Abstract
DDK (Dbf4-dependent kinase) is a serine/threonine protein kinase conserved from yeast to humans. DDK is composed of two subunits, Cdc7 protein kinase and Dbf4 regulatory subunits in 1:1 stoichiometry. Both the CDC7 and DBF4 genes were discovered in budding yeast from the analysis of conditional mutants that were defective in the initiation of DNA replication. Cdc7 and Dbf4 homologues have been identified in many eukaryotes and are important in DNA replication, indicating the role of DDK was also conserved. This knowledge has been translated medically as oncogenic DDK overexpression is currently a target of therapeutic inhibitors. DDK activity is cell cycle regulated because it is inactive in G1 phase cells due to the absence of the essential Dbf4 protein as a result of APC-dependent proteolysis. It is clear from both genetic and biochemical studies that several subunits of the hexameric MCM2–7 DNA helicase/ATPase are substrates of DDK. In an allosteric model of DDK function, DDK phosphorylates the Mcm4 protein in the misaligned MCM2–7 double hexamer bound to origins of replication to align the important catalytic residues of the enzyme and to load other proteins to form a CMG (Cdc45-MCM-GINS) holoenzyme. To complete the initiation reaction, the loading of several other replication proteins is also needed, which requires ensuing CDK (cyclin-dependent kinase) phosphorylation. DDK has other substrates important for mutagenesis by TLS (translesion synthesis), meiotic recombination, and chromosome cohesion. Because all these processes are chromatin-based, DDK may have evolved to regulate chromatin-bound proteins in DNA metabolism.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
In this review, the role of Dbf4-dependent kinase (DDK) in the initiation of DNA replication will be described. DDK, composed of a Cdc7 (cell division cycle 7) subunit and a Dbf4 (dumbbell former 4) subunit, is a critical regulator of the initiation step of DNA replication. CDC7 and DBF4 genes were initially discovered using yeast mutants that arrest the cell cycle before the onset of S phase. The two proteins are conserved from yeast through humans and combine to form an active protein kinase with Cdc7 being the catalytic subunit and Dbf4 the regulatory subunit. Protein expression of the Dbf4 subunit regulates kinase activity during the cell cycle. From both genetic and biochemical evidence, DDK was found to regulate the initiation of DNA replication by phosphorylating the Mcm protein complex during S phase. DDK also has functions in many other cellular processes including meiotic recombination and mutagenesis, which are not discussed herein. Due to its role in a wide array of processes, it has become a target of cancer research and kinase inhibitor development.
Identification of CDC7 and DBF4
DNA replication is a highly regulated process that relies on a complex array of proteins to ensure complete duplication of the genome occurs once and only once during the cell cycle. One protein complex required for proper initiation of DNA replication is DDK. DDK is a heterodimer composed of the catalytic subunit, Cdc7, and the regulatory subunit Dbf4. CDC7 was originally discovered in Saccharomyces cerevisiae (budding yeast) as part of Hartwell’s collection of temperature sensitive mutants that arrest at unique points of the cell cycle [1]. cdc7 mutants have a dumbbell phenotype, large budded cells with the nucleus stuck at the neck, and cause a cell cycle arrest immediately before the onset of S-phase. Neither Cdc7 protein nor total protein synthesis is required for completion of S phase after Cdc7’s function is executed at the G1–S boundary [2]. These two attributes provide evidence that Cdc7 controls a late step immediately at the start of DNA replication initiation. Therefore, it was hypothesized that all proteins necessary for DNA replication are present at the G1–S boundary and Cdc7’s role is to activate them [3], which is consistent with Cdc7 being a protein kinase (see below).
DBF4 was isolated in a screen using the collection of cell cycle mutants to only analyze mutants that produced the dumbbell forming phenotype. This specific subset of cdc mutants, all having a similar phenotype, were thought to be indicative of a defect in DNA synthesis and thus, closely tied together [4]. DBF4, like CDC7, was further shown to be an essential gene; and dbf4 mutants have a defect in DNA replication initiation [5]. A direct interaction between the two genes was first suggested by suppression of cdc7 by DBF4 overexpression. Likewise, overexpressed CDC7 could suppress dbf4 mutations and double mutants of the two genes are synthetically lethal and not viable [6]. Dbf4 is essential and is likely an activation subunit for Cdc7 kinase activity.
CDC7 Encodes a Protein Kinase
CDC7 was cloned through yeast genomic DNA plasmid complementation of temperature sensitive mutant alleles of cdc7 revealing a genomic fragment of 1.5 kb located on Chromosome IV. This genomic fragment encodes the approximately 58 kDa Cdc7 protein and based on the predicted amino acid sequence compared to known protein sequences, suggests the protein is a serine/threonine protein kinase [7]. The homology between Cdc7 and other protein kinases was not randomly distributed across the protein, but confined to functionally important kinase domains. Cdc7 contains the conserved ATP binding site as well as phosphorylation acceptor site domains of kinases, but unlike other protein kinases, Cdc7 has longer regions of non-conserved sequences between the conserved kinase domains. DBF4 was cloned using similar methods to identify a 2.4 kb genomic fragment on Chromosome IV of the yeast genome that encodes a 81 kDa protein [8].
Cdc7 was later confirmed to be a protein kinase using mammalian histone H1 as an exogenous substrate and immunoprecipitates of Cdc7, which also contain Df4 protein [9, 10]. Serine and threonine residues within the histone H1 protein were phosphorylated by Cdc7. An in vivo substrate of Cdc7 has yet to be discovered, but because Cdc7 is found in high concentration within the nucleus, the in vivo substrates are likely nuclear proteins [9, 10]. Mutations made in key residues within the kinase domains abolished all in vivo functions of the Cdc7 protein.
Identification of CDC7-DBF4 in Other Eukaryotes
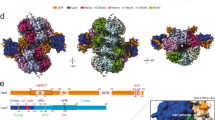
Discovery of DDK’s role in budding yeast DNA replication initiation allowed for identification of similar proteins and functions in the evolutionary distant yeast, Schizosaccharomyces pombe. Degenerate oligonucleotide directed PCR (polymerase chain reaction was used to identify a homolog of Cdc7 in S. pombe, Hsk1 (a putative homolog of Cdc7 (seven) kinase 1). Hsk1 was found to share amino acid sequence homology with Cdc7 in the critical conserved kinase domains as well as other regions [11] (Fig. 14.1). Hsk1 shares approximately 60 % identity with Cdc7 from budding yeast, but does not rescue a cdc7ts mutant of budding yeast [12]. Furthermore, Hsk1 protein phosphorylated several substrates as well as autophosphorylated; and hsk1ts mutants exhibited inhibition of DNA replication. Null mutants of hsk1 completed mitosis but failed to replicate DNA. Like Cdc7, disruption of the hsk1 gene is lethal to cell growth and therefore, likely carries out a very similar function. Surprisingly, an hsk1 deletion is conditional and is viable at 37 °C, indicating DDK is not required at this temperature [13]. Discovery of hsk1 as a yeast Cdc7 homolog within an evolutionary distant organism paved the way to identify Cdc7 homologs in other eukaryotes, especially vertebrates (Table 14.1). Shortly after Hsk1 was identified in S. pombe, Cdc7 homologs were discovered in humans, Xenopus and mouse. These proteins do not have exact protein sequence homology, but they all maintain the critical catalytic kinase residues that are required for Cdc7 function [14–16].
Comparison of DDK subunits from human, S. cerevisiae, and S. pombe. (a) Comparison of Cdc7 homologs. Red segments represent conserved kinase domains. Blue segments represent less conserved kinase insert sequences. (b) Comparison of Dbf4 homologs. Green, Blue, and Orange segments represent conserved Dbf4 motifs (Adapted from Masai et al. 2002)
Dbf4 homologs have been isolated from other eukaryotes as well providing evidence for the strong evolutionary link between the Cdc7 catalytic subunit and the regulatory subunit (Table 14.1). In S. pombe, Hsk1 copurified with a protein, Dfp1. Independent experiments identified the same molecule as Him1, and these were later identified as a homolog of budding yeast Dbf4. Cdc7 association with Dfp1/Him1 was shown to have a similar effect as the budding yeast proteins in stimulating phosphorylation of an exogenous substrate, but does not enhance autophosphorylation of Hsk1 [17]. Dbf4 homologs discovered in human, Xenopus, and mouse have all been shown to physically interact with Cdc7 to regulate initiation of DNA replication [18, 19]. The majority of Dbf4 conservation between species is restricted to three different motif regions: M, N, and C. Using the yeast two-hybrid system for protein–protein interactions, it has been shown that Cdc7 and Dbf4 interact directly with each other both in vitro and in vivo to create a functional complex [20–22]. Importantly, both subunits of DDK are conserved and thus preserve the function of the complex from yeast through humans [23]. The essential function of DDK in DNA replication is conserved in evolution as human DDK can complement deletions in yeast CDC7 and DBF4 if both human Cdc7 and Dbf4 cDNAs are co-expressed [24].
Regulation of CDC7-DBF4 Protein Kinase during the Cell Cycle
Cdc7 and Dbf4 proteins are regulated differentially throughout the cell cycle. In budding yeast, the Cdc7 catalytic subunit is stably expressed and subsequently bound to chromatin throughout the majority of the cell cycle [25]. However, Dbf4 protein expression oscillates throughout the cell cycle as a result of protein stability. Dbf4 stability is regulated by the APC (anaphase promoting complex ). The protein is rapidly degraded in G1 phase of the cell cycle and mutations in APC subunits or the Dbf4 N-terminus region resembling a destruction box eliminates the APC-dependent degradation [26]. Once Dbf4 is expressed in the cell, it is likely that it immediately associates with chromatin. Dbf4 protein expression oscillation within the cell cycle is equivalent to the role of cyclins in regulating the activity of cyclin-dependent kinases (CDK) [27, 28].
Although the Cdc7 subunit is constitutively expressed during the cell cycle, the kinase activity changes depending on expression of Dbf4 in the cell. Cdc7 kinase activity is low during G1 phase of the cell cycle, increases at the G1–S transition, maintained at high activity through S phase, and then decreases again as S phase in completed [29]. This pattern of kinase activity closely mirrors the expression of the Db4 protein [30]. Similar studies performed using human and Xenopus homologs of Cdc7 and Dbf4 also show DDK activity relies on the expression of the Dbf4 subunit within the cell at the G1–S transition [31, 32].
While Dbf4 has long been known to be the main regulatory subunit of the DDK complex, human and Xenopus Cdc7 are also regulated by Drf1 (Dbf4-related factor 1) [33]. In Xenopus egg extracts, Drf1 is in far more abundance than the Dbf4 protein and removal of the protein by immunodepletion causes the characteristic inhibition of replication. However, as gastrulation completes, Drf1 levels decline and Dbf4 becomes more abundant [34]. Human Drf1 displays similar temporally regulated kinetics as Dbf4, but the function has yet to be established. Drf1 and Dbf4 may bind to different pools of Cdc7 in order to initiate replication from different origin subsets [33]. Another proposed role of human Drf1 is to restart replication forks that have arrested for various reasons throughout S phase [35]. This second regulator of Cdc7 has not been found in the genome of yeast and Drosophila indicating another layer of complexity in vertebrate replication initiation that may have developmental or timing implications.
Dbf4 is a limiting factor in DNA replication, along with Dpb11 (DNA polymerase B subunit 11), Sld2 (synthetic lethal with Dpb11), and Sld3. These proteins are found in low abundance in the cell, but overexpression of all four proteins is sufficient to advance the replication timing of late origins and heterochromatic regions [36].
DDK Structure and Function
Crystal structure of human DDK has provided insight into how DDK subunits are held together and lead to activation of the kinase (Fig. 14.2) [37]. Cdc7 has a bilobal structure with a deep cleft between the two domains that houses the active site of the kinase. The N lobe of Cdc7 is comprised of an antiparallel β-sheet as well as α-helices, while the C lobe of Cdc7 is comprised almost entirely of α-helices that encase the catalytic loop of the kinase as well as a Mg2+ ion. Dbf4 motif regions M and C are conserved between species and are necessary for binding to Cdc7 in a bipartite manner. Motif C of Dbf4 contains α-helices and a β-strand that forms an essential zinc finger used for binding the N-terminal lobe of Cdc7. Meanwhile, motif M of Dbf4 uses β-strands to pair with β-strands within the kinase insert three domain to create an antiparallel sheet necessary for packing and binding against the C-terminal lobe of Cdc7. Although it is conserved across species, motif N of Dbf4 is not used for binding nor is it required for kinase activation. Motif M and Motif C are necessary to pack Dbf4 against the lobes of Cdc7 and are needed for activation of the kinase. Upon Cdc7 and Dbf4 binding, the kinase subunit maintains a closed nucleotide (Mg-ATP) bound conformation similar to other protein kinases. Mutations in regions of Dbf4 or Cdc7 that disrupt the binding of the two subunits result in extensively reduced activity of the kinase [37].
Human DDK atomic crystal structure. Atomic crystal structure of Cdc7-Dbf4 dimer from RCSB PDB (Protein Data Bank): http://www.rcsb.org/pdb/explore/images.do?structureId=4 F99 using coordinates from reference [37]. Note that protein fragments of Cdc7 and Dbf4 were used and not the full-length molecules and the resulting smaller DDK enzyme had about 40–50 % activity. Cdc7 kinase N-terminal Beta-Sheet rich ATP binding domain at top and C-terminal alpha-helical phospho-acceptor domain at bottom. Dbf4-M and Dbf4-C domains are as shown and also depicted in Fig. 14.1. Zn2+ and Mg2+ ions are shown as gray spheres
Cdc7 lacks a threonine residue in the active site present in most other protein kinases. This residue is generally phosphorylated and leads to activation of the serine/threonine kinase. In order to obtain a crystal structure of Cdc7 and the binding interface, the expanded kinase insert region 2 was deleted. Therefore, it cannot be ruled out that this region contains other regulatory elements that make up for the loss of the critical threonine residue found in other kinases [37].
Budding yeast Cdc7 has a unique C-terminal domain consisting of 55 amino acids that has been shown to be required for binding to Dbf4 [20]. This region is not found in homologous Cdc7 proteins indicating there is redundancy that allows for Cdc7 to bind Dbf4 without this specialized region in other eukaryotes.
The Cdc7 subunit forms dimers as shown both in vivo and with recombinant proteins in vitro [38]. These Cdc7-Cdc7 dimers have low kinase activity which increases substantially when one Dbf4 molecule associates with one Cdc7 molecule to disrupt the Cdc7-Cdc7 dimer and form a Cdc7-Dbf4 heterodimer [39].
Cell Cycle Activity of DDK
All budding yeast replication origins that fire throughout S phase require DDK function [40, 41]. Cdc7 may control the transition from G1 to S as well as activation and initiation from individual origins. Dbf4 has been found to associate directly with origins of replication and thus provides a foundation that Cdc7 functions directly at replication initiation complexes at the origins [21, 42]. However, there is no direct in vivo evidence that Cdc7 also binds to origins. A first indication of the identity of the endogenous substrates of DDK came from studies in which the mcm5-bob1 (Mcm5 P83L) mutation in the Mcm5 (mini chromosome maintenance) protein was found to be a bypass suppressor of deletions in both CDC7 and DBF4, but not other cell cycle division genes [43]. The mcm5-bob1 mutation is able to bypass the need for DDK within the cell and allows for the cell to enter S phase earlier. In these studies, a model was proposed in which Mcm5 structure is modified by DDK. An important function of Mcm5 is to therefore inhibit DNA replication and a modification of the protein allows the cell to initiate replication. There is precedent in phage λ for a model where inhibition is removed to allow assembly of DNA replication proteins [44]. The mcm5-bob1 bypass of DDK indicates that essential functions of the kinase are inherently linked to the Mcm complex.
The Mcm complex was initially identified as being required for stable maintenance of ARS (autonomous replicating sequence ) plasmids in budding yeast [45]. It is made up of six homologous proteins and loaded onto origins as a double hexamer in late M phase to early G1 phase. Functions of Mcm2–7 within the cell cycle are likely regulated by the phosphorylation state of the proteins [46, 47]. Comparison of Mcm protein sequences show that Mcm2, 4, and 6 contain serine and threonine residues within long N-terminal domains. Mcm3, 5 and archaeal Mcm proteins do not have this extra region. A mutation within the MCM2 gene reduces efficiency of DNA replication at origins of DNA replication. To understand how Mcm2 is regulated it is necessary to understand what other genes may interact with the MCM2 gene. It was determined that a dbf4 mutation was able to suppress the defects seen in mcm2-1 mutant. Furthermore, Mcm2 and Dbf4 show a strong physical interaction with each other that is compromised by the mcm2-1 mutation. From these genetic data, a number of investigations focused on the Mcm complex as a substrate of DDK. Recombinant GST-Mcm fusion proteins were made for use in DDK protein kinase assay in vitro [48]. GST-Mcm2, GST-Mcm3, GST-Mcm4, and GST-Mcm6 were all shown to be phosphorylated by DDK in vitro. Surprisingly, GST-Mcm5 was not phosphorylated as was thought from the mcm5-bob1 mutation. Additionally, a GST-mcm2-1 fusion was shown to reduce phosphorylation of mcm2-1 by DDK compared to wild type indicating that the kinase is directly tied to the function of the Mcm complex. Dbf4 is the main subunit required for binding Mcm2, which recruits Cdc7 to phosphorylate amino acids S164 and S170 and leads to DNA replication and cell growth [49].
Mcm2 is phosphorylated during S phase by DDK at two N-terminal serines (Fig. 14.3). Conversion of these two residues to non-phosphorylatable alanines (S164A, S170A) results in a severe growth defect that can be partially rescued by mcm5-bob1. Upon mutating these residues, the amount of origin ssDNA is significantly reduced compared to wild type based on the amount of RPA (replication protein A) bound to the origins. These mutations reduced binding of Sld3 and GINS to the Mcm complex. It was also determined that Mcm2 phosphorylation destabilized the interaction with Mcm5 promoting opening of the Mcm2–7 hexameric ring to allow extrusion of ssDNA [50].
Identification of DDK Target sites on Mcm2–7. Schematic of DDK phosphorylation sites on each Mcm subunit. Blue triangles represent DDK phosphorylation sites as determined by mass spectrometry. Extended N-terminal domains of Mcm2, Mcm4, and Mcm6 contain phosphorylated serine and threonine residues (Adapted from Randall et al. 2010)
Consensus DDK phosphorylation sites are usually S or T followed by an acidic residue (S/T D/E) [51]. These sites are similar to sites preferred by CK2 (casein kinase 2) [52], the most similar kinase to Cdc7 in the human kinome [53]. There are also hierarchal DDK sites (S/T-S/T-P) where the second S/T residue is first phosphorylated by CDK creating a priming phosphor-acidic site (S/T-S/T(Phos)-P) and hence DDK phosphorylation will occur at the first S/T residue as seen in human Mcm2 [51, 54] and in yeast Mer2 [55]. Hierarchal sites primed by CDK or by Mec1 (S/T-S/T-Q) are also seen in the N-terminal region of yeast Mcm4 and Mcm6 (Fig. 14.3).
Mcm4 has also been analyzed as a DDK substrate due to the discovery of hierarchal phosphorylation sites. The serine/threonine rich N-terminal domain inhibits DNA replication. Phosphorylation of this region removes the inhibitory effect and cells can transition into S phase. In vitro experiments confirm DDK is the kinase responsible for phosphorylating the N-terminal region of Mcm4 and relieving the inhibitory effect [56, 57]. When the inhibitory region of Mcm4 is deleted, replication can proceed in the absence of DDK similar to the mcm5-bob1 bypass mutation. Mutating DDK specific serine/threonine residues to alanine within Mcm4 confers lethality since Mcm4 inhibition cannot be removed by DDK. However, if phosphomimetic mutations (S/T to D/E) are made in the same residues, the cell is not only viable but can also bypass the need for DDK. Cell cycle regulation of DNA replication can still occur in this case as CDK (Cdk1-Clb5) function is still required.
If both DDK and CDK steps are made constitutive, lethality occurs [58, 59]. Mcm4 phosphorylation also stimulates association of other replication proteins, such as Cdc45 (cell-division cycle 45), with the chromatin [60]. The lack of Mcm5 phosphorylation may be explained by allosteric effects by Mcm4 phosphorylation such that relief of N-terminal inhibition may result in a conformation change of Mcm5. Mcm5, as evidence from the mcm5-bob1 mutation, may be an important inhibitor of DNA replication that is removed by phosphorylation of other Mcm subunits such as Mcm2, Mcm4, or Mcm6. While Mcm2, Mcm4, and Mcm6 appear to be the direct targets of DDK, there is evidence to suggest that other residues within the Mcm proteins need to be phosphorylated first in order to prime the protein for DDK phosphorylation [61]. Once substrates were identified in S. cerevisiae, efforts were made to confirm them in other organisms, specifically, Mcm2 was shown to be a direct target of DDK in human cells [62].
Sequential Order of DDK and CDK Function in the Cell Cycle
There is controversy about the sequence of events that lead to phosphorylation of the Mcm proteins by DDK. As described above, characterization of cdc7 mutants in yeast demonstrate that protein synthesis is no longer required to complete S phase after Cdc7 executes its function indicating that it may be the final factor required for replication initiation. St udies in S. cerevisiae also suggest that CDK carries out its phosphorylation function first while DDK phosphorylates the Mcm complex second [63]. However, in vivo studies in S. pombe, and in vitro studies in Xenopus extracts suggest that DDK acts first to phosphorylate the Mcm complex and then CDK acts second to initiate DNA replication [64, 65]. Both models agree that phosphorylation of the Mcm proteins by DDK is necessary to load Cdc45 onto chromatin at replication origins.
In Xenopus and S. pombe, DDK loads Cdc45 onto chromatin before CDK carries out its catalytic function and brings in the remaining proteins necessary for DNA replication including the GINS complex. In G1 phase of the cell cycle, CDK activity is low due to the absence of S phase cyclins necessary to transition the cell into S phase [28]. Dbf4 is also low in G1, thus Cdc45 is not loaded onto chromatin [26, 27, 29]. However, in the presence of the mcm5-bob1 mutation, Cdc45 is loaded onto chromatin in G1 phase of the cell cycle when S phase CDK is inactive. The loading of Cdc45 in the absence of CDK correlates with premature ARS1 origin unwinding as seen with in vivo genomic footprinting in the mcm5-bob1 mutant arrested in G1 phase [66]. Thus, origin structural changes are directly linked to Cdc45 loading. Furthermore, in an in vitro replication system from S. cerevisiae extracts, DDK alone produced Cdc45 loading onto origins in the absence of CDK. In the latter study, there was a clear order of events in that DDK must act before CDK in order to initiate DNA replication as seen in the Xenopus in vitro studies.
DDK as a Target of Cancer Therapy
Deregulation of normal cell cycle progression has been a target of previous therapies for cancer treatment. Given that DDK plays such an important role in DNA replication, DNA damage repair and cell cycle progression, it has become a focus of new cancer therapeutics [67–69]. Increased expression of Cdc7 has become a marker for cancer and overexpression of Cdc7 has been shown to inhibit apoptosis and lead to survival of oral squamous cell cancer cells [70]. Cancer cases that show increased Cdc7 expression tend to have poor clinical outcomes [71]. In breast cancer, increased Cdc7 is often correlated with increased Dbf4 that may be linked to the proliferative activity of cancer cells. While increased Cdc7 is not always correlated with proliferative status, it is always correlated with the number of cells that enter S phase. An explanation of this may be that since Cdc7 is involved in the DNA damage repair pathway, it aids in recovery of stalled replication forks to enhance survival of tumor cells. Additionally, p53 mutations or even protein loss are a result of increased Cdc7 and Dbf4 expression [72]. Conversely, loss of Cdc7 in cancer cells induces a p53-independent apoptotic response that leads to cell death without activating the standard checkpoint pathway [73].
The first inhibitors of Cdc7 were developed at Nerviano Medical Sciences as small molecule ATP-competitors in the pyrrolopyridones class [74]. One compound from this screen, PHA-767491, emerged as the leading candidate for Cdc7 inhibition. This compound impairs Mcm2 phosphorylation at DDK-dependent sites and blocks origin firing, but does not prevent progression of replication forks or activate a DNA damage cascade [75]. Antitumor activity of this compound has been seen in AML(acute myeloid leukemia), breast and colon cancer models. Another inhibitor that has been used in cancer models is XL413. However, unlike the robust anti-proliferative and apoptotic effects seen in PHA-767491, XL413 only has limited activity in a smaller percentage of tester cancer cell types [76].
Model of DDK Action
A simplified model of DDK action is DDK phosphorylates the Mcm complex, which produces a structural change that activates the helicase and allows for the binding of important replication proteins (Fig. 14.4). The model is bolstered by atomic crystal structures and functional assays of the Mcm complex as deduced from the studies of Archaeal Mcm homomultimeric proteins together with yeast genetic studies [77, 78]. In the Archaeal Mcm protein, the alpha helical bundle (A domain) located in the N-terminus, anchors the proline that is converted to the leucine in the context of the mcm5-bob1 mutation. Leucine has a large side chain that pushes this A domain out and creates a structural change in the protein. It is possible that this structural change removes an inhibition and allows for other loading proteins that recognize this change. Phosphorylation of the wild type Mcm2 or Mcm4 may cause a structural change in the Mcm complex as a whole and simulate this expansion of the A domain in Mcm5. Thus, the mcm5-bob1 mutation mimics a conformational change brought about by phosphorylation within the complex. In S. pombe, this structural change may occur spontaneously at higher temperatures allowing for DDK bypass [13].
Model of DDK action in replication. (1) In late M/early G1 phase of the cell cycle, Mcm2–7 complex is loaded onto origins as a twisted double hexamer. (2) At the G1–S transition, DDK phosphorylates the N-terminal serine/threonine residues of Mcm2, Mcm4, and Mcm6. (3) The double hexamer untwists to allow additional protein loading. (4) Cdc45, GINS and other required proteins bind to activate the helicase. (5) A gate between Mcm2 and Mcm5 opens to allow extrusion of ssDNA and form the actively replicating CMG complex
As further evidence of the model, other amino acids were substituted in place of the proline residue within the yeast Mcm5 protein [78, 79]. Only residues with large side chains (K, W) that would give similar expansion of the A domain and not smaller substitutions (A, G) produced a mcm5-bob1 phenotype and bypassed DDK. Surprisingly, the P to L change in the Archaeal protein only resulted in a 1 Å structural change but the corresponding yeast genetics confirmed that amino acids with large side chains caused a significant effect in the cell. The small structural difference can be explained by the fact the crystallization pushes the domains inward to pack it together tightly, whereas a protein in solution will have more room to expand. Ultracentrifugation of the Archaeal protein with the mcm5-bob1 mutation showed increased heterogeneity in the sedimentation of the protein [79]. An explanation of this anomaly is that in vivo pushes out the A domain to produce a unique conformation, while the mcm5-bob1 mutation allows for a greater range in conformations, only a few of which are active. It is possible that the homogeneity due to phosphorylation is preventing the structural studies from seeing the domain push out. Using a construct that contains the N-terminal domain of Mcm from one archaeon fused to the C-terminal of Mcm of another archaeon, the A domain does in fact push out much further than the 1 Å seen previously [80]. Once this A domain of the Mcm is pushed out, the complex can now act as a landing pad for other DNA replication proteins to create the helicase holoenzyme Cdc45-Mcm-GINS complex (CMG). Mcm2 phosphorylation also destabilizes the interaction with Mcm5 promoting opening of the Mcm2–7 hexameric ring to allow extrusion of ssDNA and to allow elongation of DNA replication [50].
Concluding Remarks
Since over 50 years after DDK was initially discovered in yeast, a great deal has been learned about the function and roles DDK plays throughout the cell cycle. Its scope goes well beyond its role in replication initiation to include DNA translesion synthesis and mutagenesis [81, 82], kinetochore function [42], and meiotic recombination [55, 83] (Fig. 14.5). We now know DDK is a serine/threonine protein kinase with the most widely known substrates being the Mcm complex but more substrates will probably be identified in the future. During evolution, DDK may have been recruited for a number of cellular roles that all involve regulating the binding of proteins to chromatin [81]. Clearly, DDK’s most well studied and understood role is to initiate DNA replication from individual origins of replication. At the atomic level, crystallography has been used to capture both the binding of the catalytic subunit Cdc7 to its regulatory subunit Dbf4 as well as possible binding to substrates. Furthermore, the substrate specificity of DDK for the Mcm complex has been shown using cryo-EM as the two hexamers in the inactive Mcm complex in the pre-RC are misaligned, which acts as a unique binding site for DDK [84]. Phosphorylation by DDK then aligns the double hexamer correctly to produce the active DNA helicase. The knowledge about DDK has already yielded important translational information for the clinic in that a number of recent studies have identified potential inhibitors of DDK function during cancer development and progression.
DDK (Cdc7-Dbf4) regulates four different chromatin-bound substrates. DDK (Cdc7-Dbf4) phosphorylates Rev7 to regulate TLS (translesion synthesis) in mutagenesis, MCM complex in the initiation of DNA replication, Mer2 in meiotic recombination, and possibly Scc2-Scc4 in chromosome cohesion (see text for references)
References
Hartwell LH. Genetic control of the cell division cycle in yeast II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59:183–94.
Hartwell LH. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J Bacteriol. 1973;115:966–74.
Hollingsworth RE Jr. Molecular Biology of the CDC7 gene and protein of Saccharomyces cerevisiae [PhD]. University of Colorado; 1990.
Johnston LH, Thomas AP. The isolation of new DNA synthesis mutants in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186(3):439–44.
Johnston LH, Thomas AP. A further two mutants defective in initiation of the S phase in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186:445–8.
Kitada K, Johnston LH, editors. The Saccharomyces cerevisiae DBF4 gene, required for the G1Æ S phase transition, suppresses temperature-sensitive cdc7 mutations. 1991 Yeast Genetics and Molecular Biology Meeting; 1991; San Francisco, CA: Genetics Society of America.
Patterson M, Sclafani RA, Fangman WL, Rosamond J. Molecular characterization of cell cycle gene CDC7 from Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:1590–8.
Chapman JW, Johnston LH. The yeast gene, DBF4, essential for entry into the S phase is cell cycle regulated. Exp Cell Res. 1989;180:419–28.
Hollingsworth Jr RE, Sclafani RA. DNA metabolism gene CDC7 from yeast encodes a serine (threonine) protein kinase. Proc Natl Acad Sci U S A. 1990;87:6272–6.
Yoon H-J, Campbell JL. The CDC7 protein of Saccharomyces cerevisiae is a phosphoprotein that contains protein kinase activity. Proc Natl Acad Sci U S A. 1991;88:3574–8.
Masai H, Miyake T, Arai K-I. hsk1 +, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14(13):3094–104.
Masai H, Arai K. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J Cell Physiol. 2002;190(3):287–96.
Matsumoto S, Hayano M, Kanoh Y, Masai H. Multiple pathways can bypass the essential role of fission yeast Hsk1 kinase in DNA replication initiation. J Cell Biol. 2011;195(3):387–401.
Sato N, Arai K-I, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homolgue of Cdc7. EMBO J. 1997;16:4340–51.
Hess GF, Drong RF, Weiland KL, Sligthom JL, Sclafani RA, Hollingsworth Jr RE. A human homolog of the yeast CDC7 gene is overexpressed in some tumors and transformed cell lines. Gene. 1998;211:133–40.
Faul T, Staib C, Nanda I, Schmid M, Grummt F. Identification and characterization of mouse homologue to yeast Cdc7 protein and chromosomal localization of the cognate mouse gene Cdc7l. Chromosoma. 1999;108(1):26–31.
Brown GW, Kelly TJ. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem. 1998;273(34):22083–90.
Jares P, Luciani MG, Blow JJ. A Xenopus Dbf4 homolog is required for Cdc7 chromatin binding and DNA replication. BMC Mol Biol. 2004;5:5.
Lepke M, Putter V, Staib C, Kneissl M, Berger C, Hoehn K, et al. Identification, characterization and chromosomal localization of the cognate human and murine DBF4 genes. Mol Gen Genet. 1999;262(2):220–9.
Jackson AL, Pahl PMB, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13(5):2899–908.
Dowell SJ, Romanowski P, Diffley JFX. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast regulatory origins in vivo. Science. 1994;265:1243–6.
Harkins V, Gabrielse C, Haste L, Weinreich M. Budding yeast Dbf4 sequences required for Cdc7 kinase activation and identification of a functional relationship between the Dbf4 and Rev1 BRCT domains. Genetics. 2009;183(4):1269–82.
Roberts BT, Ying CY, Gautier J, Maller JL. DNA replication in vertebrates requires a homolog of the Cdc7 protein kinase. Proc Natl Acad Sci U S A. 1999;96(6):2800–4.
Davey MJ, Andrighetti HJ, Ma X, Brandl CJ. A synthetic human kinase can control cell cycle progression in budding yeast. G3 (Bethesda). 2011;1(4):317–25.
Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18(19):5334–46.
Cheng L, Collyer T, Hardy CF. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol Cell Biol. 1999;19(6):4270–8.
Ferreira MF, Santocanale C, Drury LS, Diffley JF. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol Cell Biol. 2000;20(1):242–8.
Morgan DO (2007) The Cell Cycle. Primers In Biology. New Science Press Ltd., London UK.
Oshiro G, Owens JC, Shellman Y, Sclafani RA, Li JJ. Cell cycle control of cdc7p kinase activity through regulation of dbf4p stability. Mol Cell Biol. 1999;19(7):4888–96.
Takeda T, Ogino K, Matsui E, Cho MK, Kumagai H, Miyake T, et al. A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol Cell Biol. 1999;19(8):5535–47.
Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci U S A. 1997;94:14320–5.
Furukohri A, Sato N, Masai H, Arai K, Sugino A, Waga S. Identification and characterization of a Xenopus homolog of Dbf4, a regulatory subunit of the Cdc7 protein kinase required for the initiation of DNA replication. J Biochem. 2003;134(3):447–57.
Montagnoli A, Bosotti R, Villa F, Rialland M, Brotherton D, Mercurio C, et al. Drf1, a novel regulatory subunit for human Cdc7 kinase. EMBO J. 2002;21(12):3171–81.
Takahashi TS, Walter JC. Cdc7-Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 2005;19(19):2295–300.
Yoshizawa-Sugata N, Ishii A, Taniyama C, Matsui E, Arai K, Masai H. A second human Dbf4/ASK-related protein, Drf1/ASKL1, is required for efficient progression of S and M phases. J Biol Chem. 2005;280(13):13062–70.
Mantiero D, Mackenzie A, Donaldson A, Zegerman P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011;30(23):4805–14.
Hughes S, Elustondo F, Di Fonzo A, Leroux FG, Wong AC, Snijders AP, et al. Crystal structure of human CDC7 kinase in complex with its activator DBF4. Nat Struct Mol Biol. 2012;19(11):1101–7.
Shellman YG, Schauer IE, Oshiro G, Dohrmann P, Sclafani RA. Oligomers of yeast Cdc7/Dbf4 protein kinase exist in the cell. Mol Gen Genet. 1998;259:429–36.
Kaplan DL, Bruck I. Methods to study kinase regulation of the replication fork helicase. Methods. 2010;51:358–62.
Bousset K, Diffley JFX. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–90.
Donaldson AD, Fangman WF, Brewer BJ. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;491:491–501.
Natsume T, Muller CA, Katou Y, Retkute R, Gierlinski M, Araki H, et al. Kinetochores coordinate pericentromeric cohesion and early DNA replication by cdc7-dbf4 kinase recruitment. Mol Cell. 2013;50(5):661–74.
Hardy CFJ, Dryga O, Seematter S, Pahl PMB, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci U S A. 1997;94:3151–5.
Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–80.
Tye B-K. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–86.
Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–45.
Young MR, Tye BK. Mcm2 and Mcm3 are constitutive nuclear proteins that exhibit distinct isoforms and bind chromatin during specific cell cycle stages of Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1587–601.
Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–74.
Bruck I, Kaplan D. Dbf4-Cdc7 phosphorylation of Mcm2 is required for cell growth. J Biol Chem. 2009;284(42):28823–31.
Bruck I, Kaplan DL. The Dbf4-Cdc7 kinase promotes Mcm2-7 ring opening to allow for single-stranded DNA extrusion and helicase assembly. J Biol Chem. 2015;290:1210–21.
Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci U S A. 2006;103(31):11521–6.
Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17(3):349–68.
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34.
Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, et al. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem. 2006;281(15):10281–90.
Wan L, Niu H, Futcher B, Zhang C, Shokat KM, Boulton SJ, et al. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 2008;22(3):386–97.
Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24(1):101–13.
Sheu Y-J, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463(7277):113–7.
Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445(7125):328–32.
Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445(7125):281–5.
Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem. 2006;281(51):39249–61.
Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, et al. Mec1 Is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40(3):353–63.
Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 1999;18(20):5703–13.
Nougarede R, Della Seta F, Zarzov P, Schwob E. Hierarchy of S-phase-promoting factors: yeast Dbf4-cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol Cell Biol. 2000;20(11):3795–806.
Walter JC. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem. 2000;275(50):39773–8.
Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146(1):80–91.
Geraghty DS, Ding M, Heintz NH, Pederson DS. Premature structural changes at replication origins in a yeast minichromosome maintenance (MCM) mutant. J Biol Chem. 2000;275(24):18011–21.
Hollingsworth RE, Ostroff RM, Klein MB, Niswander LA, Sclafani RA. Molecular genetic studies of the Cdc7 protein kinase and induced mutagenesis in yeast. Genetics. 1992;132:53–62.
Sclafani RA, Patterson M, Rosamond J, Fangman WL. Differential regulation of the yeast CDC7 gene during mitosis and meiosis. Mol Cell Biol. 1988;8(1):293–300.
Pessoa-Brandão L. Genetic and molecular studies of Saccahromyces cerevisiae Cdc7-Dbf4 kinase function in DNA damage-induced mutagenesis [Dissertation/Thesis]. Aurora, Colorado: University of Colorado Health Sciences Center; 2005.
Cheng AN, Jiang SS, Fan CC, Lo YK, Kuo CY, Chen CH, et al. Increased Cdc7 expression is a marker of oral squamous cell carcinoma and overexpression of Cdc7 contributes to the resistance to DNA-damaging agents. Cancer Lett. 2013;337(2):218–25.
Choschzick M, Lebeau A, Marx AH, Tharun L, Terracciano L, Heilenkotter U, et al. Overexpression of cell division cycle 7 homolog is associated with gene amplification frequency in breast cancer. Hum Pathol. 2010;41(3):358–65.
Bonte D, Lindvall C, Liu H, Dykema K, Furge K, Weinreich M. Cdc7-Dbf4 kinase overexpression in multiple cancers and tumor cell lines is correlated with p53 inactivation. Neoplasia. 2008;10(9):920–31.
Montagnoli A, Moll J, Colotta F. Targeting cell division cycle 7 kinase: a new approach for cancer therapy. Clin Cancer Res. 2010;16(18):4503–8.
Vanotti E, Amici R, Bargiotti A, Berthelsen J, Bosotti R, Ciavolella A, et al. Cdc7 kinase inhibitors: pyrrolopyridinones as potential antitumor agents. 1. Synthesis and structure-activity relationships. J Med Chem. 2008;51(3):487–501.
Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, et al. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol. 2008;4(6):357–65.
Sasi NK, Tiwari K, Soon FF, Bonte D, Wang T, Melcher K, et al. The potent Cdc7-Dbf4 (DDK) kinase inhibitor XL413 has limited activity in many cancer cell lines and discovery of potential new DDK inhibitor scaffolds. PLoS One. 2014;9(11):e113300.
Hoang ML, Leon RP, Pessoa-Brandao L, Hunt S, Raghuraman MK, Fangman WL, et al. Structural changes in Mcm5 protein bypass Cdc7-Dbf4 function and reduce replication origin efficiency in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27(21):7594–602.
Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10(3):160–7.
Fletcher RJ, Chen XS. Biochemical activities of the BOB1 mutant in Methanobacterium thermoautotrophicum MCM. Biochemistry. 2006;45(2):462–7.
Miller JM, Arachea BT, Epling LB, Enemark EJ. Analysis of the crystal structure of an active MCM hexamer. Elife. 2014;3:e03433.
Brandao LN, Ferguson R, Santoro I, Jinks-Robertson S, Sclafani RA. The role of Dbf4-dependent protein kinase in DNA polymerase zeta-dependent mutagenesis in Saccharomyces cerevisiae. Genetics. 2014;197:1111–22.
Pessoa-Brandao L, Sclafani RA. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics. 2004;167(4):1597–610.
Sasanuma H, Hirota K, Fukuda T, Kakusho N, Kugou K, Kawasaki Y, et al. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 2008;22(3):398–410.
Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, et al. Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Genes Dev. 2014;28(20):2291–303.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Rossbach, D., Sclafani, R.A. (2016). Role of DDK in Replication Initiation. In: Kaplan, D. (eds) The Initiation of DNA Replication in Eukaryotes. Springer, Cham. https://doi.org/10.1007/978-3-319-24696-3_14
Download citation
DOI: https://doi.org/10.1007/978-3-319-24696-3_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-24694-9
Online ISBN: 978-3-319-24696-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)