Abstract
Humans with obesity and type 2 diabetes exhibit the classic triad of hyperinsulinemia, hyperglycemia, and hypertriglyceridemia. The paradox of selective insulin resistance in the liver, in which the gluconeogenic pathway becomes insensitive to insulin but the lipogenesis pathway remains sensitive to insulin, leads to an elevation in hepatic and plasma levels of fatty acids and triglyceride and makes detrimental contributions to the development of insulin resistance. However, the precise mechanism for selective insulin resistance remains largely unknown. AMP-activated protein kinase (AMPK) is an energy sensor that regulates metabolic homeostasis. Recently, elucidating the role of AMPK leads to surprising findings and helps identify novel downstream effectors of AMPK. Cellular and molecular biological approach and obese, diabetic mouse models are utilized to characterize that sterol regulatory element binding protein (SREBP), a family of the transcription regulator of lipid synthesis, functions as a conserved substrate of AMPK. AMPK specifically interacts with and phosphorylates SREBP-1c and SREBP-2. AMPK and its pharmacological activators, such as metformin and polyphenols, inhibit the cleavage processing of SREBP-1c and SREBP-2, decrease the nuclear translocation, and reduce the transcription of target genes involved in the biosynthesis of fatty acid, triglyceride, and cholesterol at least in part through AMPK-dependent inhibition of SREBP in hepatocytes. Strikingly, integrated inhibition of AMPK and stimulation of SREBP are implicated on hepatic lipogenesis and steatosis. In contrast, suppression of the de novo lipogenesis by AMPK in the liver results from an increase in SREBP-1 phosphorylation and a reduction in its cleavage processing and transcriptional activity in insulin resistance. These studies provide mechanistic insight into the development of potential therapeutic strategies to target the nutrient sensing AMPK-SREBP pathway for treating type 2 diabetes and related metabolic disorders.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Fatty acid synthesis
- Hepatic lipogenesis
- Sterol regulatory element binding protein 1 (SREBP-1)
- AMP-activated protein kinase (AMPK)
- Non-alcoholic fatty liver disease
- Hepatic selective insulin resistance
- Type 2 diabetes
- Obesity
1 Introduction
In mammals, the liver is the principle organ responsible for the regulation of glucose and lipid metabolism . Fatty acids are substrates for cellular metabolism and are essential components of cellular membranes. In cellular metabolism , sources of fatty acids are exogenously-derived (dietary) fatty acids and endogenously-synthesized fatty acids.
The biosynthesis of endogenous fatty acids, known as de novo lipogenesis, is the metabolic pathway that converts excess carbohydrates (glucose) into fatty acids, which are ultimately esterified with glycerol 3-phosphate to form triglyceride in the liver (Fig. 2.1). During the de novo lipogenic process, glucose is first converted to pyruvate, which enters the tricarboxylic acid (TCA) cycle in the mitochondria to yield citrate. Citrate is then transported into the cytosol and broken down by ATP citrate lyase to yield acetyl-CoA and oxaloacetate. In the rate-limiting step in the lipogenesis pathway, the conversion of acetyl-CoA to malonyl-CoA is catalyzed by acetyl-CoA carboxylase (ACC). Fatty acid synthase (FAS) can synthesize long-chain fatty acids by using acetyl-CoA as a primer, malonyl-CoA as a two-carbon donor, and NADPH as a reducing equivalent. Palmitic acid (C16:0), the predominant fatty acid, is generated by FAS. Palmitic acid is desaturated by stearoyl-CoA desaturase-1 (SCD-1) to produce palmitoleic acid or it can be elongated to yield stearic acid (C18:0). SCD-1 catalyzes the conversion of stearoyl-CoA to oleoyl-CoA, which is a major substrate for triglyceride synthesis. Oleic acid (C18:1) is formed as a result of desaturation of stearic acid and it is thought to be the end product of de novo fatty acid synthesis (Dentin et al. 2005).
De novo lipogenesis is regulated by insulin and glucose in hepatocytes . Metabolic flux in the liver reflects the activation of major pathways including glycolysis and lipogenesis. Acetyl-CoA is produced from glucose through a glycolytic process; it can be used as a substrate for fatty acid synthesis. The major function of glycolysis in the liver provides carbons from glucose for promoting de novo lipogenesis. The transcription of SREBP-1c and its target genes involving lipogenic enzymes is induced by high concentrations of glucose and insulin
The activity of the lipogenic pathway is regulated upon nutritional conditions (Kim et al. 1998). A diet rich in carbohydrates stimulates the lipogenic pathway in the liver, whereas starvation decreases hepatic lipogenesis. Activities of lipogenic enzymes are controlled by post-translational and transcriptional mechanisms. It is established that the transcription of lipogenic enzymes is regulated by insulin and glucose (Hasty et al. 2000; Foretz et al. 1999a). Sterol regulatory element binding protein-1c (SREBP-1c) is implicated in the transcriptional regulation of multiple lipogenic enzymes such as acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), and stearoyl-CoA desaturase-1 (SCD-1). Aberrant activation of hepatic de novo lipogenesis is observed in obesity-induced insulin resistance in animals. In this chapter, we discuss nutrient sensing mechanisms and signaling crosstalk responsible for the regulation of de novo lipogenesis in the liver and discuss how these nutrient sensing pathways become deregulated in metabolic disease.
2 Mammalian AMPK
2.1 The Regulation and Function of AMPK
AMP-activated protein kinase (AMPK) is an intracellular energy and nutrient sensor that has been implicated in the regulation of glucose and lipid homeostasis (Kahn et al. 2005; Hardie et al. 2012). AMPK serine/threonine protein kinase is a heterotrimeric protein complex that consists of α catalytic subunit and β and γ regulatory subunits (Hardie et al. 2012). Phosphorylation of a conserved Thr-172 site within the activation loop of the kinase domain of the α subunit is required for AMPK activity. The regulation of AMPK is complex: it involves allosteric activation by elevated AMP or ADP concentrations; and AMPK activity is increased by ATP depletion. AMPK activity can also be regulated by upstream kinases such as LKB1, CaMKKβ, and TAK1 (Hardie et al. 2012; Kahn et al. 2005; Hawley et al. 2005; Momcilovic et al. 2006). The protein kinase LKB1 , the Peutz-Jeghers syndrome tumor-suppressor, can directly phosphorylate and activate AMPK (Shaw et al. 2004; Sakamoto et al. 2004; Lizcano et al. 2004). Both LKB1 and CaMKKβ phosphorylate the same residue (Thr-172) on the α subunit of AMPK. AMPK is activated by LKB1 in response to increased intracellular levels of AMP or ADP and depleted intracellular levels of ATP. Binding of AMP to the γ subunit of AMPK allosterically activates the kinase while preventing dephosphorylation of the α subunit of AMPK. On the other hand, AMPK is also activated by CaMKKβ in response to elevated intracellular concentrations of Ca2+ (Fig. 2.1).
AMPK activity is induced by physiological stimuli such as exercise and adiponectin . AMPK is also activated by pharmacological agents such as metformin and xenobiotics . Additionally, AMPK can be activated by metabolic stresses that inhibit mitochondrial ATP production or accelerate ATP consumption. In numerous systems, AMPK serves as a protective response to energy stress. Once activated, AMPK restores cellular energy balance by switching off ATP-consuming anabolic pathways including the synthesis of fatty acid, triglyceride and cholesterol as well as by switching on ATP-generating catabolic pathways, such as fatty acid oxidation and lipolysis (Fig. 2.2) . Notably, AMPK is thought to be a checkpoint that maintains energy balance in both cells and organisms. AMPK also plays a role in the regulation of whole-body energy metabolism through decreased energy production and increased energy expenditure. Specifically, AMPK can increase whole-body energy expenditure via actions on the hypothalamus . However, the dysregulation of AMPK is implicated in the pathogenesis of diabetes as well as its related metabolic disorders and some cancers.
2.2 AMPK Activators
Pharmacological activation of AMPK by metformin and other compounds holds promising application that may improve metabolic abnormalities associated with obesity and type 2 diabetes . Given the fact that the correction of the deregulated LKB1/AMPK pathway can reduce the Warburg effect in cancer (Faubert et al. 2013), these studies suggest that AMPK has the potential as a drug target for cancer prevention and treatment (Luo et al. 2010; Shackelford and Shaw 2009). Activation of AMPK by naturally occurring compounds also prevents the development of chronic diseases related to aging (Martin-Montalvo et al. 2013). These findings raise the possibility of metformin-based interventions that extend lifespan and promote healthy aging.
2.2.1 AMPK and Metformin
Type 2 diabetes is associated with insulin resistance, accompanied by elevated hepatic glucose production, hyperglycemia, and hyperlipidemia. Since the 1950s, one of the few classes of therapeutics effective in reducing hepatic glucose production has been the biguanides (Viollet et al. 2012). Biguanide class of antidiabetic drugs contains two linked guanidine rings that are originally derived from galegine (isoamylene guanidine), a guanidine derivative found in the French lilac Galega officinalis. The biguanide drugs include phenformin and metformin (N, N-dimethylbiguanide). Particularly, metformin is the most widely used antidiabetic agent and is currently recommended as the first line drug for patients with type 2 diabetes worldwide.
Metformin is an oral antidiabetic drug that improves systemic insulin sensitivity with reduced plasma glucose levels in patients with type 2 diabetes. Metformin may increase hepatic insulin sensitivity through the upregulation of insulin signaling by increased tyrosine phosphorylation of the insulin receptor and insulin receptor substrate 1, leading to reduced hepatic gluconeogenesis and glucose output (Viollet et al. 2012).
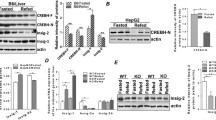
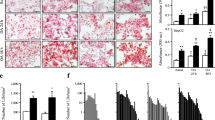
Beyond its effect on glucose metabolism , metformin has been reported to improve fatty liver and cardiovascular complications associated with type 2 diabetes. Metformin has also emerged as an adjunct treatment for cancer due to its role in cancer metabolism. However, the precise mechanism by which metformin lowers lipids is not fully understood. To explore the mechanism of metformin action, an in vitro cell model of high glucose-induced insulin resistance and triglyceride accumulation is initially developed (Zang et al. 2004). An in vitro insulin-resistant state , as characterized by a decrease in insulin-stimulated phosphorylation of Akt and its downstream effector, glycogen synthase kinase 3α/β in hepatocytes, is induced by high concentrations of glucose (30 mm), which mimics an in vivo hyperglycemia state. Under these conditions, metformin dose- and time-dependently increases phosphorylation of AMPK at Thr-172, the major stimulatory phosphorylation site of the AMPK α subunit, and its downstream target acetyl-CoA carboxylase 1 (ACC1) at Ser-79 and ACC2 at Ser-212, major inhibitory phosphorylation sites of ACC. A decrease in AMPK activity is correlated with an elevation in hepatocellular lipid content in an insulin-resistant state. These results indicate that the inhibition of AMPK may represent a critical mechanism for hepatocellular lipid accumulation associated with insulin resistance (Zang et al. 2004). Strikingly, the impairment of AMPK and accumulation of lipids caused by high glucose are prevented by metformin treatment. The protective effects of metformin are mimicked by overexpression of the constitutively active form of AMPKα and blocked by expressing the kinase-inactive form of AMPKα. These studies provide biochemical evidence that while AMPK activation is required for the lipid-lowering effect of metformin in insulin-resistant hepatocytes, it is up for debate whether the glucose-lowering effect of metformin is dependent on AMPK (Zhou et al. 2001; Shaw et al. 2005; Foretz et al. 2010; Fullerton et al. 2013; Shaw 2013), Given that the salutary effect of metformin on hepatocellular lipid accumulation is primarily mediated by AMPK in hepatocytes exposed to high glucose, it suggests that this cell model not only provides a useful tool to screen or develop potential therapeutic agents to target AMPK for the treatment of fatty liver and insulin resistance, but also enables us to seek new drug targets for these diseases.
Previous studies have shown that AMPK activation by either AICAR or metformin stimulates fatty acid oxidation, because AICAR reduces [14C]-oleate and [3H]-glycerol incorporation into triacylglycerol in rat hepatocytes (Zhou et al. 2001; Muoio et al. 1999). ACC, the major downstream target of AMPK, is a key enzyme that catalyzes the conversion of acetyl-CoA to malonyl-CoA. Malonyl-CoA serves as a potent inhibitor of carnitine palmitoyltransferase 1 α (CPT-1α) , which is the rate-limiting enzyme for mitochondrial fatty acid oxidation, as evidenced by increased fatty acid oxidation and leanness in mice deficient in ACC2 (Abu-Elheiga et al. 2001). On the other hand, malonyl-CoA produced by ACC acts as the initial substrate for fatty acid biosynthesis. Interestingly, the ability of metformin to lower triglyceride levels coincides with an increase in inhibitory phosphorylation of ACC1 and ACC2 by AMPK in vitro. The lipid-lowering effect of metformin can be explained by increased fatty acid oxidation and decreased fatty acid synthesis possibly in part through phosphorylation and inactivation of ACC1 and ACC2 (Zang et al. 2004). Recent studies from the Steinberg and Kemp group confirm these early findings in a novel mouse model with alanine knock-in mutations in both ACC1 at Ser79 and ACC2 at Ser212 (ACC double knock-in, ACC-DKI) (Fullerton et al. 2013). Compared to wild-type mice, the ACC-DKI mice fed a control diet display a complete loss of phosphorylation of ACC1 and ACC2 as well as have lowered fatty acid oxidation and elevated lipogenesis. Strikingly, ACC-DKI mice challenged with high fat feeding are refractory to the lipid-lowering and insulin-sensitizing effects of metformin. Taken together, these in vitro and in vivo findings illustrate that AMPK-dependent phosphorylation and inactivation of ACC at least partially contributes to metformin-induced improvements in insulin resistance in the setting of obesity (Fullerton et al. 2013).
Glucose production in the liver is essential for providing a substrate for the glucose utilization and survival of critical tissues, such as the brain and heart, during prolonged fasting. The inability of insulin to suppress hepatic glucose output contributes to the pathological process of hyperglycemia in type 2 diabetes. Over a decade ago, metformin was demonstrated to activate AMPK by increasing phosphorylation of the AMPK catalytic α subunit at Thr-172 in primary hepatocytes (Zhou et al. 2001; Zang et al. 2004; Kahn et al. 2005). A subsequent study showed that hepatic knockout of LKB1, the upstream kinase for AMPK, abolishes metformin-mediated suppression of hepatic glucose production. These studies also indicate that LKB1 mediates hepatic AMPK activation and therapeutic effects of metformin in mice (Shaw et al. 2005). Recent studies provide a novel mechanism by which metformin antagonizes the action of glucagon and thus reduces fasting glucose levels. Metformin is also shown to reduce levels of cyclic AMP and protein kinase A (PKA) activity that abrogates phosphorylation of critical protein targets of PKA and that blocks glucagon-dependent glucose output in hepatocytes (Miller et al. 2013).
2.2.2 AMPK and Polyphenols
Many of polyphenols are widely reported to have a favorable effect on type 2 diabetes, dyslipidemia, and cardiovascular disease in animal models with obesity (Bradamante et al. 2004; Vita 2005), but their mechanism(s) remain a mystery, limiting their therapeutic potential. In addition to galegine, a number of natural polyphenolic products—many derived from plants used as herbal medicines in Asian countries—are reported to activate AMPK. These include resveratrol from red grapes, quercetin present in many fruits and vegetables, berberine from Coptis chinensis (used in the Chinese herbal medicine Huanglian), epigallocatechin gallate from green tea, theaflavin from black tea, ginsenoside from Panax ginseng, and curcumin from Curcuma longa (Manach et al. 2004). Resveratrol, one of natural polyphenols, has multiple beneficial activities similar to those associated with caloric restriction (CR), such as increased life span and delayed onset of diseases associated with aging (Howitz et al. 2003). Elucidating the role of AMPK in diabetes makes some surprising discoveries and identifies novel agents that directly or indirectly activate AMPK (Kahn et al. 2005; Steinberg and Kemp 2009; Zhang et al. 2009). In vitro studies demonstrate that a synthetic polyphenol, S17834, strongly and persistently stimulates AMPK phosphorylation and activity at concentrations 50–200 times lower than those of AMPK-activating compounds, 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (AICAR) or metformin (Zang et al. 2006). Natural polyphenols such as resveratrol, a key component in red wine, have similar but less potent effects on hepatic AMPK activity. Mechanistic studies demonstrate that the activation of AMPK by polyphenols is dependent on LKB1, but not on CaMKKβ (Hou et al. 2008). These findings suggest that LKB1 functions as an upstream regulator for AMPK signaling in response to polyphenols. As a consequence, S17834 and resveratrol suppress hepatocellular lipid accumulation caused by high glucose largely through an AMPK-dependent mechanism (Zang et al. 2006). Further animal studies show decreased phosphorylation and activity of hepatic AMPK, elevated hepatic lipids, increased hyperlipidemia, and accelerated aortic atherosclerosis in type 1 diabetic low density lipoprotein receptor (LDLR) knockout mice. In contrast, treatment with S17834 in these animals prevents impaired hepatic AMPK and elevated hepatic and serum lipids and thereby suppresses the acceleration of atherosclerosis caused by diabetes. These studies define that AMPK activation may represent a novel molecular mechanism by which polyphenols, like S17834 and resveratrol, attenuate the deleterious effect of diabetic milieu on hepatic steatosis, hyperlipidemia, and atherogenesis (Zang et al. 2006).
These early and important findings have been confirmed by several independent groups. Results from Sinclair’s group indicate that resveratrol activates hepatic AMPK, improves insulin sensitivity, and modulates known longevity pathways (Baur et al. 2006). By using the mice deficient in the catalytic subunit of AMPK α1 or α2, Chung’s group indicates that the effect of resveratrol on insulin sensitivity, glucose tolerance, and physical endurance are diminished in the absence of either AMPK α1 or α2. Consistent with this, expression of genes involved in mitochondrial biogenesis was not induced by resveratrol in AMPK-deficient mice (Um et al. 2010). Strikingly, a clinical trial study shows calorie restriction-like effects of resveratrol supplementation on energy metabolism and on the metabolic profile in obese humans, where healthy subjects and obese patients are given with either a placebo or 150 mg/day resveratrol in a randomized double-blind crossover study for 30 days. Mechanistically, administration of resveratrol to obese patients leads to activated AMPK, increased SIRT1 and PGC-1α protein levels, and improved mitochondrial respiration in skeletal muscle (Timmers et al. 2011). Taken together, these animal experiments and clinical studies provide strong biochemical evidence that AMPK is the central target for the metabolic effects of resveratrol.
Berberine, a natural plant product, activates AMPK and benefits metabolic disorder in obese, insulin-resistant mice (Lee et al. 2006; Turner et al. 2008). Berberine treatment results in increased AMPK activity in 3T3-L1 adipocytes and L6 myotubes. Clinical trials of berberine in newly diagnosed type 2 diabetic subjects reveal favorable effects on plasma glucose, lipids, and HbA1c, although plasma berberine levels and AMPK activity are not assessed (Zhang et al. 2008; Yin et al. 2008).
3 Mammalian SREBP
3.1 The Structure of SREBP
Sterol regulatory element binding protein (SREBP) is a key regulator of intracellular lipid metabolism that belongs to the basic helix-loop-helix–leucine zipper (bHLH-Zip) family of transcription factors (Brown and Goldstein 1997, 2009; Horton et al. 2002; Goldstein and Brown 2008). SREBP differs from other bHLH-Zip proteins because it is synthesized as an inactive precursor bound to the endoplasmic reticulum (ER). The SREBP precursor with about 1150 amino acids consists of three domains: (i) an NH2-terminal domain of about 480 amino acids that contains the transactivation domain—a region of the bHLH-Zip domain for DNA binding and dimerization; (ii) two hydrophobic transmembrane-spanning segments interrupted by a short loop of about 30 amino acids that projects into the lumen of the ER; and (iii) a COOH-terminal domain of about 590 amino acids—a regulatory region that binds to the COOH-terminal domain of an escort protein known as SREBP cleavage activating protein (SCAP). The SREBP family has three isoforms: SREBP-1a, SREBP-1c, and SREBP-2. SREBP-1c is also referred to as adipocyte determination and differentiation dependent factor 1/ADD1. SREBP-1a and SREBP-1c are derived from a single gene by using alternative transcription start sites that produce alternate forms of the first exon. SREBP-1c is expressed in most adult tissues, with especially high expression in the liver; it is considered to be more physiologically relevant. Interestingly, SREBP-1c and SREBP-2 are highly expressed in the liver and also expressed in other tissues of adult animals. SREBP-1a appears to be constitutively expressed at low levels in almost all tissues of adult animals.
3.2 The Function of SREBP on Lipid Biosynthesis
SREBP family controls lipid metabolism by stimulating expression of more than 30 genes encoding a range of enzymes required for the synthesis or uptake of fatty acids, triacylglycerol, cholesterol, and phospholipid (Brown and Goldstein 1997; Horton et al. 2002; Goldstein and Brown 2008). Three SREBP isoforms have isoform-specific functions on lipid biosynthetic processes. In vivo studies using transgenic and knockout mice demonstrate that SREBP-1c, the predominant isoform in the liver, preferentially promotes a de novo lipogenic process by activating genes involved in fatty acid and triglyceride synthetic pathways, whereas SREBP-2 primarily activates a cholesterol biosynthetic pathway by stimulating expression of genes governing cholesterol synthetic pathways and uptake. In contrast, the SREBP-1a isoform, which is highly expressed in cell lines and tissues with a high capacity for cell proliferations such as intestine and spleen (Shimomura et al. 1997), is likely involved in both pathways.
3.3 The Regulation of SREBP Activity
The regulation of SREBP activity occurs at posttranscriptional and transcriptional levels (Brown and Goldstein 1997; Horton et al. 2002). The sterol-sensitive feedback inhibition process is mainly involved in the regulation of SREBP-2. Accumulating evidence suggests that the SREBP-1c isoform appears to be mainly regulated by glucose and insulin .
3.3.1 The Posttranscriptional Regulation of SREBP Activity
Sterols such as cholesterol are fundamental components of cellular membranes and precursors of molecules such as steroid hormones. Cholesterol can be obtained from the diet as well as endogenously synthesized de novo. Hence, energetically demanding cholesterol biosynthetic pathway is only active when external supply and internal levels of sterols are low. Adequate sensing of internal cholesterol levels involves the posttranscriptional regulation of SREBP. The control occurs through SREBP cleavage processing and movement from the endoplasmic reticulum (ER) to the Golgi apparatus. Proteolytic activation of SREBP is controlled by sterols through two intracellular sterol sensors: SCAP and Insulin-induced gene (Insig) (Ye and DeBose-Boyd 2011). SCAP contains an NH2-terminal domain with eight transmembrane helices and a cytosolic COOH-terminal domain that mediates the complex formation with the COOH-terminal regulatory domain of SREBP (Matsuda et al. 2001; Moon et al. 2012). Insig is a polytopic membrane protein with six transmembrane helices that binds to SCAP for the retention of the SCAP-SREBP complex in the ER membrane (Yang et al. 2002; Engelking et al. 2004; Flury et al. 2005; Rawson 2003b).
SREBP protein is initially synthesized as an inactive precursor that is inserted into ER membrane. The retention of SCAP-SREBP complex in the ER membrane is mediated by an increase in the binding of SCAP to the Insig protein and a decrease in the interaction of SCAP with the COPII vesicle-formation proteins Sar1, Sec23 and Sec24 (Rawson 2003a, b; Osborne and Espenshade 2009). Upon activation, the newly synthesized precursor form of SREBP (~125 kDa) migrates from the ER membrane to the Golgi apparatus where SREBP precursor undergoes a sequential two-step proteolytic processing mediated by the site 1 (S1P) and site 2 (S2P) proteases. This cleavage process can lead to the release of the transcriptionally active NH2-terminal domain of the protein (~68 kDa) in the Golgi apparatus. Once the mature, active nuclear form of SREBP, designated nuclear SREBP, is translocated into the nucleus, the nuclear SREBP can bind to the sterol regulatory element (SRE) sequence (5′-TCACNCCAC-3′) present in promoters/enhancers of SREBP and its target genes that activates the transcription of SREBP-responsive genes and promotes the lipid synthetic process in the liver (Horton et al. 2002; Raghow et al. 2008; Amemiya-Kudo et al. 2000).
Since the discovery of SREBP in 1993, the molecular mode of SREBP-2 regulation is well characterized. The proteolytic processing of SREBP-2 is mainly controlled by cellular sterols through the feedback inhibition (Fig. 2.3). When intracellular sterols rise, cholesterol binds to its sensor SCAP in the ER that leads to the conformation change of SCAP and promotes the binding of SCAP to the ER-resident protein Insig. Under this condition, the SCAP-SREBP complex, which is not incorporated into ER transport vesicles, cannot migrate to the Golgi apparatus. Because the bHLH-Zip domain cannot be released from the membrane, the transcription of SREBP-2 target genes is suppressed. When intracellular sterols decline, the precursor of SREBP-2 can be cleaved to produce the nuclear, active form of SREBP-2, which in turn induces cholesterogenic gene expression . Consequently, newly synthesized cholesterol leads to the feedback inhibition of SREBP-2 activity in order to maintain cholesterol homeostasis (Nohturfft et al. 2000; Yang et al. 2002). Additionally, sterol-regulated ubiquitination and degradation of Insig-1 create a convergent mechanism for the negative feedback control of SREBP-2 activity and its function on cholesterol synthesis and uptake (Gong et al. 2006).
The nutrient regulation of SREBP in the liver. (a) In the presence of cholesterol, cholesterol binds to the intracellular sterol sensor SCAP. The SCAP–SREBP complex binds to the Insig protein in the ER membrane and remains anchored in the ER. (b) In the absence of cholesterol, the SCAP–SREBP complex does not interact with the Insig protein, and the complex traffics to the Golgi where the cytoplasmic tail of SREBP is released by the proteolytic cleavage. For instance, the nuclear, active form of SREBP-2 triggers a transcriptional program in the nucleus through its binding to the SRE motif on promoters of its target genes such as HMG-CoA synthase and HMG-CoA reductase
SREBP-1 is likely regulated by fatty acids through the negative feedback regulation. In mice, polyunsaturated fatty acids (PUFA) inhibit mRNA expression of hepatic SREBP-1c, but they are not affect SREBP-1a gene expression (Yoshikawa et al. 2002). The transcription of SREBP-1c is antagonized by PUFA through the effect of liver X receptor (LXR), because two LXR-responsive elements (LXREs) are identified on the promoter of the SREBP-1c gene, but not on that of the SREBP-1a gene (Repa et al. 2000). Strikingly, unsaturated fatty acids also inhibit proteolytic activation of SREBP-1 (Hannah et al. 2001). Further studies indicate that unsaturated fatty acids can have the greatest inhibitory effect on SREBP-1 (Hannah et al. 2001), whereas sterols have the greatest inhibitory effects on SREBP-2.
Mature nuclear SREBP is highly unstable owing to its ubiquitin-dependent degradation. Results from Ericsson’s group indicate that the nuclear form of SREBP is rapidly degraded by the ubiquitin-proteasome pathway through the action of an E3 ubiquitin ligase Fbw7 (Sundqvist et al. 2005). Fbw7 negatively regulates the stability and function of nuclear SREBP-1 and SREBP-2 by promoting the ubiquitination and proteasome-mediated degradation via GSK3 dependent-phosphorylation of SREBP (Sundqvist et al. 2005). Because GSK3 activity is shown to be inhibited by insulin signaling, it suggests this non-transcriptional mechanism may be involved in insulin-induced SREBP. Moreover, the coactivator PGC-1β activity can bind to SREBP-1c, which can explain SREBP-induced lipogenesis in response to a fat-rich diet (Lin et al. 2005). Importantly, these factors permit the modulation of SREBP-1c expression independently of sterol-regulated proteolytic processing of SREBP-1.
3.3.2 The Transcriptional Regulation of SREBP Activity
The central role of SREBP in controlling lipid synthesis has recently been highlighted by the multiple inputs to regulate SREBP activity from different nutrient sensing pathways . In addition to the posttranscriptional regulation of SREBP, the transcriptional regulation of the SREBP is more complex (Eberle et al. 2004; Osborne and Espenshade 2009). SREBP-1a appears to be constitutively expressed at low levels in most tissues. SREBP-1c and SREBP-2 in the liver are transcriptionally regulated through at least three mechanisms.
In the first mechanism, a feed-forward transcriptional regulation of SREBP-1c or SREBP-2 acts through the binding of nuclear SREBP to the SRE motif (5′-TCACNCCAC-3′) present in the promoter/enhancer region of each gene (Dif et al. 2006). Elevated nuclear SREBP-1c and SREBP-2 activate the transcription of their own genes via the auto-regulatory loop (Horton et al. 2002). The feed-forward stimulation may explain increased hepatic mRNA levels of SREBP-1c and SREBP-2 observed in transgenic animals with overexpression of nuclear SREBP-1a and in obesity-induced insulin-resistant mice (Horton et al. 2003; Li et al. 2011). In contrast, when the nuclear SREBP declines, a secondary decline in mRNA expression of SREBP-1c and SREBP-2 is also seen in the liver of S1P- and SCAP-deficient animals (Yang et al. 2001; Matsuda et al. 2001).
In the second mechanism, the transcription of SREBP-1c is selectively stimulated by nutrients and hormones (Horton et al. 2002; Brown and Goldstein 2008). This concept can be illustrated by examining the effects of nutrient availability and deprivation on SREBP-1c. Prolonged fasting or food deprivation leads to a decrease in plasma glucose and insulin levels and an increase in plasma-free fatty acid concentrations. Hyperglycemia stimulates lipogenesis via three possible mechanisms: (i) glucose itself could be a substrate for the de novo lipogenesis in hepatocytes, because glucose promotes fatty acid synthesis through its conversion to acetyl-CoA via a glycolytic pathway; (ii) glucose induces the expression of lipogenic genes, the mechanisms of which are explained by activating SREBP-1c; and (iii) glucose increases lipogenesis by stimulating the release of insulin and inhibiting the release of glucagon from the pancreas. The metabolic adaption to fasting is associated with changes in plasma hormone concentrations, such as a decrease in plasma insulin levels and an increase in plasma glucagon levels . This notion is supported by the observation that the transcription of SREBP-1c is controlled positively by insulin and negatively by glucagon and cyclic AMP in primary hepatocytes, establishing a link between this transcription factor and carbohydrate availability (Foretz et al. 1999b)
In the third mechanism, the transcription of SREBP-1c is selectively regulated through the cooperation of nuclear receptors with DNA-binding properties. Nuclear receptors , such as liver X receptor (LXR) and retinoid X receptor (RXR), function as a heterodimer to selectively upregulate SREBP-1c (Liang et al. 2002; Dif et al. 2006). The induction of SREBP-1c by LXR is supported by the observation that expression of hepatic SREBP-1c and fatty acid synthetic genes is increased by synthetic LXR agonists and these effects are reduced in LXRα- and LXRβ-deficient mice (Repa et al. 2000). Similarly, critical fatty acid synthetic genes are not upregulated by LXR agonists in SREBP-1c knockout mice (Liang et al. 2002). One major function of the liver is to convert excess carbohydrates to fatty acids for storage as triglycerides . It has been shown that insulin stimulates fatty acid synthesis in response to excess carbohydrates and that the lipogenic effect of insulin in the liver is mediated by SREBP-1c. Although a complete understanding of the regulation of the SREBP-1c by insulin requires further investigation, a study shows that the stimulatory effect of insulin on SREBP-1c transcription is mediated by LXR and liver X receptor (LXR)-responsive elements (LXREs) in the promoter region of SREBP-1c, but not that of SREBP-1a (Okazaki et al. 2010; Chen et al. 2004). Insulin activates the transcription of SREBP-1c promoter primarily by increasing LXR activity, possibly through production of a ligand that activates LXR or their heterodimerizing partner, the retinoid X receptor. Interestingly, polyunsaturated fatty acids (PUFA) inhibit SREBP-1c and fatty acid synthesis by antagonizing LXR-dependent activation of SREBP-1c (Yoshikawa et al. 2002).
The unique regulation and functions of each SREBP isoform facilitate the coordinate control of lipid metabolism ; however, the signaling pathways that specifically regulate each SREBP isoform are not fully understood.
The dysregulation of SREBP-1c has been implicated in the pathogenesis of hepatic steatosis and dyslipidemia in type 2 diabetes (Raghow et al. 2008). This transcription factor is identified as an attractive target for the development of new pharmaceutical interventions for metabolic disorders such as hypertriglyceridemia and obesity (Tang et al. 2011). Elucidating this mechanism for the regulation of SREBP will be fundamental to understanding the molecular basis of de novo lipogenesis and selective insulin resistance.
4 Role of AMPK-SREBP Nutrient Signaling in the Regulation of Hepatic Lipid Synthesis
Previous studies show that there is an inverse correlation between AMPK activity and SREBP-1c mRNA expression in livers of mice following re-feeding (Zhou et al. 2001; Foretz et al. 2005). However, little is known about how SREBP is downregulated by AMPK and how this regulation plays a role in the regulation of liver and systemic lipid homeostasis and slows the progression of insulin resistance and type 2 diabetes . Another important but unanswered question is how elevated hepatocellular cholesterol levels, a condition that is known to cause the negative feedback regulation of SREBP in physiological stimuli such as refeeding , lead to the aberrant activation of SREBP in pathological conditions such as insulin resistance and type 2 diabetes.
4.1 AMPK Suppresses the Transcriptional Activity of SREBP-1 and SREBP-2
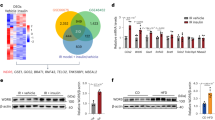
Recent studies demonstrate that hepatic AMPK activity, as determined by phosphorylation of AMPK and ACC, is repressed in insulin-resistant mice fed on a high fat, high sucrose diet. This impairment is reversed by treatment with S17834, a polyphonic AMPK activator (Li et al. 2011), which is consistent with increased hepatic AMPK activity seen in S17834-treated type 1 diabetic mice (Zang et al. 2006). Importantly, this study shows that AMPK activation by S17834 leads to a decrease in proteolytic processing of SREBP-1c and in mRNA expression of SREBP-1c in the liver of insulin-resistant mice, without affecting gene expression of SREBP-1a (Li et al. 2011). Consistently, mRNA expression of SREBP-1c target genes including acetyl-CoA carboxylase (ACC1), fatty acid synthase (FAS), and stearoyl-CoA desaturase-1 (SCD1) is suppressed, suggesting decreased rates of fatty acid and triglyceride synthesis in the liver. As a consequence, hepatic and plasma triglyceride concentrations are significantly reduced by the AMPK activator. These in vivo studies suggest that increasing AMPK activity reduces hepatic fatty acid and triglyceride synthesis and ameliorates hepatic steatosis by inhibiting SREBP-1c- dependent lipogenesis (Li et al. 2011).
SREBP-2 has been implicated in the regulation of cholesterol synthesis and uptake in the liver . SREBP-2 drives a cholesterol synthetic process by stimulating gene expression of cholesterol biosynthetic enzymes including rate-limited enzymes, 3’-hydroxy-methylglutaryl coenzyme A synthase (HMG-CoA synthase) and 3’-hydroxy-methylglutaryl coenzyme A reductase (HMG-CoA reductase) (Shimano et al. 1997). To further elucidate the precise mechanism by which AMPK regulates lipid homeostasis, recent studies indicate that high levels of nuclear active form and mRNA expression of hepatic SREBP-2 are associated with AMPK inhibition in obesity-induced insulin-resistant mice. Interestingly, nuclear accumulation of SREBP-2 is reduced by the AMPK activator, which correlates with a decrease in hepatic expression of SREBP-2 and its target genes, HMG-CoA synthase and HMG-CoA reductase. The suppression of SREBP-2 causes a reduction in hepatic cholesterol synthesis and content. The decrease in hepatic cholesterol synthesis in the liver is likely the major driving force leading to the lipid-lowering effect of AMPK (Li et al. 2011). Collectively, AMPK-mediated inhibition of SREBP-1 and SREBP-2 in the liver may explain the favorable effect of pharmacological activation of AMPK on obesity-induced hepatic steatosis, insulin resistance, and hyperlipidemia (Li et al. 2011).
To support the hypothesis that AMPK is an upstream regulator to inhibit SREBP-1 activity at the posttranslational level, in vitro mechanistic experiments are performed to obtain several important observations in freshly isolated mouse hepatocytes or human HepG2 hepatoma cells (Li et al. 2011). First, hepatic triglyceride content is dramatically increased in hepatocytes exposed to high glucose or high glucose plus insulin that mimic hyperglycemia and insulin resistance in vivo (Zang et al. 2004, 2006; Hou et al. 2008; Li et al. 2011). Under this condition, cleavage processing of SREBP-1 and mRNA expression of SREBP-1c are increased, whereas gene expression of SREBP-1a is rarely affected. This suggests the feed-forward stimulation of SREBP-1c caused by high concentrations of glucose and insulin. Interestingly, polyphenols and metformin , two pharmacological activators of AMPK (Zhou et al. 2001; Zang et al. 2006; Um et al. 2010), suppress nuclear translocation and accumulation of SREBP-1 and reduce gene expression of SREBP-1c, but not SREBP-1a, leading to the inhibition of a feed-forward regulation of SREBP-1c. Second, the ability of AMPK activators such as resveratrol to repress the cleavage processing of SREBP-1 and expression of lipogenic genes are largely abrogated by overexpression of a dominant negative form of AMPK, which encodes a catalytically inactive AMPKα2 bearing a mutation altering lysine 45 to arginine (K45R) in the kinase domain of AMPKα2 subunit that exerts a dominant negative effect on AMPKα1 and AMPKα2 activities (Zang et al. 2004). These results suggest that resveratrol represses SREBP activity in an AMPK-dependent manner. Third, overexpression of the constitutively active mutant of AMPK, which contains a carboxyl-terminal truncated form of AMPKα2 lacking the auto-inhibitory domain and the region interacting with β and γ regulatory subunits of AMPK (Zang et al. 2004), increases AMPK activity in hepatocytes . The constitutively active form of AMPK also decreases mRNA expression of SREBP-1c and FAS. Fourth, unlike other bHLH-Zip transcription factors that contain a well conserved arginine residue in the bHLH-Zip domain, SREBP has a conserved tyrosine residue that enables SREBP to bind on both E-boxes, like all bHLH proteins, and SRE sequences on target genes of SREBP-1c (Kim et al. 1995). To determine whether SREBP-1c is required for AMPK-medicated inhibition of the lipogenic process, a dominant negative mutant of SREBP-1c, which disrupts the binding of SREBP-1c to the SRE motif owning to a point mutation of tyrosine 320 to arginine on a carboxyl-terminal truncated SREBP-1c (1-403) (Kim et al. 1995), is used to be transduced into hepatocytes. In vitro experiments demonstrate that AMPK suppresses the transcriptional activity of FAS promoter in a SREBP-1c-dependent manner. Lastly, to precisely elucidate the mechanism by which AMPK inhibits SREBP transcription, in vitro experiments is performed using luciferase reporters e ncoding different lengths of human SREBP-1c promoters (Dif et al. 2006). AMPK activators decrease the transcriptional activity of the wild-type SREBP-1c promoter (−1470/+90 and −257/+90). Conversely, disruption of the SRE motif present on the SREBP-1c promoter reduces the basal transcription of SREBP-1c and prevents the effect of AMPK activators on SREBP-1c promoter activity. These results indicate that the SRE motif is responsible for AMPK inhibiting the auto-regulation of SREBP-1c. Taken together, these biochemical studies provide strong evidence that AMPK inhibits de novo lipogenesis by downregulating the cleavage processing of SREBP-1c and suppressing its feed-forward regulation in hepatocytes and in vivo insulin-resistant states (Li et al. 2011).
4.2 AMPK Interacts with and Directly Phosphorylates SREBP
Recent studies provide evidence that SREBP serves as a conserved substrate of AMPK (Li et al. 2011). The modulation of protein-protein interaction dynamically regulates protein phosphorylation, activity, subcellular localization, and stability. To delineate the mechanism by which AMPK downregulates SREBP activity, co-immunoprecipitation experiments show that the α catalytic subunit of AMPK physically interacts with the precursor and nuclear form of SREBP-1. In addition to the repression of SREBP-2 by AMPK in mouse livers, biochemical experiments also demonstrate that the AMPKα subunit physically associates with the SREBP-2 precursor (Li et al. 2011). Because the formation of the AMPK-SREBP complex is also increased by a constitutively active form of AMPK, it postulates that this interaction makes the kinase domain of the AMPKα subunit more accessible to putative substrates, SREBP-1 and SREBP-2, for the phosphorylation modification. It has been characterized that phosphorylation at Thr-172 in the activation loop of the kinase domain of the AMPKα subunit can trigger an increase in AMPK kinase activity by converting this enzyme from an inactive conformation to a catalytically active conformation (Hardie et al. 2012). AMP or ADP can bind to the AMPK γ regulatory subunit that promotes AMPK phosphorylation by upstream kinases as well as protects the AMPK enzyme against dephosphorylation by phosphatases. AMP can also cause allosteric activation of AMPK (Hardie et al. 2012). Furthermore, biochemical studies have discoverd the crystal structure of human AMPKα1 subunit and provided structure insight into that the AMPKα1 subunit as being held in an inactive conformation that renders the AMPK enzyme inactive owning to the association of the auto-inhibitory domain with the kinase domain of the AMPKα1 subunit (Chen et al. 2009). When intracellular AMP levels rise, AMP is bound to the AMPKγ subunit. Consequently, the inhibitory domain of the AMPKα1 subunit is released from its kinase domain. This disassociation leads to an active conformation of AMPK, which allows the upstream kinase such as LKB1 to phosphorylate the Thr-172 site on the kinase domain of the AMPKα subunit (Young 2009). Such a specific active conformation of the AMPKα subunit may possibly enable the kinase domain of the AMPKα subunit to be more accessible to its substrates. This possibility is supported by the fact that the active AMPKα subunit preferentially binds to and phosphorylates SREBP-1c and SREBP-2 (Li et al. 2011).
It is known that phosphorylation sites tend to be located on protein–protein binding interfaces and may modulate protein activity. Bioinformatics tools including Scansite (http://mit.scansite.edu) are used to identify candidate substrates bearing optimal AMPK motifs, in which the target serine and its critical flanking residues are conserved broadly throughout eukaryotes (Gwinn et al. 2008). To further search a consensus phosphorylation motif of AMPK on human SREBP-1c sequence, human SREBP-1c is identified to have two putative AMPK sites, Ser-336 and Ser-372 located in the N-terminal region of SREBP-1c, which match the AMPK consensus motif. Because a high degree of conservation in consensus sequences is important for AMPK phosphorylation, only one serine site of SREBP-1c—the Ser-372 on human SREBP-1c, is conserved. Strikingly, critical residues flanking Ser-372 of human SREBP-1c are highly conserved across mouse, rat, dog, bovine as well as in zebrafish (Li et al. 2011). Sequence alignments also show the evolutionary conservation of the motif sequences surrounding Ser-372 on human SREBP-1c and surrounding Ser-792 on Raptor, the best-established substrate of AMPK (Gwinn et al. 2008). The remarkable conservation in the candidate AMPK phosphorylation site on SREBP-1 may represent an AMPK target that dictates the responsiveness of AMPK across mammals.
To test whether SREBP is directly phosphorylated by AMPK, in vitro kinase assays with purified AMPK in rat livers are performed using recombinant SREBP-1 or SREBP-2 as a substrate in the presence of AMP as described previously (Inoki et al. 2006; Greer et al. 2007; Gwinn et al. 2008). Active AMPK substantially phosphorylates SREBP-1c or SREBP-2 in the presence of [γ-32P]-ATP in vitro. Mutagenic studies demonstrate that a point mutation of Serine 327 to alanine (S3721A) on human SREBP-1c, but not wild-type SREBP-1c, eliminates AMPK-triggered phosphorylation of SREBP-1c. To characterize the biological importance for the phosphorylation of SREBP-1c, a specific phospho-specific antibody against Ser-372 on human SREBP-1c is generated. Active AMPK potently stimulate Ser-372 phosphorylation on wild-type SREBP-1c, but not that on the S372A mutant, indicating that the Ser-372 on SREBP-1c is a major phosphorylation target of AMPK in vitro (Li et al. 2011).
To determine the functional consequence of the Ser-327 phosphorylation of SREBP-1c, further studies have demonstrated that AMPK is necessary for the induction of phosphorylation at Ser-372 in response to pharmacological activators of AMPK in cultured cells, since the ability of metformin and polyphenols to increase the phosphorylation at Ser-372 is largely diminished by a dominant-negative AMPK form in hepatocytes (Li et al. 2011). Moreover, like phosphorylation of AMPK and ACC, phosphorylation of Ser-327 is stimulated by AMPK activators in AMPK+/+ MEFs. In contrast, phosphorylation of SREBP-1c is diminished in cells lacking AMPKα1 and AMPKα2 subunits. These results strongly demonstrate that Ser-372 is potently phosphorylated by AMPK in vitro and in intact cells. To define whether AMPK-stimulated phosphorylation of SREBP-1c is physiologically relevant, mechanistic studies are performed using the phosphorylation-defective S372A mutant. Luciferase reporter assays show that the S372A mutant strongly diminishes the effect of AMPK activators on SREBP-1c activity. The results presented above indicate that AMPK inhibits the transcriptional activity of SREBP-1c in a Ser-372 phosphorylation-dependent manner. Furthermore, consistent with the changes in hepatic AMPK activity, hepatic phosphorylation of SREBP-1c at Ser-372 is reduced in insulin-resistant mice, and defective AMPK-SREBP signaling is prevented by treatment with the AMPK-activating polyphenol, revealing the in vivo physiological significance of AMPK-dependent phosphorylation of SREBP-1c (Li et al. 2011). Although AMPK can cause phosphorylation of SREBP-1c at Ser-372 in intact cells, it cannot rule out that other phosphorylation sites on SREBP-1c may be involved in the metabolic action of AMPK.
An additional mechanism for understanding the role of AMPK in the regulation of cholesterol metabolism has recently emerged (Li et al. 2011). The induction of the cleavage processing and gene expression of hepatic SREBP-2 is reduced in insulin-resistant mice treated with the AMPK activator in a similar manner to that of SREBP-1. Because both SREBP-1c and SREBP-2 exert similar cleavage processing and feed-forward regulation, phosphorylation of SREBP-2 is possibly involved in AMPK actions. Further work is required for the identification of potential phosphorylation sites present on SREBP-2 and confirm that SREBP-2 is a direct target of AMPK in intact cells. While previous studies demonstrate that AMPK directly phosphorylates acetyl-CoA carboxylase and HMG-CoA reductase , the rate-limited enzymes that regulate fatty acid and cholesterol biosynthesis, the phosphorylation regulation of SREBP by AMPK highlights the biological importance for the multilayered control of hepatic lipid biosynthesis under conditions of metabolic stresses. Therefore, the identification of the nutrient signaling crosstalk between AMPK and SREBP represents an important advance in our understanding of the molecular mechanisms linking hepatic lipogenesis to insulin resistance, hepatic steatosis, and hyperlipidemia (Fig. 2.4) (Li et al. 2011).
A proposed model of the phosphorylation regulation of SREBP by AMPK in the liver: potential therapeutic implication in hepatic steatosis and insulin resistance. AMPKα subunit binds to precursors of SREBP-1c and SREBP-2 and makes them better substrates for phosphorylation. Phosphorylation of SREBP may trigger a conformation change of SREBP that inhibits its proteolytic cleavage and the release of the transcriptionally active N-terminal bHLH-Zip domain and thereby reduces its nuclear translocation in hepatocytes. Consequently, AMPK suppresses the feed-forward activation of SREBP-1c and SREBP-2 and represses the transcription of their target lipogenic genes at least in part by reduced binding of SREBP to SRE in the promoters of SREBP-responsive genes including fatty acid synthase (FAS), HMG CoA reductase (HMGCR), and LDLR. AMPK-dependent phosphorylation and repression of SREBP in the liver may represent a novel mechanism for AMPK activators, such as polyphenols and metformin, to protect against hepatic steatosis and type 2 diabetes. Figure modified from Li et al, Cell Metabolism, 2011, 13(4): 376–388
4.3 Role of Other Nutrient Sensors in the Regulation of Hepatic SREBP Activity
The molecular mechanisms linking obesity to insulin resistance are complex. The increase in hepatic glucose production in the liver, is possibly attributed to the inhibition of insulin-mediated suppression of gluconeogenesis . Paradoxically, insulin-induced nuclear accumulation and expression of hepatic SREBP-1c contribute substantially to hepatic lipogenesis observed in obesity-induced type 2 diabetic mice (Laplante and Sabatini 2010). However, the molecular mechanisms underlying hepatic selective insulin resistance have not yet been characterized. Brown, Goldstein and co-workers have reported that the mammalian target of rapamycin complex 1 (mTORC1) plays an essential role in mediating the ability of hyperinsulinemia to stimulate SREBP-1c and lipogenesis in type 2 diabetes (Li et al. 2010). Interestingly, inhibition of mTORC1 by rapamycin reduces the mRNA expression of SREBP-1c in primary hepatocytes, whereas hepatic gluconeogenesis is unaffected (Li et al. 2010). The role of mTORC1 in controlling insulin-regulated hepatic lipogenesis, but not in insulin-mediated suppression of gluconeogenesis , provides a molecule basis for understanding the selective nature of hepatic insulin resistance. Recently, Sabatin’s group has reported that mTORC1-induced nuclear translocation of Lipin 1, a phosphatidic acid phosphatase, controls the transcriptional activity of SREBP-1 in hepatocytes and that activation of mTORC1-Lipin 1 signaling contributes to diet-induced hepatic steatosis and hypercholesterolemia in mice (Peterson et al. 2011).
Selective insulin resistance is a very challenging question in anti-diabetic therapies, because treatment of type 2 diabetes patients with large doses of insulin can overwhelm the insulin resistance and control the blood glucose levels, but insulin further enhances hepatic lipid synthesis and secretion while increasing lipotoxicity . To better understand the mechanism of selective insulin resistance, recent studies suggest that dysregulation of AMPK signaling may contribute to the induction of hepatic SREBP-1c and lipogenesis, since the transcription of SREBP-1c and FAS is upregulated in cells lacking AMPKα1 and α2 (Li et al. 2011). Given the demonstration that AMPK is a key kinase that negatively regulates mTORC1 (Inoki et al. 2006; Gwinn et al. 2008), it would be important to evaluate whether SREBP-1c processing is affected by AMPK through the inhibition of mTORC1 in hepatocytes . Alternatively, the possibility that mTORC1 could interfere with the action of LXR on SREBP-1c gene is considered because activation of AMPK suppresses SREBP-1c activity in a LXR-dependent fashion (Yap et al. 2011).
Kemper’s group have reported that SREBP-1c is an in vivo target of SIRT1 (Ponugoti et al. 2010). SIRT1 interaction with SREBP-1c is increased by fasting. Consistently, decreased SREBP-1c acetylation levels in mouse livers are associated with decreased lipogenic gene expression during fasting. In vivo knockdown of hepatic SIRT1 increases the acetylation of SREBP-1c, accompanied with elevated lipogenic gene expression. Tandem mass spectrometry and mutagenesis studies further reveal that SREBP-1c is acetylated by p300 at Lys-289 and Lys-309. Mechanistic studies using acetylation-defective mutants indicate that SIRT1 deacetylates and inhibits SREBP-1c transactivation by decreasing its stability and its occupancy at the lipogenic genes. Näär’s group has also reported a conserved role of SIRT1 orthologs in fasting-dependent inhibition of SREBP. These results demonstrate that deacetylation and degradation of nuclear SREBP-1c and SREBP-2 by SIRT1 occur in response to fasting, which in turn inhibits hepatic lipogenic and cholesterol synthetic processes (Walker et al. 2010). It has been shown that SIRT1 inhibits fatty acid synthesis and lipid accumulation caused by high glucose through activation of AMPK in cultured hepatocytes (Hou et al. 2008). According to the results presented above, AMPK inhibits SREBP-1c activity in a phosphorylation-dependent fashion. Further work is of interest to determine whether the effect of SIRT1 on SREBP-1c is possibly mediated by AMPK-dependent phosphorylation of SREBP-1c.
AMPK phosphorylates SREBP-1c at Ser-372, which is different from the residue regulated by other protein kinases. For instance, protein kinase A has been previously identified to attenuate the binding of nuclear SREBP-1c to SRE-containing promoters of SREBP-1c-responsive genes through phosphorylation of nuclear SREBP-1c at Ser-314, without altered its cleavage processing (Lu and Shyy 2006). GSK3 directly phosphorylates nuclear SREBP-1a at Thr-426/Thr-420 and SREBP-2 at Ser-433 that mediate Fbw7-induced ubiquitination and degradation of their nuclear forms (Sundqvist et al. 2005). Recent study provides additional insight that unlike the effect of these kinases on the nuclear form of SREBP-1c, AMPK binding to SREBP-1c precursor triggers the Ser-327 phosphorylation. The phosphorylation resistant S372A mutant abrogates the ability of AMPK to inhibit SREBP-1c cleavage and nuclear translocation (Li et al. 2011).
Recent studies indicate that hepatic AMPK regulates triglyceride and cholesterol metabolism at least partially through the downregulation of proteolytic processing of SREBP-1c and SREBP-2 in obese, insulin-resistant mice (Zang et al. 2006). These animals share some features with SCAP knockout mice (Matsuda et al. 2001) and with transgenic mice overexpressing Insig-1 in the liver (Engelking et al. 2004) where nuclear amounts of both SREBP-1 and SREBP-2 decline, due to the interruption of proteolytic cleavage of their precursors. Since SCAP and Insig play a key role in sterol-mediated negative feedback regulation of SREBP, it would be of interest to further determine whether SCAP and Insig are involved in the ability of AMPK to regulate SREBP proteolytic processing.
5 Conclusion
Over a decade ago, AMPK is identified as a fundamental regulator of cellular metabolism and coordinates several metabolic responses in different cells. Subsequent work has shown that AMPK plays important roles in different biological processes and influences various metabolic disorders associated with diabetes. Though our understanding of nutrient-sensing mechanisms is far from complete, significant progress has been achieved. Recent studies provide compelling biochemical evidence that AMPK-dependent phosphorylation and inactivation of SREBP lead to inhibited cleavage processing and nuclear translocation of SREBP and suppressed its auto-regulatory loop and lipogenic gene's transcription. The underlying mechanism plays an important role in controlling hepatocyte and whole-body lipid homeostasis, as well as in mediating the therapeutic effects of AMPK activators on insulin resistance. While some of metabolic consequences of the nutrient-sensing AMPK-SREBP, such as those that occur in obese states, have been defined, we have yet determined whether exercise modulates this nutrient sensing pathway or how ageing affects nutrient-sensing abilities. Additionally, nutrient excess not only affects the onset of diabetes but also influences cancer development and the ageing process. Nutrient sensing and metabolism in cancer cells have received a new attention, partly due to advances in metabolomics and next-generation sequencing. Therefore, understanding nutrient-sensing mechanisms is a prerequisite for designing better interventions against human diseases such as diabetes, non-alcoholic fatty liver disease, and cancer.
References
Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ (2001) Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science 291(5513):2613–2616
Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Osuga J, Harada K, Gotoda T, Sato R, Kimura S, Ishibashi S, Yamada N (2000) Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem 275(40):31078–31085. doi:10.1074/jbc.M005353200
Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang MY, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444(7117):337–342
Bradamante S, Barenghi L, Villa A (2004) Cardiovascular protective effects of resveratrol. Cardiovasc Drug Rev 22(3):169–188
Brown MS, Goldstein JL (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89(3):331–340. doi:10.1016/s0092-8674(00)80213-5
Brown MS, Goldstein JL (2008) Selective versus total insulin resistance: a pathogenic paradox. Cell Metab 7(2):95–96. doi:10.1016/j.cmet.2007.12.009
Brown MS, Goldstein JL (2009) Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res 50(Suppl):S15–S27. doi:10.1194/jlr.R800054-JLR200
Chen G, Liang G, Ou J, Goldstein JL, Brown MS (2004) Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci U S A 101(31):11245–11250. doi:10.1073/pnas.0404297101
Chen L, Jiao ZH, Zheng LS, Zhang YY, Xie ST, Wang ZX, Wu JW (2009) Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature 459(7250):1146–1149. doi:10.1038/nature08075
Dentin R, Girard J, Postic C (2005) Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie 87(1):81–86
Dif N, Euthine V, Gonnet E, Laville M, Vidal H, Lefai E (2006) Insulin activates human sterol-regulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. Biochem J 400:179–188
Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F (2004) SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86(11):839–848
Engelking LJ, Kuriyama H, Hammer RE, Horton JD, Brown MS, Goldstein JL, Liang G (2004) Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J Clin Invest 113(8):1168–1175
Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong ZF, Dupuy F, Chambers C, Fuerth BJ, Viollet B, Mamer OA, Avizonis D, DeBerardinis RJ, Siegel PM, Jones RG (2013) AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab 17(1):113–124. doi:10.1016/j.cmet.2012.12.001
Flury I, Garza R, Shearer A, Rosen J, Cronin S, Hampton RY (2005) INSIG: a broadly conserved transmembrane chaperone for sterol-sensing domain proteins. EMBO J 24(22):3917–3926
Foretz M, Guichard C, Ferre P, Foufelle F (1999a) Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci U S A 96(22):12737–12742
Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Le Liepvre X, Berthelier-Lubrano C, Spiegelman B, Kim JB, Ferre P, Foufelle F (1999b) ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol 19(5):3760–3768
Foretz M, Ancellin N, Amdreelli F, Saintillan Y, Grondin P, Kahn A, Thorens B, Vaulont S, Viollet B (2005) Short-term overexpression of a constitutively active form of AMP-activated protein kinase in the liver leads to mild hypoglycemia and fatty liver. Diabetes 54(5):1331–1339
Foretz M, Hebrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, Sakamoto K, Andreelli F, Viollet B (2010) Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest 120(7):2355–2369. doi:10.1172/jci40671
Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen ZP, O’Neill HM, Ford RJ, Palanivel R, O’Brien M, Hardie DG, Macaulay SL, Schertzer JD, Dyck JR, van Denderen BJ, Kemp BE, Steinberg GR (2013) Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med 19(12):1649–1654. doi:10.1038/nm.3372
Goldstein JL, Brown MS (2008) From fatty streak to fatty liver: 33 years of joint publications in the JCI. J Clin Invest 118(4):1220–1222. doi:10.1172/jci34973
Gong Y, Lee JN, Lee PC, Goldstein JL, Brown MS, Ye J (2006) Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab 3(1):15–24. doi:10.1016/j.cmet.2005.11.014
Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A (2007) The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem 282(41):30107–30119
Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30(2):214–226
Hannah VC, Ou J, Luong A, Goldstein JL, Brown MS (2001) Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J Biol Chem 276(6):4365–4372. doi:10.1074/jbc.M007273200
Hardie DG, Ross FA, Hawley SA (2012) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13(4):251–262. doi:10.1038/nrm3311
Hasty AH, Shimano H, Yahagi N, Amemiya-Kudo M, Perrey S, Yoshikawa T, Osuga J, Okazaki H, Tamura Y, Iizuka Y, Shionoiri F, Ohashi K, Harada K, Gotoda T, Nagai R, Ishibashi S, Yamada N (2000) Sterol regulatory element-binding protein-1 is regulated by glucose at the transcriptional level. J Biol Chem 275(40):31069–31077. doi:10.1074/jbc.M003335200
Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG (2005) Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab 2(1):9–19. doi:10.1016/j.cmet.2005.05.009
Horton JD, Goldstein JL, Brown MS (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109(9):1125–1131
Horton JD, Shimomura I, Ikemoto S, Bashmakov Y, Hammer RE (2003) Overexpression of sterol regulatory element-binding protein-1a in mouse adipose tissue produces adipocyte hypertrophy, increased fatty acid secretion, and fatty liver. J Biol Chem 278(38):36652–36660
Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, Cohen RA, Zang M (2008) SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem 283(29):20015–20026
Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425(6954):191–196
Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL (2006) TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126(5):955–968
Kahn BB, Alquier T, Carling D, Hardie DG (2005) AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab 1(1):15–25
Kim JB, Spotts GD, Halvorsen YD, Shih HM, Ellenberger T, Towle HC, Spiegelman BM (1995) Dual DNA-binding specificity of ADD1/SREBP1 controlled by a single amino-acid in the basic helix-loop-helix domain. Mol Cell Biol 15(5):2582–2588
Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM (1998) Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest 101(1):1–9
Laplante M, Sabatini DM (2010) mTORC1 activates SREBP-1c and uncouples lipogenesis from gluconeogenesis. Proc Natl Acad Sci U S A 107(8):3281–3282. doi:10.1073/pnas.1000323107
Lee YS, Kim WS, Kim KH, Yoon MJ, Cho HJ, Shen Y, Ye JM, Lee CH, Oh WK, Kim CT, Hohnen-Behrens C, Gosby A, Kraegen EW, James DE, Kim JB (2006) Berberine, a natural plant product, activates AMP-activated protein kinase with beneficial metabolic effects in diabetic and insulin-resistant states. Diabetes 55(8):2256–2264. doi:10.2337/db06-0006
Li S, Brown MS, Goldstein JL (2010) Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A 107(8):3441–3446. doi:10.1073/pnas.0914798107
Li Y, Xu SQ, Mihaylova MM, Zheng B, Hou XY, Jiang BB, Park O, Luo ZJ, Lefai E, Shyy JYJ, Gao B, Wierzbicki M, Verbeuren TJ, Shaw RJ, Cohen RA, Zang MW (2011) AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab 13(4):376–388
Liang GS, Yang J, Horton JD, Hammer RE, Goldstein JL, Brown MS (2002) Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem 277(11):9520–9528
Lin JD, Yang RJ, Tarr PT, Wu PH, Handschin C, Li SM, Yang WL, Pei LM, Uldry M, Tontonoz P, Newgard CB, Spiegelman BM (2005) Hyperlipidemic effects of dietary saturated fats mediated through PGC-1 beta coactivation of SREBP. Cell 120(2):261–273
Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 23(4):833–843
Lu M, Shyy JYJ (2006) Sterol regulatory element-binding protein 1 is negatively modulated by PKA phosphorylation. Am J Physiol Cell Physiol 290(6):C1477–C1486
Luo ZJ, Zang MW, Guo W (2010) AMPK as a metabolic tumor suppressor: control of metabolism and cell growth. Future Oncol 6(3):457–470
Manach C, Scalbert A, Morand C, Remesy C, Jimenez L (2004) Polyphenols: food sources and bioavailability. Am J Clin Nutr 79(5):727–747
Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, Schwab M, Pollak M, Zhang YQ, Yu YB, Becker KG, Bohr VA, Ingram DK, Sinclair DA, Wolf NS, Spindler SR, Bernier M, de Cabo R (2013) Metformin improves healthspan and lifespan in mice. Nat Commun 4:2192. doi:10.1038/ncomms3192
Matsuda M, Korn BS, Hammer RE, Moon YA, Komuro R, Horton JD, Goldstein JL, Brown MS, Shimomura I (2001) SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev 15(10):1206–1216
Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ (2013) Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494(7436):256–260. doi:10.1038/nature11808
Momcilovic M, Hong SP, Carlson M (2006) Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J Biol Chem 281(35):25336–25343
Moon YA, Liang GS, Xie XF, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Brown MS, Goldstein JL, Horton JD (2012) The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab 15(2):240–246
Muoio DM, Seefeld K, Witters LA, Coleman RA (1999) AMP-activated kinase reciprocally regulates triacylglycerol synthesis and fatty acid oxidation in liver and muscle: evidence that sn-glycerol-3-phosphate acyltransferase is a novel target. Biochem J 338(Pt 3):783–791
Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ (2000) Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102(3):315–323
Okazaki H, Goldstein JL, Brown MS, Liang G (2010) LXR-SREBP-1c-phospholipid transfer protein axis controls very low density lipoprotein (VLDL) particle size. J Biol Chem 285(9):6801–6810. doi:10.1074/jbc.M109.079459
Osborne TF, Espenshade PJ (2009) Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev 23(22):2578–2591. doi:10.1101/gad.1854309
Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM (2011) mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146(3):408–420
Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang MW, Wu SY, Chiang CM, Veenstra TD, Kemper JK (2010) SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem 285(44):33959–33970
Raghow R, Yellaturu C, Deng X, Park EA, Elam MB (2008) SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol Metab 19(2):65–73
Rawson RB (2003a) Control of lipid metabolism by regulated intramembrane proteolysis of sterol regulatory element binding proteins (SREBPs). Biochem Soc Symp 70:221–231
Rawson RB (2003b) The SREBP pathway—insights from Insigs and insects. Nat Rev Mol Cell Biol 4(8):631–640
Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ (2000) Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev 14(22):2819–2830
Sakamoto K, Goransson O, Hardie DG, Alessi DR (2004) Activity of LKB1 and AMPK-related kinases in skeletal muscle: effects of contraction, phenformin, and AICAR. Am J Physiol Endocrinol Metab 287(2):E310–E317
Shackelford DB, Shaw RJ (2009) The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 9(8):563–575. doi:10.1038/nrc2676
Shaw RJ (2013) Metformin trims fats to restore insulin sensitivity. Nat Med 19(12):1570–1572. doi:10.1038/nm.3414
Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC (2004) The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A 101(10):3329–3335
Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, DePinho RA, Montminy M, Cantley LC (2005) The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science 310(5754):1642–6
Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD (1997) Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest 100(8):2115–2124
Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS (1997) Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest 99(5):838–845
Steinberg GR, Kemp BE (2009) AMPK in health and disease. Physiol Rev 89(3):1025–1078
Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin JP, Harper JW, Ericsson J (2005) Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCFFbw7. Cell Metab 1(6):379–391
Tang JJ, Li JG, Qi W, Qiu WW, Li PS, Li BL, Song BL (2011) Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab 13(1):44–56
Timmers S, Konings E, Bilet L, Houtkooper RH, van de Weijer T, Goossens GH, Hoeks J, van der Krieken S, Ryu D, Kersten S, Moonen-Kornips E, Hesselink MK, Kunz I, Schrauwen-Hinderling VB, Blaak EE, Auwerx J, Schrauwen P (2011) Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab 14(5):612–622. doi:10.1016/j.cmet.2011.10.002
Turner N, Li JY, Gosby A, To SWC, Cheng Z, Miyoshi H, Taketo MM, Cooney GJ, Kraegen EW, James DE, Hu LH, Li J, Ye JM (2008) Berberine and its more biologically available derivative, dihydroberberine, inhibit mitochondrial respiratory complex I: a mechanism for the action of berberine to activate AMP-activated protein kinase and improve insulin action. Diabetes 57(5):1414–1418. doi:10.2337/db07-1552
Um JH, Park SJ, Kang H, Yang ST, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH (2010) AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes 59(3):554–563
Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F (2012) Cellular and molecular mechanisms of metformin: an overview. Clin Sci (Lond) 122(6):253–270. doi:10.1042/cs20110386
Vita JA (2005) Polyphenols and cardiovascular disease: effects on endothelial and platelet function. Am J Clin Nutr 81(1):292S–297S
Walker AK, Yang FJ, Jiang KR, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, Whetstine JR, Mostoslavsky R, Puigserver P, Li XL, Dyson NJ, Hart AC, Naar AM (2010) Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev 24(13):1403–1417
Yang J, Goldstein JL, Hammer RE, Moon YA, Brown MS, Horton JD (2001) Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc Natl Acad Sci U S A 98(24):13607–13612
Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS (2002) Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110(4):489–500
Yap F, Craddock L, Yang J (2011) Mechanism of AMPK suppression of LXR-dependent Srebp-1c transcription. Int J Biol Sci 7(5):645–650
Ye J, DeBose-Boyd RA (2011) Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb Perspect Biol 3(7). doi:10.1101/cshperspect.a004754
Yin J, Xing H, Ye J (2008) Efficacy of berberine in patients with type 2 diabetes mellitus. Metabolism 57(5):712–717. doi:10.1016/j.metabol.2008.01.013
Yoshikawa T, Shimano H, Yahagi N, Ide T, Amemiya-Kudo M, Matsuzaka T, Nakakuki M, Tomita S, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Takahashi A, Sone H, Osuga Ji J, Gotoda T, Ishibashi S, Yamada N (2002) Polyunsaturated fatty acids suppress sterol regulatory element-binding protein 1c promoter activity by inhibition of liver X receptor (LXR) binding to LXR response elements. J Biol Chem 277(3):1705–1711. doi:10.1074/jbc.M105711200
Young LH (2009) A crystallized view of AMPK activation. Cell Metab 10(1):5–6
Zang MW, Zuccollo A, Hou XY, Nagata D, Walsh K, Herscovitz H, Brecher P, Ruderman NB, Cohen RA (2004) AMP-activated protein kinase is required for the lipid-lowering effect of metformin in insulin-resistant human HepG2 cells. J Biol Chem 279(46):47898–47905
Zang MW, Xu SQ, Maitland-Toolan KA, Zuccollo A, Hou XY, Jiang BB, Wierzbicki M, Verbeuren TJ, Cohen RA (2006) Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes 55(8):2180–2191
Zhang Y, Li X, Zou D, Liu W, Yang J, Zhu N, Huo L, Wang M, Hong J, Wu P, Ren G, Ning G (2008) Treatment of type 2 diabetes and dyslipidemia with the natural plant alkaloid berberine. J Clin Endocrinol Metab 93(7):2559–2565. doi:10.1210/jc.2007-2404
Zhang BB, Zhou GC, Li C (2009) AMPK: an emerging drug target for diabetes and the metabolic syndrome. Cell Metab 9(5):407–416
Zhou GC, Myers R, Li Y, Chen YL, Shen XL, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, Musi N, Hirshman MF, Goodyear LJ, Moller DE (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 108(8):1167–1174
Acknowledgements
Studies described that were carried out in the authors’ laboratory were supported by the National Institutes of Health Grants (DK076942, R01DK100603, and R21 AA021181), American Diabetes Association Basic Science Award (1-15-BS-216), and Wing Tat Lee Award (1UL1TR001430).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Zang, M. (2016). The Molecular Basis of Hepatic De Novo Lipogenesis in Insulin Resistance. In: Ntambi, J. (eds) Hepatic De Novo Lipogenesis and Regulation of Metabolism. Springer, Cham. https://doi.org/10.1007/978-3-319-25065-6_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-25065-6_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-25063-2
Online ISBN: 978-3-319-25065-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)