabolism with glucose (Tsai et al. 1989). However, isocitrate lyase is detected in Chytridiomycota, Zygomycetes, Basidiomycetes and Ascomycetes showing that this cycle is very ancient in the fungal lineage. However, even among Ascomycetes the regulation, importance and use of the glyoxylate cycle differ as will be outlined below.
A. Utilisation of the Glyoxylate Cycle in Ascomycetes
The glyoxylate cycle branches from the citric acid cycle and is characterised mainly by two key enzymes: isocitrate lyase (ICL) and malate synthase (MLS) (Dunn et al. 2009). Similar to the citric acid cycle, acetyl-CoA initially condenses with oxaloacetate to form citrate, which is subsequently converted into isocitrate via aconitase de- and rehydration reactions. While the citric acid cycle would normally continue with the first decarboxylation reaction that yields α-ketoglutarate, isocitrate lyase mediates a C–C bond cleavage on isocitrate resulting in the products glyoxylate and succinate (Fig. 14.2). While succinate is regenerated to oxaloacetate in order to replenish the substrate for the initial citrate synthase reaction, glyoxylate undergoes a condensation with a second molecule of acetyl-CoA. This reaction is mediated by MLS and leads to malate, which is easily oxidised to oxaloacetate. Thus, from two acetyl units, a new molecule of oxaloacetate is formed that can be used for biosynthetic purposes. It should be mentioned that, in contrast to the mitochondrial citric acid cycle, the fungal glyoxylate cycle is generally located within peroxisomes (Peraza-Reyes and Berteaux-Lecellier 2013). This requires additional shuttling of molecules over compartmental membranes, which will not be discussed in detail.
In general, activation of expression of the glyoxylate cycle genes is under control of carbon catabolite repression, which means that glyoxylate cycle genes are not transcribed or rapidly degraded in the presence of glucose (De Lucas et al. 1994a, b). In Aspergillus spp. the main carbon catabolite repressor CreA binds to the promoter region of the target genes and interferes with initiation of gene transcription (Strauss et al. 1999). Similarly, in the yeast S. cerevisiae, glyoxylate cycle genes are under negative control of the carbon catabolite repressor Mig1p (Turcotte et al. 2010).
In the absence of glucose, the carbon catabolite repressor does no longer bind to target promoter sequences. This enables transcription factors such as the glyoxylate cycle-specific transcription factor FacB in A. nidulans (Todd et al. 1998) or the more global gluconeogenic transcription factors Cat8p, Sip4p, Adr1p and Rds2p in S. cerevisiae (Soontorngun et al. 2012) to activate gene expression. Besides this rather general carbon control mechanism of gene regulation, the dimorphic pathogenic fungus Paracoccidioides brasiliensis follows a different strategy to regulate activation and inactivation of glyoxylate cycle genes.
P. brasiliensis grows as a filamentous fungus at temperatures below 26 °C, but switches into yeast growth at elevated temperatures. Regulation of the glyoxylate cycle in the filamentous form follows the regulation mechanisms as seen for Aspergillus spp. However, regardless of the available nutrient source, glyoxylate cycle genes are constitutively transcribed when cells are growing in the yeast form.
Interestingly, although ICL, as a marker for the glyoxylate cycle, can be detected by Western blot on all nutrient sources, ICL activity is only detected on acetyl-CoA generating nutrient sources, but not on glucose (Cruz et al. 2011). Two-dimensional gel electrophoresis revealed an extensive phosphorylation of ICL on glucose, which was strongly reduced when yeast cells were shifted to acetate medium. In addition, dephosphorylation of ICL that derived from glucose medium results in strongly increased activity, indicating that a phosphorylation/dephosphorylation mechanism is responsible for glyoxylate cycle regulation in P. brasiliensis yeast cells (Cruz et al. 2011).
Besides these different strategies to regulate the activity of the glyoxylate cycle, the impact of this cycle on various nutrient sources also differs among fungi. In Aspergillus spp., the glyoxylate cycle is essential when cells are growing on nutrients exclusively producing acetyl-CoA such as ethanol, acetate, acetamide or fatty acids, and the cycle is specifically induced by FacB (Todd et al. 1997). However, the pathway is dispensable when growing on peptone or most amino acids since sufficient pyruvate is formed that can be carboxylated to oxaloacetate for gluconeogenic purposes (Brock 2009). However, this situation differs in yeasts as shown for C. albicans. Here, isocitrate lyase is induced in the presence of all gluconeogenic nutrient sources and during carbon starvation. Additionally, a C. albicans ICL mutant is unable to grow on most gluconeogenic nutrient sources, implying that the glyoxylate cycle is directly linked with gluconeogenesis (Brock 2009). How is this explained?
First, transcription of the glyoxylate cycle genes in yeasts seems to be tightly linked with transcription of genes from gluconeogenesis (Turcotte et al. 2010). In contrast, this regulation is separated in Aspergillus spp. Here, a heterodimer of transcription factors AcuM and AcuK is responsible for activation of gluconeogenesis. AcuM and AcuK are always activated in the absence of glucose, independent of an acetyl-CoA-generating nutrient source (Suzuki et al. 2012), and these transcription factors are insufficient to induce ICL and MLS. This requires FacB, the glyoxylate cycle-specific transcription factor that is only induced in the presence of nutrients generating acetyl-CoA.
Another difference between yeasts and filamentous fungi is the completeness of substrate oxidation. Filamentous fungi generally rely on complete substrate oxidation for gain of energy. This requires the transport of pyruvate and acetyl-CoA to mitochondria where oxidative phosphorylation takes place. In contrast, yeasts prefer fermentation (Tylicki et al. 2008). Thus, on glucose yeasts produce ethanol from pyruvate. Pyruvate is decarboxylated in the cytoplasm via the pyruvate decarboxylase to acetaldehyde, which is either reduced to ethanol or oxidised to acetate (Fig. 14.2). The key enzyme pyruvate decarboxylase is highly active on glucose, but expression analyses with a luciferase reporter in C. albicans revealed that the gene is also strongly transcribed on gluconeogenic substrates (Jacobsen et al. 2014). Thus, cytosolic pyruvate, deriving both from glycolysis and also from the degradation of gluconeogenic substrates, is rapidly decarboxylated to acetaldehyde and might not be amenable to carboxylation by a mitochondrial pyruvate carboxylase. By this strategy large amounts of cytosolic acetyl-CoA can be generated, which makes an ATP citrate lyase dispensable. However, in turn, it requires the glyoxylate cycle to generate oxaloacetate from acetyl-CoA units for gluconeogenesis.
In conclusion, the glyoxylate cycle appears well conserved among fungi, but its regulation and use differ among fungal species and are dependent on the set of enzymatic activities from accompanying metabolic pathways.
IV. Amino Acid Biosynthesis and Utilisation as Nutrient Sources
Most fungi have specifically adapted to sense, transport and utilise amino acids, as de novo synthesis of amino acids is very energy consuming. Additionally, amino acids can provide the cell with important macroelements such as nitrogen and sulphur and can serve as carbon sources. Since environmental amino acids are frequently bound in proteins that are too large for direct import, fungi are rich in extracellular proteases and peptidases that cleave proteins in smaller subunits that can be taken up by various peptide and amino acid transporters (Yike 2011). Besides that, proteases and peptidases can support the virulence of pathogenic fungi, because these enzymes not only provide nutrients from the host environment but also ease tissue penetration, inactivate components of the immune system or cause inflammation and allergies (Gropp et al. 2009; Yike 2011).
The uptake of amino acids as nitrogen sources in fungi follows a specific preference. The nitrogen-rich amino acids glutamine and arginine as well as ammonia generally constitute preferred nitrogen sources in fungi, whereas other amino acids, urea, purines, nitrate and nitrite are less preferred. Thus, similar to carbon catabolite repression, a nitrogen catabolite repression system is involved in sensing and utilisation of the available nitrogen sources. A key player in this regulatory path is a GATA-type transcription factor. One of the best studied members is AreA from Aspergillus spp. (Gonzalez et al. 1997), which has close relatives in other fungi such as NIT-2 from Neurospora crassa (Tao and Marzluf 1999) or Gat1 in the basidiomycete Cryptococcus neoformans (Kmetzsch et al. 2011). Fungal nitrogen metabolism is reviewed in great depth in Chap. 11. The AreA-type GATA factors sense the intracellular glutamine levels and interact with other transcription factors that are specific for a given nitrogen source as exemplified for nitrate assimilation (Berger et al. 2008) or proline utilisation (Garcia et al. 2004).
Interestingly, some amino acids cannot be used as carbon sources and a few even do not serve as a nitrogen sources in selected fungi as will be exemplified by the degradation of histidine. However, regardless the ability to utilise external amino acids as carbon, nitrogen or sulphur sources, fungi are able to import all proteinogenic amino acids to replenish the intracellular amino acid pool. Even more, amino acids can be stored in vacuoles and mobilised from these compartments under conditions of starvation (Sekito et al. 2014).
Although fungi prefer to procure amino acids from the environment, there is a strong competition for nutrients in microbial communities and some or all amino acids may become limited. To cope with this limitation, fungi are able to synthesise all amino acids de novo. This contrasts the anabolic capacity of mammals, which have lost pathways for the biosynthesis of several amino acids and therefore must obtain amino acids from the diet. Examples are the biosynthesis of the basic amino acids histidine, lysine and arginine, the aromatic amino acids tryptophane and phenylalanine or the aliphatic branched-chain amino acids isoleucine, leucine and valine. Thus, these amino acids are called “essential” amino acids, although it should be mentioned that also the uptake of other amino acids for which biosynthetic pathways still exist is required by humans (Wu 2014). Therefore, it is of great interest to modify microorganisms for amino acid production in order to use the amino acids as food and feed additives. While most amino acids are produced from bacterial sources such as Corynebacterium glutamicum and E. coli (Wendisch 2014), especially the synthesis of the amino acid lysine has been studied in detail in several fungal species. This amino acid not only is important as a food additive but also provides precursors for penicillins and cephalosporins. Therefore, knowledge on its biosynthesis is indispensable for industrial production of β-lactam antibiotics (Fazius et al. 2013). Fungal lysine biosynthesis seems to have evolved independently of bacterial and plant lysine biosynthesis, and at least the use of different isoforms of one enzyme is dependent on the fungal lifestyle as will be outlined later.
A. Histidine Degradation in Fungi
Histidine is an important amino acid frequently found in the catalytic centre of enzymes because it is the only amino acid from which a protonation and deprotonation of the side chain at neutral pH is possible (Kulis-Horn et al. 2014).
Examples for enzymes with essential functional histidine residues are the bisphosphoglycerate mutase from glycolysis/gluconeogenesis (White et al. 1993), the catalytic triad of serine proteases (Betzel et al. 2001) or transcription factors like the carbon catabolite repressor Mig1 from S. cerevisiae that contains zinc coordinating Cys2–His2 zinc fingers (De Vit et al. 1997). Histidine has been assumed as a very ancient amino acid (Kulis-Horn et al. 2014) since its synthesis starts from phosphoribosyl pyrophosphate and ATP.
All genes identified in bacteria to contribute to histidine biosynthesis are also found in yeast (Alifano et al. 1996).
Although histidine can be synthesised by S. cerevisiae de novo, this yeast is unable to proliferate with histidine as sole nitrogen source (Brunke et al. 2014). However, HIS3 codes for the imidazole glycerol-phosphate dehydratase and deletions are frequently used as a selection marker in yeast transformations (Baudin et al. 1993). Importantly, his3 mutants are viable in the presence of externally supplied histidine, indicating that S. cerevisiae can utilise external histidine to replenish the intracellular histidine pool. This is in agreement with the presence of high- and low-affinity histidine transport systems (Bajmoczi et al. 1998).
However, in contrast to the inability of S. cerevisiae to utilise histidine as a nitrogen source, studies on the filamentous fungus A. nidulans showed that histidine serves at least for some fungi as nitrogen source, although it cannot be used as a sole source of carbon (Polkinghorne and Hynes 1982). When grown with histidine as sole nitrogen source, A. nidulans most likely releases ammonium by a histidase (histidine ammonia-lyase) since the reaction product urocanate accumulated in the culture broth (Fig. 14.3). Urocanate is a well-known intermediate from bacterial and most likely also from the mammalian histidine degradation pathway that leads to the end product glutamate. Urocanate is subsequently hydrated by an urocanase yielding 4-imidazolone-5-propionate. Due to the accumulation of urocanate in the medium and the lack of urocanase activity in A. nidulans, the enzymatic set for glutamate synthesis appears to be lost (Polkinghorne and Hynes 1982) explaining the inability of A. nidulans to use histidine as carbon source. Furthermore, a BLAST search on fungal genomes against urocanase from bacteria or mammals revealed no positive hit from any fungal species. This implies that (1) either the amino acid sequence for urocanases is not highly conserved and therefore not identified by sequence analyses, (2) the degradation of urocanate follows a different pathway or (3) histidine can generally not be used as a carbon source by fungi. However, due to the presence of histidase-like enzymes in many fungal species (Brunke et al. 2014), degradation to urocanate might provide a general mechanism for utilisation of histidine as a nitrogen source. Nevertheless, even for A. nidulans an alternative degradation pathway might exist (Fig. 14.3).
Postulated fungal pathways for the use of histidine as nitrogen source. A histidase releases ammonia from histidine resulting in urocanate, which is secreted to the medium. Alternatively, an l-amino acid oxidase such as SarA from A. nidulans releases ammonia from histidine by producing hydrogen peroxide and imidazole pyruvate. The fate of imidazole pyruvate in filamentous fungi and the impact of this pathway are unclear, since only low l-amino acid oxidase activity is detected in wild-type cells. Finally, yeasts use aromatic amino acid aminotransferases such as Aro8 or Aro9 to transfer the amino group from histidine to an α-keto acid resulting in imidazole pyruvate. Via reactions of the Ehrlich pathway, imidazole pyruvate is further converted to the fusel alcohol histaminol
A point mutation in the areA gene of A. nidulans, denoted areA102, causes increased growth rates on several amino acids, among them histidine (Davis et al. 2005). It has been shown that this mutation leads to increased expression of sarA, which encodes an l-amino acid oxidase with broad substrate specificity. Thus, histidine might be converted to imidazole pyruvate under the formation of H2O2 and the release of ammonia. However, in wild-type cells this oxidase activity is hardly detectable (Davis et al. 2005), leaving room for speculation on the normal in vivo function of this reaction, since it would not lead to the formation of urocanate as detected in the culture broth of A. nidulans wild-type cells (Polkinghorne and Hynes 1982).
However, a third pathway might account for histidine utilisation in Candida glabrata, a yeast that is closely related to S. cerevisiae. Growth of C. glabrata on histidine was well supported, although no histidase was found in its genome. In contrast, two genes coding for aromatic amino acid aminotransferases can be detected (Fig. 14.3). One of these aromatic aminotransferases, called Aro8, was essentially required for growth on aromatic amino acids, whereas the second aromatic aminotransferase, Aro9, was dispensable. Unexpectedly, deletion of C. glabrata ARO8 also resulted in the inability to utilise histidine as nitrogen source (Brunke et al. 2014). In agreement with a requirement of Aro8 for growth on histidine, purified Aro8 protein catalysed the conversion of histidine to imidazole pyruvate when α-ketoglutarate was added as acceptor for the amino group. Thus, C. glabrata most likely acquires nitrogen from histidine via an aminotransferase reaction.
Surprisingly, Aro8 and Aro9 are also conserved in S. cerevisiae and ScARO8 shows a similar expression pattern as CgARO8. However, while in C. glabrata CgARO9 cannot be induced -most likely due to a genome inversion upstream CgARO9 - S. cerevisiae ScARO9 is strongly induced by aromatic amino acids and also by histidine. Due to the simultaneous activation of ScARO8 and ScARO9 this might result in aminotransferase levels that produce toxic amounts of intermediates from histidine degradation. In agreement, when S. cerevisiae was cultivated with aromatic amino acids that strongly induce ScARO9 expression, histidine supplementation completely abolished growth. In contrast, histidine only had a moderate growth inhibitory effect when added in combination with ammonium as nitrogen source, which does not induce ScARO8 or ScARO9 expression (Brunke et al. 2014). Thus, S. cerevisiae and C. glabrata, can, in principle, acquire ammonium from histidine via an aminotransferase mechanism. However, the co-expression of ScARO9 might result in accumulation of toxic amounts of intermediates from imidazole pyruvate, which might be circumvented by C. glabrata either due to lower aromatic aminotransferase levels or due to an optimised detoxification of intermediates. It can be assumed that the final product of histidine degradation from imidazole pyruvate in yeasts are fusel alcohols that are generated via the Ehrlich pathway (Hazelwood et al. 2008). This is supported by studies on the fate of histidine during alcoholic fermentation of grape must. Added histidine was slowly consumed and resulted in the formation of imidazole ethanol (histaminol) (Lopez-Rituerto et al. 2013). Histaminol can be assumed to derive from a decarboxylation of imidazole pyruvate yielding imidazole acetaldehyde, which is subsequently reduced to histaminol (Fig. 14.3). Especially the aldehyde intermediate might cause toxic effects.
In conclusion, histidine does not seem to serve as a carbon source for any fungus. Moreover, utilisation of histidine as a nitrogen source differs among fungal species in respect of the mechanism by which nitrogen is released. In addition, enzyme levels might require a strict control to avoid the production of toxic levels of intermediates as seen from the comparison of C. glabrata and S. cerevisiae.
B. Synthesis of the Amino Acid Lysine
While the degradation of amino acids in fungi has hardly been studied, more information is available on the biosynthesis of amino acids. Of special interest are those amino acids for which no biosynthetic pathway is present in humans. These “essential” amino acids are not only valuable food and feed additives but also interesting targets for new antibiotics, because interruption of their synthesis is assumed to cause only minor side effects on human metabolism (Hutton et al. 2007).
Of special interest is the biosynthesis of the amino acid lysine, for two reasons. First, it has important functions in several enzymatic reaction mechanisms, in binding proteins such as histones and transcription factors to DNA and in signal sequences for subcellular localisation of proteins. Second, lysine is essential for the structure of the bacterial cell wall and the production of β-lactam antibiotics (Fazius et al. 2013). Although lysine can be synthesised by bacteria, plants and fungi, different pathways are involved, whereby plant lysine biosynthesis is highly related to the bacterial pathway and the fungal pathway seems to have evolved independently, most likely from the citric acid cycle and leucine biosynthesis (Irvin and Bhattacharjee 1998; Xu et al. 2006).
With the exception of some thermophilic bacteria from the Deinococcus-Thermus phylum (Nishida and Nishiyama 2012), bacteria and plants generally synthesise lysine via the meso-diaminopimelate pathway, whereby different isoforms of the pathway exist (Hudson et al. 2005). In brief, synthesis starts by phosphorylation of aspartate that is converted to aspartate semialdehyde. The semialdehyde condenses with pyruvate resulting in 2,3-dihydrodipicolinate and subsequently in tetrahydrodipicolinate. This is the common intermediate in all variants of the pathway. Then, either a succinylase, an acetylase, a dehydrogenase or an aminotransferase branch leads to the formation of meso-diaminopimelate that is finally decarboxylated to lysine (Watanabe and James 2011).
In contrast to bacteria and plants, fungi and bacteria from the Deinococcus-Thermus phylum synthesise lysine via the α-aminoadipate pathway (Fig. 14.4). This pathway can be divided in two parts. The first part leads to the intermediate α-aminoadipate, which is a precursor molecule for the synthesis of β-lactam antibiotics, whereas the second part leads to the formation of the final product lysine (Fazius et al. 2013). Synthesis starts by condensation of α-ketoglutarate with acetyl-CoA via homocitrate synthase leading to the product homocitrate (Schöbel et al. 2010). While filamentous fungi only seem to possess one enzyme encoding homocitrate synthase with moderate sensitivity to feedback inhibition by lysine, S. cerevisiae possesses two isozymes of homocitrate synthase that show different expression levels depending on respiratory or fermentative growth and display different degrees of feedback inhibition by lysine (Quezada et al. 2011). In S. cerevisiae expression of lysine, biosynthesis is under control of the pathway-specific transcriptional activator Lys14, which is induced by the downstream intermediate α-aminoadipate semialdehyde (Feller et al. 1999). Thus, due to lysine feedback inhibition of homocitrate synthases, S. cerevisiae can counteract this activation by measuring the internal lysine pool. Interestingly, such regulatory circuit and the existence of a transcription factor specifically contributing to induction of lysine biosynthesis have not been described for filamentous fungi. This might be explained by the fact that yeasts are unable to produce β-lactam antibiotics and exclusively use the α-aminoadipate pathway for lysine biosynthesis. In contrast, in β-lactam-producing fungi, a significant proportion of α-aminoadipate may be required and withdrawn for antibiotic production even in the presence of high internal lysine levels. Thus, a feedback mechanism by lysine or the activation by a downstream intermediate would interfere with the dual function of this pathway.
Scheme of the fungal α-aminoadipate pathway. 1 homocitrate synthase, 2 aconitase, 3 homoaconitase, 4 homoisocitrate dehydrogenase, 5 α-aminoadipate aminotransferase, 6 α-aminoadipate reductase, 7 saccharopine reductase, 8 saccharopine dehydrogenase. Details on reactions (2) and (3) (shown in red) and the differences between yeasts and filamentous fungi are described in the text
Homocitrate undergoes a de-and rehydration reaction to form homoisocitrate (Fazius et al. 2012). Biochemical and genetic analyses have shown that this reaction is split on two separate enzymes, similar to the de- and rehydration of methylcitrate to methylisocitrate in the methylcitrate cycle described above. The enzymes involved in these reactions are an aconitase and a pathway-specific homoaconitase (Fazius et al. 2012). Since a striking difference between yeasts and filamentous fungi in the utilisation of the aconitase has been observed, this isomerisation will be explained in more detail below.
Homoisocitrate is oxidatively decarboxylated by a homoisocitrate dehydrogenase to α-ketoadipate (Lin et al. 2009) that is subsequently transaminated to α-aminoadipate by an α-aminoadipate aminotransferase. Interestingly, in S. cerevisiae this aminotransferase reaction may be carried out by Aro8 (Bulfer et al. 2013), which can be assumed to be involved in the transamination of histidine as described above. This reaction completes the first part of the α-aminoadipate pathway. The second half of the pathway mainly constitutes reversible reactions that may also be used for the degradation of lysine, in which lysine is degraded via α-aminoadipate and α-ketoadipate to acetoacetyl-CoA (Pink et al. 2011).
Interestingly, while the initial reactions of the α-aminoadipate pathway are shared between the thermophilic bacteria and fungi, the subsequent synthesis of lysine branches between those groups of organisms. In the thermophilic bacteria, the α-aminoadipate becomes N-protected by addition of the molecule to a small 54-amino acid protein, at which all intermediates maintain attached until the final release of lysine (Horie et al. 2009). In contrast, no N-protection has been observed in fungal lysine biosynthesis, and subsequent reactions involve the reduction of α-aminoadipate to the semialdehyde by an aminoadipate reductase (Yan et al. 2007). The α-aminoadipate semialdehyde is subsequently condensed with a glutamate via saccharopine reductase yielding saccharopine (Vashishtha et al. 2009). The final step in the synthesis is performed by a saccharopine dehydrogenase catalysing an oxidative deamination of saccharopine, which results in the products α-ketoglutarate and lysine (Kumar et al. 2012).
While the α-aminoadipate pathway is well conserved among fungi, it involves at least one enzyme that is shared by several metabolic pathways. This enzyme is the aconitase that is involved in the citric acid cycle, in the methylcitrate cycle and in lysine biosynthesis.
In the citric acid cycle the enzyme performs the de- and rehydration of citrate to isocitrate by binding the substrate cis-aconitate either in citrate mode or 180° rotated in the isocitrate mode. In the methylcitrate cycle, cis-methylaconitate can only be bound in the isocitrate mode due to sterical hindrance of the methyl-group with an aspartate residue (Lauble and Stout 1995). Finally, in lysine biosynthesis cis-homoaconitate can only bind in the citrate mode, because the enhanced aliphatic chain in cis-homoaconitate does not allow correct positioning in the isocitrate mode (Fazius et al. 2012).
Both yeasts and filamentous fungi use an aconitase for the dehydration of homocitrate to homoaconitate. Genome analyses revealed that fungi contain at least two aconitase-like enzymes (AcoA and AcoB in Aspergillus spp., Aco1 and Aco2 in S. cerevisiae). In Aspergillus spp. AcoB seems to have lost its function, since no phenotype is observed when the gene is deleted, and AcoB cannot complement aconitase-negative mutants from S. cerevisiae. However, deletion of AcoA is lethal in A. fumigatus, confirming its essential function, and AcoA is the only aconitase contributing to lysine biosynthesis (Fazius et al. 2012). In contrast, S. cerevisiae has specifically adapted Aco2 to serve for lysine biosynthesis. Aco1 is the main citric acid cycle aconitase in S. cerevisiae and a deletion leads to glutamate auxotrophy (Gangloff et al. 1990). However, although a lysine auxotrophy would be simultaneously expected, growth in the absence of external lysine is only slightly retarded. When Aco2 is deleted, no glutamate auxotrophy is observed, but, again, growth is retarded in the absence of external lysine. Thus, a deletion of both ACO1 and ACO2 is required to cause a lysine auxotrophic phenotype in S. cerevisiae (Fazius et al. 2012). Biochemical characterisation additionally revealed that Aco2 is highly specific for homocitrate as substrate and hardly active with citrate. This confirms that S. cerevisiae specifically adapted Aco2 to serve in lysine biosynthesis, which is not the case for AcoB in Aspergillus spp.
The reason for the difference between filamentous fungi and yeast might be due to the different mode of glucose utilisation. While filamentous fungi rely on a functional citric acid cycle for substrate oxidation and energy gain, yeasts tend to fermentation and can grow independent from a citric acid cycle when glutamate is supplied. Thus, due to the adaptation of Aco2 to serve lysine biosynthesis, yeast can grow by fermentation without disturbing accompanying metabolic pathways (Fazius et al. 2012).
In conclusion, lysine biosynthesis is highly conserved among fungi. However, differences exist in the regulation of the pathway. This is caused by the fact that yeasts use the α-aminoadipate solely for lysine biosynthesis, whereas several filamentous fungi also require the intermediate α-aminoadipate for production of β-lactam antibiotics. Additionally, the adaptation of an aconitase isozyme in S. cerevisiae appears due to its ability of decoupling energy metabolism and anaplerosis, a mechanism that is not possible for oxidatively growing filamentous fungi.
V. Conclusions
Investigation of fungal metabolism has mainly been performed on Ascomycetes such as the yeast S. cerevisiae or the filamentous fungus A. nidulans. In case of bakers’ yeast, this is mainly due to the early use of this organism in food and ethanol production and its easy cultivation (Nevoigt 2008). Thus, early investigations of cellular metabolism based on S. cerevisiae from which important metabolic pathways such as glycolysis had been identified (Buchholz and Collins 2013). Besides yeast, A. nidulans established as a model due to its homothallic sexual mating and the early development of sexual crossing systems that allowed the identification of complementation groups in developmental and metabolic mutants (Nierman et al. 2005). In contrast, due to the lack of transformation systems or problems in laboratory cultivation, studies on the metabolism of Basidiomycetes or Zygomycetes are limited. However, as exemplified in this chapter, even relatively closely related Ascomycetes show a great diversity in their metabolic properties. These differences appear related to fungal morphology (yeast against hyphae), nutritional niches, gene loss and acquisition and organismal interaction. Taking into account that even closely related fungal species already display a great variety in use and occurrence of metabolic pathways, it can be expected that future analyses of metabolism and physiology of Basidiomycota, Zygomycota and Chytridiomycota provide a treasure chest of new pathways that might become important for several biotechnological processes.
References
Abdeljalil S et al (2013) Improvement of cellulase and xylanase production by solid-state fermentation of Stachybotrys microspora. Biotechnol Appl Biochem. doi: 10.1002/bab.1195
Alam S, Shah HU, Magan N (2010) Effect of calcium propionate and water activity on growth and aflatoxins production by Aspergillus flavus. J Food Sci 75:M61–M64
Alifano P et al (1996) Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev 60:44–69
Aschenbach JR, Kristensen NB, Donkin SS, Hammon HM, Penner GB (2010) Gluconeogenesis in dairy cows: the secret of making sweet milk from sour dough. IUBMB Life 62:869–877
Bajmoczi M, Sneve M, Eide DJ, Drewes LR (1998) TAT1 encodes a low-affinity histidine transporter in Saccharomyces cerevisiae. Biochem Biophys Res Commun 243:205–209
Balestrini R, Bonfante P (2014) Cell wall remodeling in mycorrhizal symbiosis: a way towards biotrophism. Front Plant Sci 5:237
Ballhausen D, Mittaz L, Boulat O, Bonafe L, Braissant O (2009) Evidence for catabolic pathway of propionate metabolism in CNS: expression pattern of methylmalonyl-CoA mutase and propionyl-CoA carboxylase alpha-subunit in developing and adult rat brain. Neuroscience 164:578–587
Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res 21:3329–3330
Berger H et al (2008) Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol Microbiol 69:1385–1398
Betzel C et al (2001) Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 A resolution. Biochemistry 40:3080–3088
Bigner DR, Goff JP, Faust MA, Tyler HD, Horst RL (1997) Comparison of oral sodium compounds for the correction of acidosis. J Dairy Sci 80:2162–2166
Brock M (2009) Fungal metabolism in host niches. Curr Opin Microbiol 12:371–376
Brock M, Buckel W (2004) On the mechanism of action of the antifungal agent propionate. Eur J Biochem 271:3227–3241
Brock M, Fischer R, Linder D, Buckel W (2000) Methylcitrate synthase from Aspergillus nidulans: implications for propionate as an antifungal agent. Mol Microbiol 35:961–973
Brock M, Darley D, Textor S, Buckel W (2001) 2-Methylisocitrate lyases from the bacterium Escherichia coli and the filamentous fungus Aspergillus nidulans: characterization and comparison of both enzymes. Eur J Biochem 268:3577–3586
Brock M, Maerker C, Schutz A, Volker U, Buckel W (2002) Oxidation of propionate to pyruvate in Escherichia coli. Involvement of methylcitrate dehydratase and aconitase. Eur J Biochem 269:6184–6194
Brunke S et al (2014) Histidine degradation via an aminotransferase increases the nutritional flexibility of Candida glabrata. Eukaryot Cell 13:758–765
Buchholz K, Collins J (2013) The roots--a short history of industrial microbiology and biotechnology. Appl Microbiol Biotechnol 97:3747–3762
Bulfer SL, Brunzelle JS, Trievel RC (2013) Crystal structure of Saccharomyces cerevisiae Aro8, a putative alpha-aminoadipate aminotransferase. Protein Sci 22:1417–1424
Chi Z, Chi Z, Liu G, Wang F, Ju L, Zhang T (2009) Saccharomycopsis fibuligera and its applications in biotechnology. Biotechnol Adv 27:423–431
Chittori S, Simanshu DK, Savithri HS, Murthy MR (2010) Preliminary X-ray crystallographic analysis of 2-methylcitrate synthase from Salmonella typhimurium. Acta Crystallogr Sect F: Struct Biol Cryst Commun 66:467–470
Coblentz WK, Coffey KP, Young AN, Bertram MG (2013) Storage characteristics, nutritive value, energy content, and in vivo digestibility of moist, large rectangular bales of alfalfa-orchardgrass hay treated with a propionic acid-based preservative. J Dairy Sci 96:2521–2535
Creighton DJ, Murthy NSRK (1990) Stereospecificity in enzymatic reactions. In: Sigman DS, Boyer PD (eds) The enzymes, 3rd edn. Academic, San Diego
Crittenden PD, Davis JC, Hawksworth DL, Campbell FS (1995) Attempted isolation and success in culturing of a broad spectrum of lichen forming and lichenicolous fungi. New Phytol 130:267–297
Cruz AH et al (2011) Phosphorylation is the major mechanism regulating isocitrate lyase activity in Paracoccidioides brasiliensis yeast cells. FEBS J 278:2318–2332
Cuomo CA et al (2012) Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res 22:2478–2488
Davis MA, Askin MC, Hynes MJ (2005) Amino acid catabolism by an areA-regulated gene encoding an L-amino acid oxidase with broad substrate specificity in Aspergillus nidulans. Appl Environ Microbiol 71:3551–3555
De Lucas JR, Gregory S, Turner G (1994a) Analysis of the regulation of the Aspergillus nidulans acuD gene, encoding isocitrate lyase, by construction of a hybrid promoter. Mol Gen Genet 243:654–659
De Lucas JR, Valenciano S, Laborda F, Turner G (1994b) Glucose-induced inactivation of isocitrate lyase in Aspergillus nidulans. Arch Microbiol 162:409–413
De Vit MJ, Waddle JA, Johnston M (1997) Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell 8:1603–1618
Domin N, Wilson D, Brock M (2009) Methylcitrate cycle activation during adaptation of Fusarium solani and Fusarium verticillioides to propionyl-CoA-generating carbon sources. Microbiology 155:3903–3912
Dunn MF, Ramirez-Trujillo JA, Hernandez-Lucas I (2009) Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155:3166–3175
Emmanuel B, Kennelly JJ (1984) Effect of propionic acid on ketogenesis in lactating sheep fed restricted rations or deprived of food. J Dairy Sci 67:344–350
Fazius F, Shelest E, Gebhardt P, Brock M (2012) The fungal alpha-aminoadipate pathway for lysine biosynthesis requires two enzymes of the aconitase family for the isomerization of homocitrate to homoisocitrate. Mol Microbiol 86:1508–1530
Fazius F, Zaehle C, Brock M (2013) Lysine biosynthesis in microbes: relevance as drug target and prospects for beta-lactam antibiotics production. Appl Microbiol Biotechnol 97:3763–3772
Fellbaum CR et al (2014) Fungal nutrient allocation in common mycorrhizal networks is regulated by the carbon source strength of individual host plants. New Phytol 203:646–656
Feller A, Ramos F, Pierard A, Dubois E (1999) In Saccharomyces cerevisae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur J Biochem 261:163–170
Free SJ (2013) Fungal cell wall organization and biosynthesis. Adv Genet 81:33–82
Gangloff SP, Marguet D, Lauquin GJ (1990) Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol Cell Biol 10:3551–3561
Garcia I, Gonzalez R, Gomez D, Scazzocchio C (2004) Chromatin rearrangements in the prnD-prnB bidirectional promoter: dependence on transcription factors. Eukaryot Cell 3:144–156
Garvey GS, Rocco CJ, Escalante-Semerena JC, Rayment I (2007) The three-dimensional crystal structure of the PrpF protein of Shewanella oneidensis complexed with trans-aconitate: insights into its biological function. Protein Sci 16:1274–1284
Gonzalez R, Gavrias V, Gomez D, Scazzocchio C, Cubero B (1997) The integration of nitrogen and carbon catabolite repression in Aspergillus nidulans requires the GATA factor AreA and an additional positive-acting element, ADA. EMBO J 16:2937–2944
Graybill ER, Rouhier MF, Kirby CE, Hawes JW (2007) Functional comparison of citrate synthase isoforms from S. cerevisiae. Arch Biochem Biophys 465:26–37
Grimek TL, Escalante-Semerena JC (2004) The acnD genes of Shewenella oneidensis and Vibrio cholerae encode a new Fe/S-dependent 2-methylcitrate dehydratase enzyme that requires prpF function in vivo. J Bacteriol 186:454–462
Gropp K, Schild L, Schindler S, Hube B, Zipfel PF, Skerka C (2009) The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol Immunol 47:465–475
Halarnkar PP, Blomquist GJ (1989) Comparative aspects of propionate metabolism. Comp Biochem Physiol B 92:227–231
Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74:2259–2266
Hers HG, Hue L (1983) Gluconeogenesis and related aspects of glycolysis. Annu Rev Biochem 52:617–653
Hori C et al (2013) Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycologia 105:1412–1427
Horie A et al (2009) Discovery of proteinaceous N-modification in lysine biosynthesis of Thermus thermophilus. Nat Chem Biol 5:673–679
Horswill AR, Escalante-Semerena JC (2001) In vitro conversion of propionate to pyruvate by Salmonella enterica enzymes: 2-methylcitrate dehydratase (PrpD) and aconitase enzymes catalyze the conversion of 2-methylcitrate to 2-methylisocitrate. Biochemistry 40:4703–4713
Hudson AO et al (2005) Biosynthesis of lysine in plants: evidence for a variant of the known bacterial pathways. Biochim Biophys Acta 1721:27–36
Hutton CA, Perugini MA, Gerrard JA (2007) Inhibition of lysine biosynthesis: an evolving antibiotic strategy. Mol Biosyst 3:458–465
Hynes MJ, Murray SL (2010) ATP-citrate lyase is required for production of cytosolic acetyl coenzyme A and development in Aspergillus nidulans. Eukaryot Cell 9:1039–1048
Irvin SD, Bhattacharjee JK (1998) A unique fungal lysine biosynthesis enzyme shares a common ancestor with tricarboxylic acid cycle and leucine biosynthetic enzymes found in diverse organisms. J Mol Evol 46:401–408
Jacobsen ID, Lüttich A, Kurzai O, Hube B, Brock M (2014) In vivo imaging of disseminated murine Candida albicans infection reveals unexpected host sites of fungal persistence during antifungal therapy. J Antimicrob Chemother 69(10):2785–96
Jiang W, Nakayama Y, Sequeira JM, Quadros EV (2013) Mapping the functional domains of TCblR/CD320, the receptor for cellular uptake of transcobalamin-bound cobalamin. FASEB J 27:2988–2994
Kanamasa S, Dwiarti L, Okabe M, Park EY (2008) Cloning and functional characterization of the cis-aconitic acid decarboxylase (CAD) gene from Aspergillus terreus. Appl Microbiol Biotechnol 80:223–229
Klement T, Buchs J (2013) Itaconic acid – a biotechnological process in change. Bioresour Technol 135:422–431
Kmetzsch L et al (2011) The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet Biol 48:192–199
Kobayashi K, Hattori T, Honda Y, Kirimura K (2013) Gene identification and functional analysis of methylcitrate synthase in citric acid-producing Aspergillus niger WU-2223L. Biosci Biotechnol Biochem 77:1492–1498
Kulis-Horn RK, Persicke M, Kalinowski J (2014) Histidine biosynthesis, its regulation and biotechnological application in Corynebacterium glutamicum. Microb Biotechnol 7:5–25
Kumar VP, West AH, Cook PF (2012) Supporting role of lysine 13 and glutamate 16 in the acid-base mechanism of saccharopine dehydrogenase from Saccharomyces cerevisiae. Arch Biochem Biophys 522:57–61
Lauble H, Stout CD (1995) Steric and conformational features of the aconitase mechanism. Proteins 22:1–11
Ledley FD, Crane AM, Klish KT, May GS (1991) Is there methylmalonyl CoA mutase in Aspergillus nidulans? Biochem Biophys Res Commun 177:1076–1081
Lin Y, West AH, Cook PF (2009) Site-directed mutagenesis as a probe of the acid-base catalytic mechanism of homoisocitrate dehydrogenase from Saccharomyces cerevisiae. Biochemistry 48:7305–7312
Liu S, Lu Z, Han Y, Melamud E, Dunaway-Mariano D, Herzberg O (2005) Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution. Biochemistry 44:2949–2962
Lopez-Rituerto E, Avenoza A, Busto JH, Peregrina JM (2013) NMR study of histidine metabolism during alcoholic and malolactic fermentations of wine and their influence on histamine production. J Agric Food Chem 61:9464–9469
Maerker C, Rohde M, Brakhage AA, Brock M (2005) Methylcitrate synthase from Aspergillus fumigatus. Propionyl-CoA affects polyketide synthesis, growth and morphology of conidia. FEBS J 272:3615–3630
Marin S et al (1999) Control of growth and fumonisin B1 production by Fusarium verticillioides and Fusarium proliferatum isolates in moist maize with propionate preservatives. Food Addit Contam 16:555–563
Müller S, Fleck CB, Wilson D, Hummert C, Hube B, Brock M (2011) Gene acquisition, duplication and metabolic specification: the evolution of fungal methylisocitrate lyases. Environ Microbiol 13:1534–1548
Nehls U, Grunze N, Willmann M, Reich M, Kuster H (2007) Sugar for my honey: carbohydrate partitioning in ectomycorrhizal symbiosis. Phytochemistry 68:82–91
Nevoigt E (2008) Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev 72:379–412
Nierman WC, May G, Kim HS, Anderson MJ, Chen D, Denning DW (2005) What the Aspergillus genomes have told us. Med Mycol 43(Suppl 1):S3–S5
Nishida H, Nishiyama M (2012) Evolution of lysine biosynthesis in the phylum Deinococcus-Thermus. Int J Evol Biol 2012:745931
Otzen C, Bardl B, Jacobsen ID, Nett M, Brock M (2014) Candida albicans utilizes a modified beta-oxidation pathway for the degradation of toxic propionyl-CoA. J Biol Chem 289:8151–8169
Papagianni M (2007) Advances in citric acid fermentation by Aspergillus niger: biochemical aspects, membrane transport and modeling. Biotechnol Adv 25:244–263
Peraza-Reyes L, Berteaux-Lecellier V (2013) Peroxisomes and sexual development in fungi. Front Physiol 4:244
Pink DB et al (2011) Lysine alpha-ketoglutarate reductase, but not saccharopine dehydrogenase, is subject to substrate inhibition in pig liver. Nutr Res 31:544–554
Polkinghorne MA, Hynes MJ (1982) L-histidine utilization in Aspergillus nidulans. J Bacteriol 149:931–940
Pronk JT, van der Linden-Beuman A, Verduyn C, Scheffers WA, van Dijken JP (1994) Propionate metabolism in Saccharomyces cerevisiae: implications for the metabolon hypothesis. Microbiology 140(Pt 4):717–722
Quezada H et al (2011) The Lys20 homocitrate synthase isoform exerts most of the flux control over the lysine synthesis pathway in Saccharomyces cerevisiae. Mol Microbiol 82:578–590
Schöbel F, Jacobsen ID, Brock M (2010) Evaluation of lysine biosynthesis as an antifungal drug target: biochemical characterization of Aspergillus fumigatus homocitrate synthase and virulence studies. Eukaryot Cell 9:878–893
Sekito T, Chardwiriyapreecha S, Sugimoto N, Ishimoto M, Kawano-Kawada M, Kakinuma Y (2014) Vacuolar transporter Avt4 is involved in excretion of basic amino acids from the vacuoles of Saccharomyces cerevisiae. Biosci Biotechnol Biochem 78:969–975
Soontorngun N et al (2012) Genome-wide location analysis reveals an important overlap between the targets of the yeast transcriptional regulators Rds2 and Adr1. Biochem Biophys Res Commun 423:632–637
Steiger MG, Blumhoff ML, Mattanovich D, Sauer M (2013) Biochemistry of microbial itaconic acid production. Front Microbiol 4:23
Stocker-Wörgötter E (2008) Metabolic diversity of lichen-forming ascomycetous fungi: culturing, polyketide and shikimate metabolite production, and PKS genes. Nat Prod Rep 25:188–200
Strauss J, Horvath HK, Abdallah BM, Kindermann J, Mach RL, Kubicek CP (1999) The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol Microbiol 32:169–178
Suzuki Y, Murray SL, Wong KH, Davis MA, Hynes MJ (2012) Reprogramming of carbon metabolism by the transcriptional activators AcuK and AcuM in Aspergillus nidulans. Mol Microbiol 84:942–964
Tabuchi T, Hara S (1974) Production of 2-methylisocitric acid from n-paraffins by mutants of Candida lipolytica. Agric Biol Chem 38:1105–1106
Tabuchi T, Uchiyama H (1975) Methylcitrate condensing and methylisocitrate cleaving enzymes; evidence for the pathway of oxidation of propionyl-CoA to pyruvate via C7-tricarboxylic acids. Agric Biol Chem 39:2035–2042
Takahashi-Iniguez T, Garcia-Hernandez E, Arreguin-Espinosa R, Flores ME (2012) Role of vitamin B12 on methylmalonyl-CoA mutase activity. J Zhejiang Univ Sci B 13:423–437
Tao Y, Marzluf GA (1999) The NIT2 nitrogen regulatory protein of Neurospora: expression and stability of nit-2 mRNA and protein. Curr Genet 36:153–158
Textor S et al (1997) Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol 168:428–436
Tiwari P, Misra BN, Sangwan NS (2013) beta-Glucosidases from the fungus Trichoderma: an efficient cellulase machinery in biotechnological applications. Biomed Res Int 2013:203735
Todd RB et al (1997) The acetate regulatory gene facB of Aspergillus nidulans encodes a Zn(II)2Cys6 transcriptional activator. Mol Gen Genet 254:495–504
Todd RB, Andrianopoulos A, Davis MA, Hynes MJ (1998) FacB, the Aspergillus nidulans activator of acetate utilization genes, binds dissimilar DNA sequences. EMBO J 17:2042–2054
Trepanier M et al (2005) Dependence of arbuscular-mycorrhizal fungi on their plant host for palmitic acid synthesis. Appl Environ Microbiol 71:5341–5347
Tsai CS, Mitton KP, Johnson BF (1989) Acetate assimilation by the fission yeast, Schizosaccharomyces pombe. Biochem Cell Biol 67:464–467
Tsang AW, Horswill AR, Escalante-Semerena JC (1998) Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J Bacteriol 180:6511–6518
Tu T et al (2013) High-yield production of a low-temperature-active polygalacturonase for papaya juice clarification. Food Chem 141:2974–2981
Turcotte B, Liang XB, Robert F, Soontorngun N (2010) Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res 10:2–13
Tylicki A, Ziolkowska G, Bolkun A, Siemieniuk M, Czerniecki J, Nowakiewicz A (2008) Comparative study of the activity and kinetic properties of malate dehydrogenase and pyruvate decarboxylase from Candida albicans, Malassezia pachydermatis, and Saccharomyces cerevisiae. Can J Microbiol 54:734–741
Vashishtha AK, West AH, Cook PF (2009) Chemical mechanism of saccharopine reductase from Saccharomyces cerevisiae. Biochemistry 48:5899–5907
Watanabe N, James MN (2011) Structural insights for the substrate recognition mechanism of LL-diaminopimelate aminotransferase. Biochim Biophys Acta 1814:1528–1533
Wendisch VF (2014) Microbial production of amino acids and derived chemicals: synthetic biology approaches to strain development. Curr Opin Biotechnol 30C:51–58
White MF, Fothergill-Gilmore LA, Kelly SM, Price NC (1993) Substitution of His-181 by alanine in yeast phosphoglycerate mutase leads to cofactor-induced dissociation of the tetrameric structure. Biochem J 291(Pt 2):479–483
Woo JM, Yang KM, Kim SU, Blank LM, Park JB (2014) High temperature stimulates acetic acid accumulation and enhances the growth inhibition and ethanol production by Saccharomyces cerevisiae under fermenting conditions. Appl Microbiol Biotechnol 98:6085–6094
Wu G (2014) Dietary requirements of synthesizable amino acids by animals: a paradigm shift in protein nutrition. J Anim Sci Biotechnol 5:34
Xu H, Andi B, Qian J, West AH, Cook PF (2006) The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys 46:43–64
Yan H, He P, Cheng HR, Shen A, Jiang N (2007) Cloning, sequencing and characterization of the alpha-aminoadipate reductase gene (LYS2) from Saccharomycopsis fibuligera. Yeast 24:189–199
Yike I (2011) Fungal proteases and their pathophysiological effects. Mycopathologia 171:299–323
Zhang YQ, Brock M, Keller NP (2004) Connection of propionyl-CoA metabolism to polyketide biosynthesis in Aspergillus nidulans. Genetics 168:785–794
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Brock, M., Geib, E. (2016). 14 Special Aspects of Fungal Catabolic and Anabolic Pathways. In: Hoffmeister, D. (eds) Biochemistry and Molecular Biology. The Mycota, vol III. Springer, Cham. https://doi.org/10.1007/978-3-319-27790-5_14
Download citation
DOI: https://doi.org/10.1007/978-3-319-27790-5_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27788-2
Online ISBN: 978-3-319-27790-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)