Abstract
Moderate consumption of wine seems to produce positive health effects derived from the occurrence of bioactive polyphenols. The biological properties of polyphenols are greatly dependent on their bioavailability, which, in turn, is largely influenced by their chemical structure and degree of polymerization. There are scientific evidences that gut microbiota play a key role in the production, bioavailability, and biological activities of phenolic metabolites. Concurrently, these metabolites can modulate the colonic microbial population composition and/or activity. Therefore, the study of the benefits to host health of wine polyphenols should be carried out considering the two-way relationship “polyphenols–microbiota.” In general, studies evidence that wine polyphenols are extensively metabolized by gut bacteria into phenolic metabolites that are able to modulate the growth of selected bacterial groups. However, only a limited number of bacterial species have been identified as being implicated in the metabolism of wine polyphenols, most of them belonging to Firmicutes phylum. The emerging high-throughput-omic approaches can contribute to the identification of the bacteria responsible for the polyphenol metabolism as well as the biomarkers and functional outcomes derived from wine consumption, thereby improving the knowledge about the interactions between wine polyphenols and gut microbiota as well as their benefits to human health.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
In the context of a diet and healthy lifestyle, it is generally accepted that, in spite of its ethanol content, the moderate consumption of wine has beneficial health effects, as evidenced by numerous scientific studies (recently reviewed by Artero et al. 2015). These effects include protection against cardiovascular diseases, such as atherosclerosis and coronary heart disease (Droste et al. 2013), diabetes type 2 (Chiva-Blanch et al. 2013), and neurodegenerative diseases (Li et al. 2012), among others. To date, most of these protective effects have been linked to the presence of phenolic compounds in wine.
Polyphenols are secondary plant metabolites that in the case of grapes are located on the solid parts of the fruit, mainly in the skins, seeds, and scrapes. During the winemaking process, the phenolic compounds pass into the wine, constituting one of the major groups of compounds in this fermented food (Monagas et al. 2005). From the chemical point of view, the term “polyphenols” encompasses a heterogeneous group of compounds that are characterized by possessing a benzenic ring substituted by one or several hydroxyl groups (–OH) and a functional side chain. According to their chemical structure, they are divided into two groups of compounds: flavonoids and non-flavonoids. The non-flavonoid compounds are characterized by a single ring of six carbons (C6), the most prominent in this group being hydroxybenzoic (C6–C1) and hydroxycinnamic (C6–C3) acids, phenolic alcohols (C6), and stilbenes (C6–C2–C6). The flavonoid compounds are characterized by two rings of six carbons joined by a central heterocycle of three carbons (C6–C3–C6), differing from each other in the degree of oxidation of heterocyclic oxygen and the saturation of the central ring. Among the flavonoids, the flavonols (quercetin, myricetin, kaempferol, and their glycosides) and flavan-3-ols (monomers and oligomeric and polymeric proanthocyanidins) stand out. In the case of red wine, anthocyanins are also included, which are the compounds responsible for the characteristic red color, highlighting in this group the malvidin-3-O-glucoside. As an example, Fig. 13.1 shows the chemical structures of the major phenolic compounds present in wine.
The total polyphenol content of phenolic compounds in wine is around 50–400 mg/L for white wines, and 900–1400 mg/L for young red wines, although their concentration is conditioned by several factors related to the grape (variety, soil, geography, climate, etc.) and by enological practices. Therefore, a moderate consumption of wine (250 mL/day) would provide an intake of 60 mg of polyphenols for white wines and 210 mg for young red wines.
The role of polyphenols in human health depends largely on their bioavailability, absorption, and metabolism of polyphenolic compounds. Once ingested, polyphenols are recognized by the human body as xenobiotics, which limit their bioavailability. Besides, depending on their degree of structural complexity and polymerization, these compounds may be readily absorbed in the small intestine (i.e., low-molecular-weight polyphenols such as monomeric and dimeric structures) or reach the colon almost unchanged (oligomeric and polymeric polyphenols, such as condensed or hydrolyzable tannins) (Monagas et al. 2010). It has been estimated that 90–95 % of the total polyphenol intake may accumulate in the colon where they can be transformed by the resident microbiota into metabolites that could be even more bioactive than their precursors (Clifford 2004). Thus, the interindividual variability of microbial metabolism also impacts on the bioavailability and bioefficacy of polyphenols and their metabolites (Gross et al. 2010). In recent years, it has been reported that microbe-derived phenolic metabolites exert beneficial health effects, such as antioxidant activities (Biasi et al. 2014), antiproliferative actions and cytotoxicity (Tanaka et al. 1993), anti-inflammatory effects (Muñoz-González et al. 2014), and antithrombotic activities (Rechner and Kroner 2005), as well as having effects on the intestinal microbiota (Cueva et al. 2010). In relation to the latter, phenolic metabolites and nonabsorbed polyphenols could affect the growth of gut microbiota, thereby modifying their diversity and metabolic activity (Selma et al. 2009; Requena et al. 2010). Therefore, studies of wine polyphenols are expected to be carried out using a dual approach that includes the formation of bioactive polyphenol-derived metabolites and the modulation of colonic microbiota, at the framework of what has been called a two way “wine polyphenols-gut microbiota interaction (Requena et al. 2010; Dueñas et al. 2015) (Fig. 13.2). In this chapter, after describing some general aspects concerning gut microbiota, we have summarized the current knowledge about the modulation of gut microbiota by wine polyphenols as well as the intrinsic metabolism of wine polyphenols by intestinal bacteria, with special emphasis on the phenolic-metabolizer bacteria identified so far.
Microbiota-polyphenols two-way interaction at intestinal level (adapted from Muñoz-González 2014)
2 Gut Microbiota
The microbial content of the gastrointestinal tract changes along its length, ranging from a narrow diversity and low numbers of microbes in the stomach to a wide diversity and high numbers in the large intestine (Sekirov et al. 2010) (Fig. 13.3). The dominant bacterial phyla are the Firmicutes (including Clostridium, Enterococcus, Lactobacillus, and Ruminococcus genera) and Bacteroidetes (including Bacteroides and Prevotella genera). Other subdominant or minor phyla include Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia (Qin et al. 2010). It is assumed that several hundred species-level bacteria assemble in each individual in highly variable proportions, resulting in an individual microbial composition that remains stable in time (Rajilić-Stojanović et al. 2013). The temporal stability of the intestinal ecosystem is likely maintained by host-encoded mechanisms in parallel with colonization resistance, as a balanced climax community is not susceptible to new (invading) species. The temporal variation of the microbiota is mostly due to an altered abundance of existing species instead of a flux in the species composition (Rajilić-Stojanović et al. 2013).
(a) Variations in microbial number across the length of gastrointestinal tract. (b) Temporal aspects of microbiota establishment and factors influencing the composition of microbiota (adapted from Sekirov et al. 2010)
The extensive development and use of molecular methodologies in recent years has led to breakthroughs in the gut microbiota composition. In this context, a pioneering study by Arumugam et al. (2011) has suggested that the microbiota of most individuals can be categorized into three predominant variants, or “enterotypes,” dominated by three different genera: Bacteroides , Prevotella , and Ruminococcus , which are independent of age, sex, nationality, and body mass index (BMI) and allow the segmentation of subjects according to their intestinal microbiome. Nevertheless, this classification is not exempt from debate, as increasingly researchers are favoring the idea of a continuum or gradient of species functionality rather than a discontinuous variation with segregated types (Jeffery et al. 2012).
2.1 Factors Affecting Intestinal Microbiota
The composition of gut microbiota is strongly influenced by a range of factors that include, among others, age, diet, and environmental factors such as antibiotic therapy. With regard to age, individuals exhibit differences in terms of microbial diversity and variation at different life stages (Fig. 13.3) (O’Toole and Claesson 2010). Immediately after birth, babies are colonized by a population characterized by instability (Scholtens et al. 2012). Babies that are solely breast-fed until weaning have a microbiota dominated by Bifidobacterium and Ruminococcus, whilst those that are formula-fed tend to have a more diverse microbiota (Roger et al. 2010). Following the introduction of solid food to an infant’s diet, a more stable community, similar to that of adult microbiota, becomes established after 2–3 years of age (Yatsunenko et al. 2012). However, this relative stability and diversity of the microbiota is reduced in old age (Claesson et al. 2012); in particular, a decrease in the total number and species diversity of bifidobacteria and Bacteroides takes place. Interestingly, a recent study has shown for the first time that phenotypic effects can be vertically transmitted through the microbiome (Moon et al. 2015), suggesting that we should extend this new factor when it comes to understanding how the microbiome influences sickness and health.
Diet has long been considered one of the major external modulators of the human intestinal microbiota. A prominent example of the role of diet in the determination of the composition of the intestinal microbiota in humans is the study conducted by Filippo et al. (2010). The comparison of the gut microbiota of Italian (consuming a “Western” diet) and African (consuming a plant-rich “rural” diet, high in fiber content) children revealed that the latter were enriched with Bacteroidetes (mainly Prevotella and Xylanibacter), at the expense of Firmicutes, and a significantly lower amount of enterobacteria. Similar dietary associations have been found in a study linking the dietary patterns of American adults, belonging to the same geographic area, and of similar cultural backgrounds, with gut microbial enterotypes. Wu et al. (2011) found that the Bacteroides enterotype was positively associated with protein and animal fat, whereas the Prevotella enterotype was associated with a diet high in carbohydrates. Hence, there is strong evidence that high levels of Prevotella, which contain genes for cellulose hydrolysis, characterize microbiomes that are exposed mainly to complex plant-derived carbohydrates.
Despite these findings, recent global analyses of sequence and HitChip data have indicated that interindividual variation played a more major role than dietary change in determining the overall species composition of the microbiota (Salonen et al. 2014). The explanation for this apparent contradiction is twofold. Firstly, many species (especially the less abundant ones) occur only in one or a few individuals. Secondly, it appears that within the microbiota only certain species are responsive to the particular dietary switches, in this case including those bacteria that are specialists.
Finally, antibiotic treatment has also been shown (Young and Schmidt 2004) to dramatically disturb the composition of the intestinal microbiota in humans. In general, antibiotic treatment leads to a decrease in the diversity of the microbiota (Jernberg et al. 2007) as well as to a change in metabolic activity. Nonetheless, the community is quite resilient and can resemble the pretreatment state in a matter of days or weeks (Dethlefsen et al. 2008).
2.2 Functions of the Intestinal Microbiota and Its Importance in Health
Understanding the long-range metabolic interdependence between human and gut microbial metabolism is of importance to health and nutrition status (Nicholson et al. 2012). Apart from the obvious role in digestion, the gut microbiota has been associated with trophic, metabolic, and protective functions (Fig. 13.2). In fact, some authors have suggested that the microbiota could act as an “organ” that interacts with the human host and performs many essential functions to maintain human health status (Tremaroli and Bäckhed 2012). Metabolic functions of the gut microbiota allow the human host to utilize many energetic sources. The breakdown of complex indigestible dietary carbohydrates and proteins is possible thanks to the metabolic activity of the gut microbiota. Moreover, the microbiota produces vitamins, synthesizes amino acids, influences ion absorption, and is involved in the conversion of dietary polyphenolic compounds and in the bile acid biotransformation process (DiBaise et al. 2008; Lefebvre et al. 2009). The main products of the substrate fermentation in the gut are short-chain fatty acids (SCFA), particularly acetate, propionate, and butyrate, which positively influence intestinal epithelial cell proliferation and differentiation and have different metabolic features (Lepage et al. 2013).
Another essential function of the intestinal microbiota is the maintenance of intestinal epithelium barrier integrity maintaining cell-to-cell junctions, promoting epithelial repair following injury, and playing a role in the regulation of enterocytes turnover (Cario et al. 2007).
But perhaps, along with the role of nutrition, the most important functions of the intestinal microbiota are the protection from external pathogenic microorganisms and the development of a functional immune system. In the first case, called “colonization resistance,” the microbiota prevents pathogenic colonization by competing for attachment sites and nutrients (Sekirov et al. 2010), and through production and secretion of antimicrobials (Chung et al. 2012). Commensal bacteria are able to regulate the production of intestinal mucins by goblet cells, capable of inhibiting bacterial adhesion to intestinal epithelial cells (Wrzosek et al. 2013). The defense barrier of commensal microbiota could also be related to bacterial metabolic products. The production of SCFA causes a reduction of intestinal pH, which could prevent the growth of potentially pathogenic bacteria such as Escherichia coli and other members of the family Enterobacteriaceae (Zimmer et al. 2012). On the other hand, the microbiota is also essential for the development of a functional immune system, affecting both innate and adaptive immunities, and in promoting immune regulation at the intestinal surface. This can be readily appreciated from studies performed on germ-free (GF) animals (Sekirov et al. 2010), which generally are more susceptible to infection and have smaller Peyer’s patches, reduced mesenteric lymph nodes, decreased cell numbers, and defects in antibody production compared to conventional animals (Lee and Mazmanian 2010). In turn, the composition of the microbiota influences individual variations in immunity, and the absence of beneficial host-specific bacteria may promote disease in genetically susceptible individuals (Blaser et al. 2013).
As the gut microbiota has a well-established role in host homeostasis, several highly prevalent gastrointestinal diseases have been associated with imbalances in microbiota composition (dysbiosis) (Robles-Alonso and Guarner 2013). These human diseases include autoimmune and autoinflammatory disorders, such as allergies, obesity, and inflammatory bowel disease (Schippa and Conte 2014). In this context, wine polyphenols and their microbial phenolic metabolites could play a key role since it has been demonstrated that they are able to exert, among others, anti-inflammatory properties through modulation of gut microbiota (Queipo-Ortuño et al. 2012), as described in Sect. 13.3.
2.3 Analytical Approaches
Until the 1990s, knowledge of the gut microbiota was limited to traditional culture-based techniques based on phenotypic identification. However, these techniques are very restrictive as there is a large number of species that are not cultivable (Eckburg et al. 2005). Recent developments in molecular biology have allowed more accurate investigation of microbial communities, as the new techniques are culture-independent. Molecular biological techniques are based on the differences in the sequence of nucleotides of the microbial genes. The majority of these techniques consist of the extraction of DNA from the sample, followed by amplification and sequencing of 16S ribosomal RNA genes, which contain conserved and variable regions that allow taxonomic identification, ranging from the domain and phylum level to the species level (Robles-Alonso and Guarner 2013), thus providing information about microbial composition and diversity of species in a given sample. “Denaturing gradient gel electrophoresis (DGGE), fluorescent in situ hybridization (FISH), quantitative Polymerase Chain Reaction (qPCR) and capillary sequencing by using the Sanger method are among the most frequently used molecular techniques.
However, to perform a more complex analysis of the intestinal microbiota, emerging technologies, such as next-generation DNA sequencing (NGS) based on real-time monitoring of DNA synthesis, are of great interest. Due to these techniques the concept of “metagenomics” has emerged, defined as the study of metagenomes, i.e., the collective genetic content of the combined genomes of the constituents of an ecological community. In addition, the metagenomic approach also provides information about biological functions present in the community.
In the context of polyphenol–microbiota interactions, these emerging high-throughput-omic approaches can be adopted to identify genes and microorganisms involved in polyphenol (in)activation and conversion, to reconstruct metabolic pathways, and to monitor how microbial communities adjust their metabolic activities upon polyphenol exposure (Kemperman et al. 2010). Application of these technologies to human fecal samples requires further investigation to determine how these samples reflect metabolism inside the gut and, ultimately, to improve the understanding of the impact of polyphenols on host health (Hervert-Hernández and Goñi 2011; Kemperman et al. 2013).
Other potential molecular approaches include metatranscriptomics, metaproteomics, and metabolomics, which analyze the RNA, proteins, and metabolites, respectively, of complex communities. Environmental metatranscriptomics retrieves and sequences environmental mRNA from a microbial ecosystem to assess what genes may be expressed in that community. Metaproteomics allows us to link the abundance and activity of enzymes to their phylogenetic origin based on proteins. Lastly, the metabolome is the terminal downstream product of the genome and consists of the total complement of all the low-molecular-weight molecules in a cell, tissue, or organism.
3 Modulation of Gut Microbiota by Wine Polyphenols
As mentioned above, most of the polyphenols that are ingested in the diet reach the colon, where they can be converted by microbiota into bioactive metabolites that can affect the intestinal ecology and influence host health. In order to assess the modulating effect of wine polyphenols, several in vitro and in vivo animal and human intervention studies have been carried out (Tables 13.1 and 13.2). For example, studies using batch culture fermentation, a model reflective of the distal region of the human large intestine, have evaluated both the polyphenols metabolism in the presence of human gut microbiota and the changes in microbial communities after incubation with pure phenolic compounds (Tzounis et al. 2008; Hidalgo et al. 2012), and with extracts rich in polyphenols, derived from grapes (Cueva et al. 2013) and wine (Barroso et al. 2013; Sánchez-Patán et al. 2012). In general, these studies have demonstrated the increase of some bacterial groups in the intestine, such as Lactobacillus, Enterococcus, and Bifidobacterium, and the decrease of others, mainly Clostridium histolyticum (Table 13.1). On the other hand, three recent studies conducted with gastrointestinal tract simulators (SHIME and SIMGI) found notable changes in certain intestinal bacterial groups after simulation with an extract of wine (Barroso et al. 2014; Kemperman et al. 2013) and with red wine (Cueva et al. 2015), the most affected bacterial groups being Bacteroides and Bifidobacterium (Table 13.1).
With regard to animal experiments, several studies have been performed using red wine and grape seed extracts (Table 13.2). The fecal bacteria composition of rats fed with red wine polyphenols shifted from a predominance of Bacteroides, Clostridium, and Propionibacterium spp. to a predominance of Lactobacillus and Bifidobacterium spp. (Dolara et al. 2005). Another animal experiment was carried out to study the effect of the inclusion of grape seed extracts in the diet of broiler chicks (Viveros et al. 2011) on intestinal microbiota. It was observed that grape extracts modified the gut microbiota, increasing E. coli, and Lactobacillus and Enterococcus species populations. Recently, two animal studies performed in pigs have demonstrated that grape seed extract administration caused an ecological shift in the microbiome. Recently, two animal studies performed in pigs have demonstrated that grape seed extract administration caused an ecological shift in the microbiome, decreasing Streptococcus spp. and Clostridium cluster XIVa counts (Fiesel et al. 2014), and increasing Lachnospiraceae, Clostridiales, Lactobacillus, and Ruminococcaceae populations during the intervention period (Choy et al. 2014).
Investigations carried out with humans potentially provide the best models for studying the interactions of food components (e.g., polyphenols) with microbiota; however to date only a few studies have been conducted. Yamakoshi et al. (2001) reported that administration of a proanthocyanidin-rich extract significantly increased the fecal number of Bifidobacterium spp., whereas a reduction in the bacteria belonging to the Enterobacteriaceae family was observed. On the other hand, Queipo-Ortuño et al. (2012) assessed the effect of the moderate intake of red wine. A significant increase in the number of Enterococcus, Prevotella, Bacteroides, Bifidobacterium, Bacteroides uniformis, Eggerthella lenta, and Blautia coccoides-E. rectale was found. Specifically, an increase of Bifidobacterium spp. has recently been correlated with an increase in microbial metabolites derived from wine anthocyanins (Boto-Ordónez et al. 2014). In contrast, concentrations of Clostridium spp. and C. histolyticum group decreased after the red wine period. In summary, all these studies confirm the modulatory capacity of wine polyphenols on intestinal microbiota, which could have positive health effects or even prevent disease.
4 Catabolism of Wine Polyphenols by Intestinal Bacteria
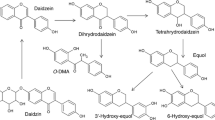
Although polyphenol metabolism starts in the mouth due to β-glycosidase activity, the colon is seen as being the main important organ for the catabolism of wine polyphenols and is widely influenced by their chemical structure. Oligomers and polymers of flavan-3-ols are the major phenolic compounds present in the wine that reaches the colon (Monagas et al. 2010; Rodriguez-Mateos et al. 2014). The catabolism of dimeric procyanidins involves the C-ring opening, followed by lactonization, decarboxylation, dehydroxylation, and oxidation reactions, among others (Selma et al. 2009). In the case of galloylated monomeric flavan-3-ols, the microbial catabolism usually starts with the rapid cleavage of the gallic acid ester moiety by microbial esterases, giving rise to gallic acid, which is further decarboxylated into pyrogallol (Kohri et al. 2003; Meselhy et al. 1997). The C-ring is subsequently opened, giving rise to 1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol, which is later converted into 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone in the case of (epi)catechin or 5-(3′,4′,5′-trihydroxyphenyl)-γ-valerolactone in the case of (epi)gallocatechin (Roowi et al. 2010). The valerolactone ring later breaks, giving rise to 5-(3′,4′-dihydroxyphenyl)valeric acid and/or 4-hydroxy-5-(3′,4′-dihydroxyphenyl)valeric acid. Subsequent biotransformations of these valeric acids give rise to hydroxyphenylpropionic and hydroxybenzoic acids by successive loss of carbon atoms from the side chain through β-oxidation (Meselhy et al. 1997). With regard to the microbial catabolism of flavonols, they are directly transformed into 3,4- or 3,5-dihydroxylated phenylacetic acids (Aura 2008). In the case of anthocyanins, they are converted into 3,4-dihydroxy-, 4-hydroxy-, 3,4-dimethoxy-, or 3-methoxyl-4-hydroxyl benzoic acids according to the substitution pattern of the B-ring of the precursor anthocyanin molecule (Aura 2008; De Ferrars et al. 2014). However, in spite of the fact that anthocyanins are abundant in wine, their circulating levels in plasma are very low, which has been attributed to anthocyanin instability under neutral pH, their extensive metabolism in vivo, and their probable catabolism by intestinal microbiota (De Ferrars et al. 2014). On the other hand, non-flavonoid compounds present in wine, hydroxycinnamic esters (i.e., caffeic acid derivatives), are mainly transformed into 3-hydroxyphenylpropionic acid, benzoic acid, and 4-ethylcatechol (Gonthier et al. 2006).
Once absorbed, the microbial metabolites are mainly metabolized in the liver by phase II enzymes as conjugated metabolites (glucuronides and sulfates), which can reach the colon via enterohepatic circulation and are also susceptible to degradation by the intestinal microbiota. Finally, the phenolic metabolites are excreted via urine and feces (Jiménez-Girón et al. 2015; Muñoz-González et al. 2013).
4.1 Bacteria Identified as Metabolizers of Certain Phenolic Groups
Intestinal bacteria play a crucial role in the metabolism of wine polyphenols and may therefore contribute to health-promoting effects. Despite the advances recently made in the knowledge of the identification of phenolic metabolites, the specific bacterial species able to metabolize wine polyphenols in the gastrointestinal tract and the anaerobic degradation pathways remain largely unknown. One of the main factors limiting the isolation and subsequent identification of polyphenol catabolic bacteria is the difficulty in growing them in commercial culture media. The reasons for this cultivation anomaly include the unknown growth requirements of the bacteria, the selectivity of the media that are used, the stress imposed by the necessity of strictly anoxic conditions, and difficulties with simulating the interactions of bacteria with other microbes and host cells. Table 13.3 shows an overview of the intestinal bacteria involved in the metabolism of wine phenolic compounds as well as the metabolites produced. The main bacteria involved in flavonol (quercetin, quercetin-3-glucoside, and kaempferol), and flavan-3-ol (catechin and epicatechin) metabolism belong to the phyla Firmicutes, and Firmicutes and Actinobacteria, respectively. This metabolic activity has been also confirmed in Lactobacillus plantarum IFPL935, which has demonstrated its ability to favor the initial metabolism of red wine polyphenols (Barroso et al. 2013, 2014). This greater phenolic metabolic activity of members of Firmicutes phylum leads us to hypothesize that this group might possess a specific function to degrade polyphenols. On the other hand, it can be expected that the large individual differences generate differences in the microbial metabolite profiles, because human microbiota contains more than 1000 different species with high individual variation (Qin et al. 2010). However, different colonic communities share general metabolic activities, which convert food components to specific metabolite profiles (Jacobs et al. 2009).
Therefore, the identification of the bacteria responsible for polyphenol metabolism is of vital importance to map functional metabolic reactions and describe the interaction between host and microorganisms in order to understand metabolism. In turn, this knowledge would help in the development of potential functional foods and ingredients with health benefits for individuals who produce low levels of these bioactive metabolites.
5 Conclusions
The bioavailability and effects of polyphenols greatly depend on their transformation by gut microbiota. Several studies have demonstrated that metabolization of wine polyphenols by gut microbiota leads to the production of a wide variety of metabolites with potential positive effects on human health. Most of the wine polyphenol-metabolizing intestinal bacteria identified belong to the phylum Firmicutes. In turn, wine polyphenols and their metabolites modulate the growth of selected bacterial groups, highlighting the importance of the two-way polyphenols–microbiota interaction in the maintenance of gut health. Even though it is well established that diet influences gut microbiota composition, recent findings suggest that interindividual variation plays a more major role than dietary change in determining the overall species composition of the microbiota. Therefore, further investigations using emerging molecular methods are necessary in order to achieve a better understanding of the underlying mechanisms in the polyphenols–microbiota–host triangle, and elucidate the implications of polyphenols for host health.
References
Artero A, Artero A, Tarín JJ, Cano A. The impact of moderate wine consumption on health. Maturitas. 2015;80:3–13.
Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473:174–80.
Aura AM. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem Rev. 2008;7:407–29.
Barroso E, Sánchez-Patán F, Martín-Álvarez PJ, Bartolomé B, Moreno-Arribas MV, Peláez C, et al. Lactobacillus plantarum IFPL935 favors the initial metabolism of red wine polyphenols when added to a colonic microbiota. J Agric Food Chem. 2013;61:10163–72.
Barroso E, Van de Wiele T, Jiménez-Girón A, Muñoz-González I, Martín-Álvarez PJ, Moreno-Arribas MV, et al. Lactobacillus plantarum IFPL935 impacts colonic metabolism in a simulator of the human gut microbiota during feeding with red wine polyphenols. Appl Microbiol Biotechnol. 2014;98:6805–15.
Biasi F, Deiana M, Guina T, Gamba P, Leonarduzzi G, Poli G. Wine consumption and intestinal redox homeostasis. Redox Biol. 2014;2:795–802.
Blaser M, Bork P, Fraser C, Knight R, Wang J. The microbiome explored: recent insights and future challenges. Nat Rev Microbiol. 2013;11:213–7.
Blaut M, Schoefer L, Braune A. Transformation of flavonoids by intestinal microorganisms. Int J Vitam Nutr Res. 2003;73:79–87.
Boto-Ordónez M, Urpi-Sarda M, Queipo-Ortuño MI, Tulipani S, Tinahones FJ, Andres-Lacueva C. High levels of Bifidobacteria are associated with increased levels of anthocyanin microbial metabolites: a randomized clinical trial. Food Funct. 2014;5:1932–8.
Braune A, Gutschow M, Engst W, Blaut M. Degradation of quercetin and luteolin by Eubacterium ramulus. Appl Environ Microbiol. 2001;67:5558–67.
Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology. 2007;132:1359–74.
Chiva-Blanch G, Urpi-Sarda M, Ros E, Valderas-Martinez P, Casas R, Arranz S, et al. Effects of red wine polyphenols and alcohol on glucose metabolism and the lipid profile: a randomized clinical trial. Clin Nutr. 2013;32:200–6.
Choy YY, Quifer-Rada P, Holstege DM, Frese SA, Calvert CC, Mills DA, et al. Phenolic metabolites and substantial microbiome changes in pig feces by ingesting grape seed proanthocyanidins. Food Funct. 2014;5:2298–308.
Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93.
Claesson MJ, Jeffery IB, Conde S, Power SE, O’connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84.
Clifford MN. Diet-derived phenols in plasma and tissues and their implications for health. Planta Med. 2004;70:1103–14.
Cueva C, Moreno-Arribas MV, Martín-Álvarez PJ, Bills G, Vicente MF, Basilio A, et al. Antimicrobial activity of phenolic acids against commensal, probiotic and pathogenic bacteria. Res Microbiol. 2010;161:372–82.
Cueva C, Sánchez-Patán F, Monagas M, Walton GE, Gibson GR, Martín-Álvarez PJ, et al. In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: changes in microbial groups and phenolic metabolites. FEMS Microbiol Ecol. 2013;83:792–805.
Cueva C, Jiménez-Girón A, Muñoz-González I, Esteban-Fernández A, Gil-Sánchez I, Dueñas M, et al. Application of a new dynamic gastrointestinal simulator (SIMGI) to study the impact of red wine in colonic metabolism. Food Res Int. 2015;72:149–59.
De Ferrars RM, Czank C, Zhang Q, Botting NP, Kroon PA, Cassidy A, et al. The pharmacokinetics of anthocyanins and their metabolites in humans. Br J Pharmacol. 2014;171:3268–82.
Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:2383–400.
DiBaise JK, Zhang H, Crowell MD, Krajmalnik-Brown R, Decker GA, Rittmann BE. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc. 2008;83:460–9.
Dolara P, Luceri C, De Filippo C, Femia AP, Giovannelli L, Caderni G, et al. Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats. Mutat Res. 2005;591:237–46.
Droste DW, Iliescu C, Vaillant M, Gantenbein M, De Bremaeker N, Lieunard C, et al. A daily glass of red wine associated with lifestyle changes independently improves blood lipids in patients with carotid arteriosclerosis: results from a randomized controlled trial. Nutr J. 2013;12:147.
Dueñas M, Cueva C, Muñoz-González I, Jiménez-Girón A, Sánchez-Patán F, Santos-Buelga C, et al. Studies on modulation of gut microbiota by wine polyphenols: from isolated cultures to omic approaches. Antioxidants. 2015;4:1–21.
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8.
Fiesel A, Gessner DK, Most E, Eder K. Effects of dietary polyphenol-rich plant products from grape or hop on pro-inflammatory gene expression in the intestine, nutrient digestibility and faecal microbiota of weaned pigs. BMC Vet Res. 2014;10:196.
Filippo C, Cavalieri D, Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6.
Gonthier MP, Remesy C, Scalbert A, Cheynier V, Souquet JM, Poutanen K, et al. Microbial metabolism of caffeic acid and its esters chlorogenic and caftaric acids by human faecal microbiota in vitro. Biomed Pharmacol. 2006;60:536–40.
Gross G, Jacobs DM, Peters S, Possemiers S, van Duynhoven J, Vaughan EE, et al. In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong interindividual variability. J Agric Food Chem. 2010;58:10236–46.
Hervert-Hernández D, Goñi I. Dietary polyphenols and human gut microbiota: a review. Food Rev Int. 2011;27:154–69.
Hidalgo M, Oruna-Concha MJ, Kolida S, Walton GE, Kallithraka S, Spencer JPE. Metabolism of anthocyanins by human gut microflora and their influence on gut bacterial growth. J Agric Food Chem. 2012;60:3882–90.
Jacobs DM, Gaudier E, van Duynhoven J, Vaughan EE. Non-digestible food ingredients, colonic microbiota and the impact on gut health and immunity: a role for metabolomics. Curr Drug Metab. 2009;10:41–54.
Jeffery IB, Claesson MJ, O’Toole PW, Shanahan F. Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol. 2012;10:591–2.
Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66.
Jiménez-Girón A, Ibáñez C, Cifuentes A, Simó C, Muñoz-González I, Martín-Álvarez PJ, et al. Faecal metabolomic fingerprint after moderate consumption of red wine by healthy subjects. J Proteome Res. 2015;14:897–905.
Kemperman RA, Bolca S, Roger LC, Vaughan EE. Novel approaches for analysing gut microbes and dietary polyphenols: challenges and opportunities. Microbiology. 2010;156:3224–31.
Kemperman RA, Gross G, Mondot S, Possemiers S, Marzorati M, Van de Wiele T, et al. Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res Int. 2013;53:659–69.
Kohri T, Suzuki M, Nanjo F. Identification of metabolites of (−)-epicatechin gallate and their metabolic fate in the rat. J Agric Food Chem. 2003;51:5561–6.
Kutschera M, Engst W, Blaut M, Braune A. Isolation of catechin-converting human intestinal bacteria. J Appl Microbiol. 2011;111:165–75.
Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–73.
Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–91.
Lepage P, Leclerc MC, Joossens M, Mondot S, Blottière HM, Raes J, Ehrlich D, Doré J. A metagenomic insight into our gut’s microbiome. Gut. 2013;62:146–58.
Li F, Gong Q, Dong H, Shi J. Resveratrol, a neuroprotective supplement for Alzheimer’s disease. Curr Pharm Des. 2012;18:27–33.
Meselhy MR, Nakamura N, Hattori M. Biotransformation of (−)-epicatechin 3-O-gallate by human intestinal bacteria. Chem Pharm Bull. 1997;45:888–93.
Moco S, Martin FPJ, Rezzi S. Metabolomics view on gut microbiome modulation by polyphenol rich foods. J Proteome Res. 2012;11:4781–90.
Monagas M, Bartolomé B, Gómez-Cordovés C. Updated knowledge about the presence of phenolic compounds in wine. Crit Rev Food Sci Nutr. 2005;45:85–118.
Monagas M, Urpi-Sarda M, Sánchez-Patán F, Llorach R, Garrido I, Gómez-Cordovés C. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010;1:233–53.
Moon C, Baldridge MT, Wallace MA, Brunham CAD, Virgin HB, Stappenbeck TS. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature. 2015. doi:10.1038/nature14139.
Muñoz-González I. Estudio del consumo moderado de vino sobre la función digestiva: metabolitos fenólicos y metaboloma fecal, microbiota oral y colónica y respuesta inmune. http://hdl.handle.net/10486/663355 (2014). Accessed 22 Oct 2014.
Muñoz-González I, Jiménez-Girón A, Martín-Álvarez PJ, Bartolomé B, Moreno-Arribas MV. Profiling of microbial-derived phenolic metabolites in human feces after moderate red wine intake. J Agric Food Chem. 2013;61:9470–9.
Muñoz-González I, Espinosa-Martos I, Rodríguez JM, Jiménez-Girón A, Martín-Álvarez PJ, Bartolomé B, et al. Moderate consumption of red wine can modulate human intestinal inflammatory response. J Agric Food Chem. 2014;62:10567–75.
Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7.
O’Toole PW, Claesson MJ. Gut microbiota: changes throughout the lifespan from infancy to elderly. Int Dairy J. 2010;20:281–91.
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Queipo-Ortuño MI, Boto-Ordóñez M, Murri M, Gómez-Zumaquero JM, Clemente-Postigo M, Estruch R, et al. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am J Clin Nutr. 2012;95:1323–34.
Rajilić-Stojanović M, Heilig HGHJ, Tims S, Zoetendal EG, De Vos WM. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. 2013;15:1146–59.
Rechner AR, Kroner C. Anthocyanins and colonic metabolites of dietary polyphenols inhibit platelet function. Thromb Res. 2005;116:327–34.
Requena T, Monagas M, Pozo-Bayón MA, Martín-Álvarez PJ, Bartolomé B, del Campo R, et al. Perspectives of the potential implications of wine polyphenols on human oral and gut microbiota. Trends Food Sci Technol. 2010;21:332–42.
Robles-Alonso V, Guarner F. Linking the gut microbiota to human health. Br J Nutr. 2013;109:S21–6.
Rodriguez-Mateos A, Vauzour D, Krueger CG, Shanmuganayagam D, Reed J, Calani L, et al. Bioavailability, bioactivity and impact on health of dietary flavonoids and related compounds: an update. Arch Toxicol. 2014;88:1803–53.
Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology. 2010;156:3329–41.
Roowi S, Stalmach A, Mullen W, Lean MEJ, Edwards C, Crozier A. Green tea flavan-3-ols: colonic degradation and urinary excretion of catabolites by humans. J Agric Food Chem. 2010;58:1296–304.
Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218–30.
Sánchez-Patán F, Cueva C, Monagas M, Walton GE, Gibson GR, Quintanilla-López JE, et al. In vitro fermentation of a red wine extract by human gut microbiota: changes in microbial groups and formation of phenolic metabolites. J Agric Food Chem. 2012;60:2136–47.
Schippa S, Conte MP. Dysbiotic events in gut microbiota: impact on human health. Nutrients. 2014;6:5786–805.
Schoefer L, Mohan R, Schwiertz A, Braune A, Blaut M. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl Environ Microbiol. 2003;69:5849–54.
Scholtens PAMJ, Oozeer R, Martin R, Amor KB, Knol J. The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol. 2012;3:425–47.
Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904.
Selma MV, Espín JC, Tomaá-Barberán FA. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem. 2009;57:6485–501.
Tanaka T, Kojima T, Kawamori T, Wang A, Suzui M, Okamoto K, et al. Inhibition of 4-nitroquinoline-induced rat tongue carcinogenesis by the naturally occurring plant phenolic acids caffeic, ellagic, chlorogenic and ferulic acids. Carcinogenesis. 1993;14:1321–5.
Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9.
Tzounis X, Vulevic J, Kuhnle GG, George T, Leonczak J, Gibson GR, et al. Flavanol monomer induced changes to the human faecal microflora. Br J Nutr. 2008;99:782–92.
Viveros A, Chamorro S, Pizarro M, Arija I, Centeno C, Brenes A. Effects of dietary polyphenol-rich grape products on intestinal microflora and gut morphology in broiler chicks. Poult Sci. 2011;90:566–78.
Wang LQ, Meselhy MR, Li Y, Nakamura N, Min BS, Qin G, et al. The heterocyclic ring fission and dehydroxylation of catechins and related compounds by Eubacterium sp. strain SDG-2, a human intestinal bacterium. Chem Pharm Bull. 2001;49:1640–3.
Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61.
Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8.
Yamakoshi J, Tokutake S, Kikuchi M, Kubota Y, Konishi H, Mitsuoka T. Effect of proanthocyanidin-rich extract from grape seeds on human fecal flora and fecal odor. Microb Ecol Health Dis. 2001;13:25–31.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7.
Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004;42:1203–6.
Zimmer J, Lange B, Frick J, Sauer H, Zimmermann K, Schwiertz A, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr. 2012;66:53–60.
Acknowledgements
This work was carried out with the financial support of the Spanish Ministry of Economy and Competitiveness (AGL2012-40172-C02-01 project).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Cueva, C., Gil-Sánchez, I., Moreno-Arribas, M.V., Bartolomé, B. (2016). Interactions Between Wine Polyphenols and Gut Microbiota. In: Moreno-Arribas, M., Bartolomé Suáldea, B. (eds) Wine Safety, Consumer Preference, and Human Health. Springer, Cham. https://doi.org/10.1007/978-3-319-24514-0_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-24514-0_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-24512-6
Online ISBN: 978-3-319-24514-0
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)