Abstract
P-glycoprotein (P-gp) was one of the first discovered, and most highly investigated, multidrug efflux pumps. P-gp was discovered in drug-resistant cancer cells and its ability to mediate adenosine triphosphate (ATP)-dependent efflux of drugs can confer resistance to cancer cells. The protein contains two sites for the binding and hydrolysis of ATP to power the active transport process. Drugs are known to bind within the transmembrane domain that comprises 12 membrane spanning α-helices. Biochemical, pharmacological and biophysical investigations continue to strive towards generating a molecular mechanism for drug transport. In addition, X-ray structures are available for the mouse and Caenorhabditis elegans isoforms at resolutions of 3–4 Å. However, one of the central issues related to the transport process remains elusive. A detailed understanding of how the protein is capable of binding its astonishing variety and number of compounds, remains unsolved. The hydrophobic vacuum cleaner and drug flippase models have been generated to describe this enigmatic property and some of their proposals remain intact. The majority of data supports the presence of a large binding domain that contains individual sites for drug interaction. These interaction sites are linked by an intricate allosteric network and binding to the sites is in close communication with the ATP hydrolytic machinery. This review provides a detailed account of our current understanding of how one membrane transporter is able to bind over 300 compounds.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Resistance to chemotherapy is either inherent to the cancer type or it is acquired in response to the chemotherapy regimen. The earliest reports of multidrug resistance used cancer cell lines selected for drug resistance by continuous exposure to anticancer drugs (Dano 1973). The resistant phenotype was characterised by a reduced accumulation of chemotherapeutic agents in cancer cells, which reduces the efficacy of cytotoxic anticancer drugs. The accumulation deficit was recognised as an ATP-dependent, protein mediated process and the culprit identified as the Permeability glycoprotein, or P-glycoprotein (Ling and Thompson 1974).
P-glycoprotein (P-gp; also known as ABCB1 or mdr1) is a polyspecific membrane transporter belonging to the ATP-Binding Cassette superfamily (Higgins 1992; Riordan and Ling 1985). It functions by actively effluxing compounds from within the cytosol and lipid bilayer to the external environment. Of the 48 known human ABC transporters, P-gp is arguably the most extensively researched as the expression of this protein in cancer cells plays a significant role in conferring multidrug resistance to cytotoxic anticancer drugs. P-gp has a molecular weight of 140 kD and is comprised of 1280 amino acids that are encoded as a single polypeptide (Chen et al. 1986; Ling et al. 1983). The protein is comprised of two homologous halves each containing a transmembrane domain (TMD) and a cytosolic nucleotide-binding domain (NBD). The former provides the conduit for drug transport and the latter generates energy from ATP hydrolysis for the process. Each transmembrane domain consists of six membrane spanning helices that display domain swapping and facilitate energetic coupling between nucleotide binding, substrate binding and nucleotide hydrolysis, by virtue of their direct contact to the catalytic sites, the NBDs.
Despite the extensive amount of information available on P-gp, there is a limited mechanistic understanding of its function. Of particular importance is how the protein can mediate the transport of such a large array of chemically and functionally unrelated drugs. P-gp appears to violate many of the “rules” of specificity that govern receptor, enzyme and transporter biology. It is by no means the only promiscuous member of the ABC superfamily of proteins and multidrug efflux systems exist throughout the biological world. The focus of this chapter is to outline the many different strategies that have been employed, in the 40 years since the discovery of P-gp, to address the key concept of how one protein is able to interact with such an extraordinary number of compounds.
Using Site-Directed Mutagenesis to Locate Drug-Binding Sites
The explosion of molecular biology techniques from the late 1980s was embraced in the search to define functionally important regions of P-gp; in particular, the drug-binding site. P-gp was mutated at specific amino-acid residues and the functional consequence measured in order to attribute a specific role in the transport process. P-gp function may be measured at multiple levels; for example, cell cytotoxicity assays reveal the effects on the overall resistant phenotype. Cellular accumulation of radiolabelled or fluorescent drugs, provided an insight into the transport process. The effects of mutations on drug-stimulated ATPase activity and photo-affinity labelling techniques both demonstrate a direct interaction of drugs with P-gp. Finally, and perhaps most relevantly, the binding of drug to mutated protein can be measured with equilibrium assays to provide the true affinity constant (KD) for the interaction.
Alanine Mutagenesis: An Early Approach
A theoretical study suggested that hydrophobic drug substrates of P-gp were likely to intercalate between phenylalanine (Phe) residues in the transmembrane (TM) helices (Pawagi et al. 1994), and in an early mutagenesis approach, the 31 intrinsic Phe residues were targeted. A total of 31 mutant versions of P-gp were generated, each containing a single Phe mutation to alanine and the proteins expressed in mouse NIH-3T3 cells (Loo and Clarke 1993). The mutant P-gp isoforms were analysed to assess whether the Phe → Ala mutation affected their ability to confer resistance to drugs (i.e. phenotypic assay) including vinblastine, actinomycin D, colchicine and adriamycin. A significantly different drug resistance profile was observed in only three of the mutants, while the other 28 resulted in protein products which were functionally and structurally indistinguishable from wild-type P-gp (Loo and Clarke 1993). The F777A (TM8) mutant exhibited a 2-fold decrease in the relative resistance to all four drugs and its effect was attributed to perturbed biosynthesis of P-gp due to lower level expression of this isoform. Mutant F335A (TM6) retained the ability to confer resistance to colchicine and adriamycin but conferred little resistance to actinomycin D or vinblastine. In contrast, mutant F978A (TM12) displayed little to no resistance to adriamycin or colchicine but was able to confer resistance to vinblastine or actinomycin D. Overall this investigation suggested that drugs may interact at distinct sites on the protein and that both halves of the polypeptide were involved in drug binding.
A number of other similar investigations were reported at this time, but with 1280 amino-acid residues, the possible permutations of amino-acid substitutions are daunting to say the least. A more systematic and directed mutagenesis approach was clearly required.
Cysteine Mutagenesis: A Widely Used Approach
The cysteine-scanning mutagenesis approach provides a systematic and flexible approach to elucidating the biochemical pharmacology of P-gp. The key requirement to adopt this approach is that mutation of the intrinsic cysteine residues does not significantly perturb the function of P-gp. The seven intrinsic cysteine residues (at positions 137, 431, 717, 956, 1074, 1125 and 1227) were each mutated to alanine and generated a cysteine-less (cys-less) P-gp that was functioned identically to wild-type protein (Loo and Clarke 1995a). Another team substituted the intrinsic cysteines to serine and produced a fully functional cys-less P-gp isoform (Taylor et al. 2001).
The use of a cysteine-mutagenesis strategy provides twofold benefit (Frillingos et al. 1998). First, the effects of replacing a residue with cysteine in a specific region of the protein may perturb the overall transport capability of P-gp. This will provide insight into the “native” function of that specific residue and establish the local region for further mutagenesis. Alternatively, the mutation of a “native” residue to cysteine does not cause significant perturbation of function. This will enable the investigator to utilise the chemistry of cysteine; in particular, the ability to covalently modify the sulphydryl moiety under physiological conditions (e.g. pH 7.4, 37 °C). There are numerous sulphydryl-reactive compounds (e.g. maleimide or methanethiosulphonate (MTS) containing) with fluorescent, radiolabelled or spin-labelled probes attached. Covalent attachment of more bulky compounds may provide further local perturbation of function and the use of spectroscopic probes will enable biophysical investigation of local environment or structural changes in the protein. The focus of the subsequent four sections is the identification of residues within several transmembrane segments that may be involved in binding of drugs by P-gp.
Dibromobimane and Protection from Inhibition
P-gp contains a central cavity (see Sect. “Can Structural Information Locate Drug Binding Sites?”) that is presumed to form the drug-binding domain and undergoes alternating access across the membrane during the transport process. The cysteine-scanning mutagenesis approach was used to identify residues lining the cavity that contribute to drug binding. In particular, the thiol-reactive compound, dibromobimane (dBBn), was reacted with a series of P-gp isoforms containing cysteine insertion into positions within TM6 and TM12 (Fig. 1) (Loo and Clarke 1997, 2000, 2002). Treatment of the mutant protein isoforms with dBBn impaired the drug-stimulated ATPase activity of P-gp, although this perturbation may be due to alteration of any of the multiple steps in the process (e.g. conformational change or drug binding). Consequently, the single-cysteine isoforms were pre-incubated with one of the substrates/modulators of P-gp prior to the addition of dBBn. If the drug pre-treatment prevented dBBn-induced inhibition of ATP hydrolysis (i.e. protection), then it may be inferred that the introduced cysteine residue is situated proximal to the binding site. Using the substrates vinblastine and colchicine, and the modulator verapamil, the authors concluded that residues mutated in TM6 (L339C and A342C) and in TM12 (L975C, V982C and A985C) were involved in the interaction of substrates with P-gp (Loo and Clarke 1997).
Construction of Cys-less P-glycoprotein and cysteine-directed mutagenesis studies with dibromobimane (dBBn). Wild-type P-gp contains seven endogenous cysteine residues which were mutated to alanine resulting in a Cys-less P-gp construct. Residues in a number of transmembrane helices were mutated to cysteine and purified using nickel affinity chromatography by virtue of a C-terminal polyhistidine tag. Inhibition of verapamil-stimulated ATPase activity in the presence of the thiol-reactive cross-linking agent dBBn was measured to identify various residues important in drug binding
These initial studies prompted larger investigations to identify potential residues within TM segments that interact with substrates of P-gp and thereby refine the topography and the environment of the drug-binding pocket. A study published in 2000 analysed 189 cysteine mutations scattered around the TM segments of P-gp predicted to line the central cavity (i.e. TMs1–5 and 7–10) (Loo and Clarke 2000). The mutant isoforms were analysed for inhibition of drug-stimulated ATP hydrolysis by dBBn. Seven mutants, Y118C and V125C (TM2), S22C (TM4), I306C (TM5), S766C (TM9), and I868C and G872C (TM10), displayed considerable inhibition of verapamil-stimulated ATPase activity upon treatment with dBBn. However, protection from inhibition by dBBn following pre-treatment with vinblastine, colchicine or verapamil was only seen for mutants S222C (TM4), I306C (TM5), I868 (TM10) and G872C (TM10). These results and related investigations (Loo and Clarke 1997, 1999, 2000, 2002) conclude that transmembrane helices 4, 5, 6, 10, 11 and 12 are implicated in drug binding by P-gp.
Methanethiosulphonate Analogues and Protection Assays
Concurrent to the investigations with dBBn, an alternative strategy to elucidate residues within the binding site adopted a thiol-reactive derivative of verapamil (MTS-verapamil). The premise was to use a compound more similar to a substrate to achieve covalent modification and ensure specificity for the drug-binding domain. The methanethiosulphonate group was attached by an ethyl linker arm to the methylamine group in verapamil and the ATPase activity was measured in cys-less P-gp. MTS-verapamil produced a similar degree of ATPase stimulation, with equivalent potency, thereby retaining the ability to interact with P-gp (Loo and Clarke 2001).
MTS-verapamil was used initially to generate a covalent modification within the drug-binding domain and the effects of 252 single-cysteine mutations on the labelling were investigated (Loo and Clarke 2001). The covalent attachment of MTS-verapamil to P-gp caused a reduction in the ATPase activity of several mutant isoforms. Specifically, the reduction in ATPase activity was found in only 15 of the 252 mutants: Y118C (TM2), V125C (TM2), S222C (TM4), L339C (TM6), A342C (TM6), A729C (TM7), A841C (TM9), N842C (TM9), I868C (TM10), A871C (TM10), F942C (TM11), T945C (TM11), V982C (TM12), G984C (TM12), and A985C (TM12). It was argued that the use of a substrate with an attached MTS moiety was more likely to identify residues specifically within the binding domain. However, the possibility of the MTS-cysteine affinity driving the labelling cannot be unequivocally excluded. Why should attachment of MTS-verapamil cause a reduction in ATPase activity? This is presumably related to the need for dissociation of drug from the TMD to enable completion of the ATP hydrolytic process.
In order to further support the specificity of this approach, the authors also used a “verapamil protection” strategy to differentiate cysteines that are proximal to drug-binding sites from those that are not (Loo and Clarke 2002; Loo et al. 2003a, 2006a, b). This strategy was possible since both verapamil and its thiol-reactive analogue have similar Km values (substrate affinity) for cys-less P-gp. Therefore, incubation with verapamil prior to addition of MTS-verapamil should protect the mutant from inactivation by MTS-verapamil if it is proximal to the binding site in question. Significant protection from inhibition by MTS-verapamil was found in four mutants (compared to 15 above); namely, S222C (TM4), L339C (TM6), A342C (TM6) and G984C (TM12). A lesser degree of protection was observed in mutants I868C (TM10), F942C (TM11) and T945C (TM11), which may indicate partial overlap of binding sites or a more peripheral location of the residues to the interaction site of verapamil. Overall, the results using MTS-verapamil have refined the number of helices putatively involved in the binding domain, but have strengthened the evidence that TMs 4, 6, 10, 11 and 12 contribute to the drug-binding pocket.
Verapamil is a well-established inhibitor of cytotoxic drug transport by P-gp and its administration with anticancer drugs is able to overcome the drug-resistant phenotype in vitro. It has been purported to be a transported substrate of P-gp, although the data is not conclusive to support this. Consequently, verapamil should be considered a “modulator” of P-gp function. In contrast, it is clear that several rhodamine derivatives are direct substrates for P-gp mediated transport (Shapiro and Ling 1997b). Moreover, it may transpire that substrates and allosteric modulators bind at distinct sites on P-gp. Therefore an MTS derivative of rhodamine was also used to identify the drug-binding domain of P-gp. In a similar approach to that outlined in the preceding paragraph, 252 single-cysteine mutants were generated, reacted with MTS-rhodamine and their ATPase activities measured (Loo and Clarke 2002). Of these mutations only 28, which were located in TMs 2–12, were markedly inhibited following covalent attachment of MTS-rhodamine. Subsequently, pre-treatment with the parent compound rhodamine significantly protected the activities of five mutants, I340C (TM6), A841C (TM9), L975C (TM12), V981C (TM12) and V982C (TM12), implicating TMs 6, 9 and 12 in the binding of rhodamine dyes.
The “Herculean” efforts involving site-directed mutagenesis of the TM domains of P-gp have significantly narrowed the search for the location of drug-binding sites on P-gp. A number of helices and their constitutive residues, have been implicated using distinct investigative strategies. None of the studies identified precise locations for drug binding, but this is hardly surprising given the promiscuity of P-gp and the large number of helices involved in lining the binding domain or cavity. However, these biochemical investigations will be vital in conjunction with structural data. There remains a clear need for further, and multiple, approaches to locate and define drug binding on this transporter.
A Proposed “Substrate-Induced Fit” Model
The data described in the previous sections clearly demonstrate that multiple helices, presumably those lining the central cavity, contribute to drug binding or the formation of a binding domain. In addition, data obtained with dBBn and MTS-cross-linking substrates have revealed a number of common residues from these helices that interact with distinct P-gp substrates. Conversely, the data also indicated that distinct classes of drugs interact with different residues. This point was also supported from a series of investigations, also using a cysteine-mutagenesis strategy, that focussed specifically on the involvement of TM helices 6 and 12 (Rothnie et al. 2004; Storm et al. 2008; Crowley et al. 2009, 2010a). In summary, these investigations potentially generate a hypothesis that drugs bind at “common, but different, sites” on the protein, or that binding sites may have some overlap of residues.
Can this rather confusing observation be reconciled to a unifying description of drug binding to P-gp? Such a model does exist and has been termed the “substrate-induced fit” hypothesis (Loo et al. 2003c). According to this hypothesis, a common binding site exists for drugs in the central cavity of P-gp. Following interaction at the common residues, a combination of residues from different transmembrane helices is moulded by the substrate to form its specific drug binding site.
Such a model would require that the cavity and its lining helices are mobile and a disulphide cross-linking study suggests helical mobility is present in P-gp and that there is a great deal of evidence to support this. For example, electron microscopy (EM)-based structural studies (Rosenberg et al. 2001, 2003, 2004) and biophysical approaches with 2H/1H exchange kinetics (Sonveaux et al. 1996, 1999) revealed that drugs and nucleotides generate considerable rearrangement of the TMD structure. It is the nature of contributing residues from each TM helix that determines the affinity for the substrate (Loo and Clarke 2002). Another investigation using cysteine mutagenesis to explore the nature of cross-linking between helices in the presence and absence of drugs provides further support (Loo et al. 2003c). P-gp isoforms were constructed, each with a pair of cysteine residues introduced at distinct helices. Cross-linking was induced by addition of the oxidant copper phenanthroline in the absence or presence of the drug, whereby the two conditions generated markedly different cross-linking patterns. For example, in the presence of colchicine and demecolcine, cross-linking was observed between residues P350C (TM6)/V991C (TM12), whereas cyclosporin A promoted cross-linking between P350C (TM6)/G939 (TM11) only. The addition of progesterone produced a number of cross-links; namely, P350C (TM6)/A935C (TM11), P350C (TM6)/G939C (TM11), and between P350C (TM6)/V991C (TM12). In contrast, other modulators and substrates such as verapamil and vinblastine did not promote cross-linking for any of the mutant pairs investigated. This study showed direct evidence of re-packing of TM segments to form an ‘induced’ drug-binding site upon addition of particular substrates, to help explain how the protein accommodates such a vast range of compounds.
Evidence of Simultaneous Binding
Evidence presented thus far has suggested that P-gp has a central cavity that mediates substrate and modulator binding. The binding domain comprises several TM helices and has a malleable structure to accommodate the interaction with drugs of distinct chemical composition. Can this binding domain interact with more than one drug simultaneously?
To address this issue several mutant isoforms of P-gp were investigated for their interaction with the thiol-reactive substrate tris-(2-maleimidoethyl)amine (TMEA), which will cross-link to any cysteine close to its binding site (Loo et al. 2003b). If a second drug was to bind at the same time at an overlapping site (e.g. competitive binding), then it would inhibit binding and cross-linking of TMEA to the protein. However, if the second substrate was to bind at a distant site (i.e. allosteric), then the cross-linking caused by TMEA would be unaffected, or possibly enhanced. Using verapamil as the second substrate, it was shown that cross-linking could be induced between TM6 (F343C) and TM12 (V982C). This is in contrast to the absence of any cross-linking in the presence of TMEA alone and evidence of inter-site allosteric communication. This cross-linking study using a mutagenesis approach therefore provided evidence in support of simultaneous binding of two substrates, each occupying a distinct region within the binding pocket (Loo et al. 2003b). However, this is not the only report of simultaneous binding of substrates to P-gp. Lugo and Sharom demonstrated that two fluorescent substrates (Rhodamine 123 and LDS-751) bound simultaneously to P-gp since they exhibited a spectral interaction upon binding to the protein (Lugo and Sharom 2005). In addition, classical receptor pharmacology studies demonstrated that the binding of one drug can accelerate the dissociation of another (Ferry et al. 1992; Martin et al. 1997). Altered dissociation rate constants can only occur with an allosteric interaction and therefore further proof of simultaneous drug interaction at the binding domain.
Photo-Affinity Labelling of Drug-Binding Sites
As described above, a vast amount of data has been acquired from the systematic insertion of cysteine mutants, followed by reaction with thiol-reactive substrates and cross-linking agents to characterise the large binding domain of P-gp. The data also identified the TM helices involved in mediating binding, provided an insight into some of the residues involved, and revealed the existence of multiple sites. However, the data on their own have not provided precise information on the sites of drug interaction. At the same time, a number of investigations used photo-activated probes and drugs in an attempt to find the drug-binding site(s) on P-gp.
The Anatomy of a Photo-Affinity Probe
The use of photo-affinity labelling to identify specific regions of a protein obviously requires a photo-active probe that binds to the target region. Following incubation of the photo-active probe with protein and binding equilibrium has been reached; the ternary complex is irradiated with light. Post-irradiation, the photo-active probe (bound at its target site) is converted to a short-lived, highly reactive nitrene intermediate that covalently attaches to the polypeptide chain. Due to a short half-life, these probes should efficiently label only specific sites that are in close proximity. The photo-affinity probes must be of high affinity to the target site, stable in an aqueous environment and display high specific radioactivity. Moreover, the photo-active compounds need to retain the activity or binding characteristics of the parent compound (Safa 1999). Once the photo-cross-linking has been achieved, there are a number of approaches to ascertain the site of labelling. However, great interpretive care must be exercised with this approach since the photo-active moiety may lie on a flexible region of the molecule and potentially label regions that lie external to the absolute binding site (Glossmann et al. 1987).
The first applications using photo-affinity labelling were to establish whether drug substrates and inhibitors bound to P-gp and to ascertain the potencies for drug binding to the protein (Greenberger 1993; Bruggemann et al. 1989; Hafkemeyer et al. 1998; Isenberg et al. 2001; Safa 1988; Zhang et al. 1995). Amongst the first photo-affinity probes to be used with P-gp were analogues of the well-characterised substrate vinblastine; N-(p-azido[3-125I]salicyl)-N′-aminoethylvindesine ([125I]NASV) and N-(p-azido[3,5-3H]benzoyl)-N′-(beta-amino-ethyl)vindesine ([3H]NABV) (Beck et al. 1988; Naito et al. 1989). The photo-affinity analogues of vinblastine were incubated with purified plasma membranes, mixed membrane vesicles or intact cells, and then irradiated for activation. Photo-affinity labelled protein samples were then analysed using SDS–PAGE followed by autoradiography to show that only resistant cell lines (overexpressing P-gp) were labelled with [125I]NASV. It was demonstrated that in the presence of 100 µM vinblastine, [125I]NASV photo-labelling was abrogated. This was the first demonstration that anticancer drugs bound directly to P-gp. In contrast to vinblastine, the anthracycline doxorubicin only produced partial inhibition of [125I]NASV labelling, whereas colchicine and methotrexate (a drug to which MDR cells are sensitive) did not show any blocking of photo-labelling. This was interpreted to suggest that colchicine binds at a separate site to the vinblastine analogue. Alternatively, it merely demonstrates that colchicine has a lower affinity for the vinblastine-binding site and a rank order of potency was defined as vinblastine > vincristine > doxorubicin > actinomycin D > colchicine (Safa 1988; Safa et al. 1989).
Photo-Labelling and Protease or Chemical Cleavage Approaches to Locating the Drug-Binding Site of P-gp
An early photo-affinity labelling study (Bruggemann et al. 1989) attempted to identify the regions of P-gp that were labelled by [3H]-azidopine. Azidopine is a photo-active dihydropyridine analogue that was initially developed to label L-type calcium channels (Ferry et al. 1985). The compound also inhibits the active transport of vinblastine in P-gp expressed in plasma membrane vesicles from the drug-resistant human carcinoma cell line, KB-V1 (Bruggemann et al. 1989). P-gp was photo-labelled with [3H]-azidopine, the protein was then digested with trypsin and V8 protease to enable the identification of the labelled fragments using specific antibodies. P-gp was digested and two major fragments were detected by immunoblotting. A 38-kDa fragment containing one-third of the labelled [3H]-azidopine was derived from the amino half, and a 30-kDa fragment containing two-thirds of the [3H]-azidopine, from the carboxyl half (Bruggemann et al. 1989). Immunoprecipitation and endoglycosidase treatments were used to define that the 38-kDa tryptic fragment contained TM helices 1–6 and that the 30-kDa V8 fragment contained TM helices 7–12. Since two different regions of P-gp were labelled by [3H]-azidopine, it is possible that the protein contains two binding sites for the compound, one in the carboxyl half and the other in the amino half. The alternative interpretation was that P-gp contains one binding site that is formed by the two homologous halves of the protein and that these two regions come together to create a single binding pocket for [3H]-azidopine.
A subsequent study used both [3H]-azidopine and [125I]-iodoaryl azidoprazosin ([125I]-IAAP) ligands and a similar protein digestion protocol (Greenberger et al. 1990). Both photo-affinity probes bound to highly related, if not identical, domain on the protein despite the chemical dissimilarity of the compounds. One explanation was that although both molecules were structurally diverse, they both contained the reactive arylazido group and thereby labelled the same protein region. Alternatively, P-gp could accommodate structurally diverse molecules in its binding site. In a follow-up study, the 40-kDa photo-affinity labelled domain in the C-terminal half of P-gp (predicted to reside within TM11) was shown to contain a major 4-kDa photo-labelling site. Its identity was deduced by antibody-based precipitation and consisted of approximately 29 residues, located between residues 979 and 1048 (Greenberger 1993). Further analysis of protein fragments using protease and chemical cleavage approaches had begun to identify numerous regions of drug attachment. For example, a 6-kDa trypsin fragment (found using antibody 1) in a predicted location within TM4 and up to (but not including) TM6, and a minor 40-kDa photo-labelled fragment between TM7 and TM8, were identified. In addition, a 25-kDa cyanogen bromide fragment was generated, containing an azidopine-labelled domain spanning the region from TM4 to the Walker A motif, (Bruggemann et al. 1992). Therefore, this molecular dissection revealed numerous attachment sites, a finding that may be related to the flexibility of [3H]-azidopine.
P-gp was also photo-labelled with [3H]-azidopine in the absence or presence of other substrates or inhibitors. [3H]-azidopine labels two distinct locations previously identified in P-gp: the site within the carboxy half and the one within the amino half between residues 198 and 440 (Bruggemann et al. 1992). Vinblastine strongly inhibited azidopine labelling of P-gp at both photo-labelling sites and the authors suggested that this supported the existence of a common binding site for azidopine and vinblastine. Given the subsequent investigations with equilibrium binding assays (Sect. “Classical Receptor-Drug Analysis of Multidrug Binding to P-gp”), the data are more likely explained by the presence of multiple binding sites linked by a negative heterotropic allostery (Ferry et al. 1992, 1995; Martin et al. 1997). These two investigations clearly demonstrated an allosteric interaction between vinblastine and several 1,4-dihydropyridines; the latter including azidopine.
To determine the binding site of paclitaxel, Wu et al. developed a photo-affinity analogue bearing a 3′-BzDC group and used site-directed antibodies to identify the labelled residues (Wu et al. 1998). The domain mapping identified residues 985–1088 in mouse mdr1b P-gp, which includes half of TM12 and terminates immediately after the Walker A motif of NBD2. Another study using a 7-DzDC group photo-affinity analogue of paclitaxel highlighted the importance of residues 683–760, a region that includes TM7 and half of TM8. In a third investigation, Safa et al. used the analogue [125I]-NAST to localise the paclitaxel-binding sites (Safa 2004). The sites were identified with site-directed antipeptide antisera raised against ten different domains. Following labelling, protease digestion revealed four major photo-labelled fragments of 12, 10, 8 and 6.5 kDa, which were immunoprecipitated by an antibody directed against amino-acid residues N-terminal to TM3. A short, five amino-acid intracellular sequence that was C-terminal to TM4 was found to be common across all these peptides. [125I]-NAST was found to be associated with another 6.5-kDa peptide that located C-terminal to TM segment 6. The data suggests an overlap in the binding domain of paclitaxel with vinblastine, verapamil and azidopine (Safa 2004) that includes amino-acid residues in TM segments 4 and 6.
It appears that three investigations using photo-active analogues of paclitaxel label P-gp at three distinct sites. A possible interpretation is that all the three regions comprise the paclitaxel-binding site and that they are proximal in the 3-D structure of P-gp. However, the orientation of the photo-active moiety on paclitaxel will dictate which of the regions is labelled. In addition, the photo-active moiety may be located on a highly flexible region of the probe and that regions distal to the “true” binding site are labelled (Glossmann et al. 1987).
P-gp was labelled with [125I]-IAAP, a photo-affinity analogue of the modulator prazosin (Dey et al. 1997) and subsequently subjected to proteolytic cleavage with trypsin to separate the two halves of P-gp. The ratio of [125I]-IAAP bound between the N-terminal and C-terminal halves of the P-gp molecule was 2:3. The effect of cis(Z)-flupentixol, an antipsychotic modulator of P-gp, on labelling of the two halves by [125I]-IAAP was also measured. There was an approximately 10-fold increase in photo-labelling of [125I]-IAAP at the C-terminal half of P-gp with no change at the N-site. This supported two separate “events” of drug interaction rather than simply two covalent attachment sites for [125I]-IAAP within a single site. Conceivably, the two photo-labelled sites are spatially distinct substrate interaction pockets that exist within a larger drug-binding domain. In addition, the concentrations of vinblastine and cyclosporin A required to inhibit [125I]-IAAP photo-labelling at the C-terminal site were increased 5–6-fold in the presence of cis(Z)-flupentixol. In contrast, the potency for reducing [125I]-IAAP labelling at the N-terminal site was unaffected by cis(Z)-flupentixol. The photo-labelling of the N-terminal site by [125I]-IAAP was also less susceptible to vanadate trapping of P-gp. This study supports the existence of at least two non-identical substrate interaction sites in P-gp, at least [125I]-IAAP (Dey et al. 1997).
Finally, these early attempts at locating the drug-binding sites suffered from an inability to generate precise and small fragments of P-gp and could thereby only proffer regions of interaction. However, collectively the data supported the hypotheses that P-gp contained a large binding domain comprising segments from multiple regions/helices.
Enhanced Detection of Photo-Affinity Labelled Regions of P-gp
Several novel photo-affinity ligands generated from propafenone were used to photo-label P-gp, aiming to identify the binding site for drugs (Ecker et al. 2002; Pleban 2004). Propafenones were previously shown to inhibit the transport of the P-gp substrates rhodamine 123, doxorubicin, vinblastine and mitoxantrone (Chiba et al. 1995; Ecker et al. 1996). In addition, a radioactive propafenone analogue [3H]-GPV51 was used to specifically photo-label plasma membrane vesicles expressing P-gp, confirming its direct interaction with the transporter. Following photo-labelling with [3H]-GPV51, membrane proteins were separated by SDS–PAGE and visualised by silver staining. The core-glycosylated 140 kDa P-gp band was excised and proteolytically degraded by chymotrypsin “in-gel”. The fragments were eluted from the gel and the ligand-labelled peptides identified with matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry (Ecker et al. 2002). This procedure gave considerable advantages to the earlier studies involving antibody precipitation. First, the exact peptide and amino-acid residues could be identified and second, the coverage of peptide fragments was ~90 % in the TMDs and ~80 % for the NBDs. The mass-spectrometry data identified significant [3H]-GPV51 labelling in TM helices 3, 5, 8 and 11, which, like the antibody studies, suggests the binding site comprises multiple protein segments.
Another investigation by the team (Pleban 2004) made use of the emerging structures of ABC proteins, in particular, the MsbA lipid transporter from Vibrio cholera (Chang 2003). The MsbA structure was used as a template to produce a homology model for P-gp. The homology model predicts that the helices labelled by [3H]-GPV51 lie at the interfacial regions between the two transmembrane domains of P-gp. One interface comprises TM3/11 and the other TM5/8. Although this homology model was eventually proven inappropriate due to retraction of the original structure, the interfacial location of the helices remains a valid assumption. The involvement of these helices has been borne out with models of P-gp based on the structure of Sav1866, mouse P-gp and a C. elegans homologue of P-gp.
An interpretation of this key finding was that the interfaces (TM3/11 and TM5/8) operate as a “gate” for substrates to enter the large binding domain in the central cavity (Pleban 2004; Crowley and Callaghan 2010). This interpretation is in broad agreement with results from double-cysteine mutant cross-linking studies (Loo and Clarke 2005). The findings are also compatible with both simultaneous binding of drugs on P-gp. Moreover, the gate model implies that any drug will bind sequentially at the interface and then within the central binding domain. However, there is no evidence from equilibrium binding studies for a two-component binding isotherm to describe drug interaction with P-gp. Potentially, the interfaces are not gates for entry to the cavity, but may exist as allosteric modulatory sites on the protein.
Multiple Fluorescence-Based Strategies to Locate Drug-Binding Sites
Spectroscopic techniques provide a key experimental tool for molecular biochemists and reveal an astonishing amount of information on protein structure and function. Fluorescence spectroscopy is widely used due to its general accessibility and the array of compounds and applications available. The technique has been widely used to facilitate our understanding of the molecular mechanism of P-gp. The field has taken advantage of the number of fluorescent substrates of P-gp, the presence of intrinsic tryptophan residues in key locations and the ability to attach fluorophores to target regions of the protein. Fluorescence spectroscopy has provided modest advances in our search to identify the location of binding sites on P-gp, but it has generated a wealth of functional data. In particular, the approach has informed on the nature of bioenergetic coupling in P-gp and the conformational changes that underlie its complex mechanism of drug translocation.
Fluorescence Quenching of Covalently Attached Probes to Characterise Drug Binding
The team headed by Frances Sharom developed a fluorescence quenching technique to investigate the direct binding of substrates and inhibitors of P-gp. One of these fluorescence techniques involves the use of the sulphydryl-reactive fluorescent probe, 2-4′-maleimidylanilino-naphthalene-6-sulphonic acid (MIANS) for site-directed labelling (Liu and Sharom 1996, 1997; Qu and Sharom 2001). In particular, MIANS has been shown to preferentially form a covalent attachment to P-gp at the cysteine residue located in the Walker A motif of each NBD. MIANS attached at the NBDs display a saturable quenching of its fluorescence upon binding of ATP, adenosine diphosphate (ADP) or its non-hydrolysable analogues. The binding of the nucleotide likely affects the local environment of the MIANS group which is located close to the ATP-binding sites in the NBDs. This results in the fluorescence quenching observed as a decrease in the emission intensity and/or the maximum emission wavelength. This approach has demonstrated the binding affinity of ATP/ADP, the co-operativity between NBDs, the ability of P-gp to bind two ATP molecules and the kinetics of ATP hydrolysis (Liu and Sharom 1997; Qu and Sharom 2001).
Quenching of MIANS fluorescence has also been demonstrated in response to the binding of P-gp substrates and modulators. The quenching profile follows a saturable and hyperbolic relationship with drug concentration, which enables an indirect estimation of an affinity for the interaction with drug at the TMDs (Liu and Sharom 1996). It is important to note that the affinity comprises two events; initial drug binding to P-gp and a component consisting of the conformational change required to alter the fluorescent signal. Therefore, the affinity from such measurements may only be considered an apparent KD value. Nonetheless, the apparent KD for MIANS quenching has been determined for over 80 structurally different P-gp drugs with values ranging from 25 nM to 260 µM, demonstrating that P-gp differentially binds substrates with an affinity over four orders of magnitude (Sharom 1997).
This long-range crosstalk between the TMDs where drugs bind, and the NBDs, where the MIANS is attached, demonstrates coupling between the two domains (Liu and Sharom 1996). Moreover, this inter-domain coupling, and subsequent conformational change, is a critical step in the ability of substrates to stimulate ATPase activity. In addition, the presence of bound drug reduces the rate and extent of MIANS labelling of P-gp, which is further proof of the specific conformation changes elicited in the NBDs. Numerous investigations have demonstrated communication in the opposite direction and the process of drug translocation is considered to be driven by the steps involved in ATP hydrolysis. Clearly, this communication occurs in both directions and is consistent with a high level of coupling.
Fluorescence of Intrinsic Tryptophan Residues to Characterise Drug Binding
Another approach to investigate binding makes use of the intrinsic fluorescence of P-gp afforded by its tryptophan residues. Both human and hamster P-gp isoforms contain 11 intrinsic tryptophan residues that are distributed throughout the protein. In addition, the tryptophan residues are highly conserved across the P-gp family and an involvement in substrate interaction and recognition has been suggested given the hydrophobic nature of substrates (Liu et al. 2000). Hydrophobic substrates such as vinblastine or rhodamine 123 contain multiple aromatic rings and these undertake π–π stacking interactions with the side chains of aromatic-rich TM helices. These stacking interactions with tryptophan residues will alter the spectral properties of the latter and may provide evidence for binding of drug to the protein (Fig. 2). Interestingly, there are several tryptophan residues (and other aromatic residues) in TM3/11 and TM5/8, located at the TMD/TMD interface (Pawagi et al. 1994). As described in Sect. “Enhanced Detection of Photo-Affinity Labelled Regions of P-gp”, these helices mediate the binding of several substrates/modulators of P-gp. Consequently, drug binding to P-gp in these helices may directly alter tryptophan fluorescence, rendering the assay a useful reporter for binding.
Using intrinsic tryptophan fluorescence to identify substrate-binding sites. a The molecular dynamic (MD) equilibrated P-gp model indicating the 11 intrinsic tryptophan residues. Quenching (decrease in intensity) of the fluorescent spectrum profile of P-gp in the presence of particular substrates can indicate drug binding to the protein. Binding results in a conformational change which likely alters the environment of the tryptophan residues changing the fluorescent profile. b A representative tryptophan fluorescence spectrum profile of intensity as a function of P-gp substrate concentration
One of the advantages of fluorescence spectroscopy is the intimate relationship between the fluorescent profile and its local physical and chemical environment. Consequently, tryptophan fluorescence kinetics may reveal whether the residue is subject to any steric hindrance to its motion and report changes in the polarity of the local environment. Measurement of fluorescence lifetime for the intrinsic tryptophan residues revealed the presence of two components (Lugo and Sharom 2009). The fast component is solvent exposed (i.e. aqueous) and likely to be located in exposed cytoplasmic loops. The location of this fast component in an aqueous environment was demonstrated by the use of various hydrophobic and hydrophilic quenching agents (e.g. acrylamide, iodide, benzene) to gauge their relative effects on fluorescence (Liu et al. 2000; Lugo and Sharom 2005). The quenching profile and the emission maximum for fluorescence suggested that the tryptophan residues located in a polar environment did not appear to contribute to the observed fluorescence. This may suggest that the tryptophans residing in an aqueous region are located within a very tightly folded domain of the protein, or that they are internally quenched (Liu et al. 2000).
The slow component of the tryptophan fluorescence decay is localised within the lipid membrane; again verified with the use of a number of chemical quenchers. In addition, the fluorescent drug LDS-751 affects the slow component of tryptophan fluorescence decay, which is achieved due to either direct intercalation with the amino-acid or via allosteric conformational changes.
Fluorescent P-gp Substrates and the Kinetics of Transport
P-gp mediated transport of the fluorescent substrate Hoechst 33342 was measured in plasma membrane vesicles isolated from Chinese hamster ovary cells (CHRB30). When sequestered in the lipid environment of the membrane, the fluorescence of Hoechst 33342 is enhanced and then lost upon moving into aqueous solution. Transport by P-gp in the inside-out plasma membrane vesicles was initiated following addition of Mg·ATP and observed as a reduction in fluorescence (Shapiro and Ling 1995). This assay system enables continuous fluorescent monitoring, thereby allowing accurate measurement of transport rates. The kinetics of Hoechst 33342 transport was measured in the presence, or absence, of another fluorescent P-gp substrate, rhodamine 123 (Shapiro and Ling 1997b). Conversely, the transport of rhodamine 123 was also measured in the P-gp-rich vesicles in the presence or absence of Hoechst 33342. Each substrate was able to stimulate the transport of the other, which is not possible if they interacted at a single binding site; such an interaction would have resulted in one substrate competitively inhibiting transport of the other. Both dyes were transported simultaneously and the stimulatory effect could not be attributed to the presence of one site for transport and the other for allosteric regulation. The positively co-operative manner in which each substrate stimulated the transport of the other is best explained by the presence of at least two transport-competent drug-binding sites on P-gp.
This is further supported by the observation that colchicine and quercetin stimulated the transport of rhodamine 123, but inhibited the transport of Hoechst 33342 (Shapiro and Ling 1997a). In contrast, the anthracyclines doxorubicin and daunorubicin inhibited rhodamine 123 transport, but stimulated Hoechst 33342 transport. The two sites were designated the H-site which preferentially binds and transports Hoechst 33342, quercetin and colchicine, and the R-site, which is the preferential binding site for rhodamine 123 and the anthracyclines. Overlapping specificity of these two distinct sites has been suggested for some drugs, given that vinblastine, actinomycin D and etoposide were all able to inhibit the transport of both dyes (Shapiro and Ling 1997a, b).
A flow-through system was used to measure daunorubicin efflux or accumulation from P-gp expressing cells and represented one of the only documented reports showing that the transporter works against a considerable concentration gradient (Lankelma et al. 1990). In a subsequent investigation, the interaction between the transported substrate daunorubicin and the modulator verapamil was examined (Spoelstra et al. 1994). At low concentrations of daunorubicin, verapamil caused a non-competitive inhibition of its transport; however, at high substrate concentration this inhibition became competitive. This curious finding can be explained with a model of P-gp wherein daunorubicin may bind at two pharmacological sites with different affinities. At low daunorubicin concentrations it bound at the high affinity site and the effects of verapamil on transport kinetics occurred through an allosterically linked process (i.e. non-competitive). However, daunorubicin binds to both the high and low affinity sites on P-gp at high concentrations. The low affinity site is also the location for verapamil binding. Consequently, under these conditions the verapamil effect on daunorubicin transport was partly due to a competitive interaction. Similar observations have been described for the interaction between anthracyclines and the transport of Hoechst 33342 (Shapiro and Ling 1997b). At lower concentrations, the anthracyclines bound at the R-site, which is linked to the H-site (Hoechst 33342) by a positive heterotropic allostery. When administered at higher concentrations, the anthracyclines also competed for binding with Hoechst 33342 at the H-site; thereby inhibiting its transport.
Further studies with the fluorescent substrates and modulators discovered the existence of a third binding site on P-gp (Shapiro et al. 1999). This was termed the P-site by virtue of the fact that prazosin and progesterone both interacted at this pharmacological location. Previous investigations had suggested that although progesterone was capable of modulating the transport function of P-gp, it was unlikely to be a substrate (Ueda et al. 1992). Prazosin and its analogues had been previously demonstrated to bind directly to P-gp, stimulate its rate of ATP hydrolysis, and alter substrate transport rates; however, it was classified as a weak transport substrate, or a modulator (Greenberger 1993). Consequently, it was suggested that the P-site acts primarily as an allosteric site and that compounds bound to it are not transported. In support of this, it was shown that drug binding to the P- and R-sites stimulated the transport of Hoechst 33342. In contrast, binding to the P-site alone stimulates the transport of either Hoechst 33342 or rhodamine 123. Finally, binding of drug to both the H- and P-sites appears to stimulate rhodamine 123 transport, although the effect is lower than that produced by binding to the sites individually. However, later studies using BODIPY-FL-prazosin indicated that this fluorescent derivative was a substrate for P-gp mediated transport (Gribar et al. 2000). Consequently, the P-site should be reclassified to a transport-competent one, although non-transported modulators (e.g. progesterone) were also capable of interaction (Shapiro et al. 1999).
These kinetic investigations with fluorescent substrates not only defined the existence of multiple substrate transport sites, but they also revealed complex allosteric interactions and the possibility that drugs may bind at more than one site on P-gp. Multiple factors such as this can synergise to produce the renowned substrate promiscuity of P-gp.
Fluorescence Resonance Energy Transfer to Characterise Drug Binding
Fluorescence resonance energy transfer (FRET), which is the transfer of energy from a fluorescent donor to the excited state of an acceptor fluorophore, depends on the distance between the two and only occurs if separated by 10–75 Å. This enables the technique to be used as a ‘spectroscopic ruler’ measuring intra- and inter-molecular distance.
The fluorescent probe 4-Chloro-7-Nitrobenz-2-Oxa-1,3-Diazole (NBD-Cl) was inferred to covalently attach to Cys428 and Cys1071, both of which are in the catalytic site of P-gp (Qu and Sharom 2001, 2002). Labelling at these two sites facilitates the examination of inter-domain communication in P-gp and the use of FRET to define intra-molecular distances. The fluorescent substrate H33342 bound to P-gp and exhibited an increase in fluorescence intensity, coupled with a large blue shift in its emission maximum (i.e. lower λmax), indicating a non-polar environment. The spectral properties of H33342 render it a FRET donor to NBD-Cl and this was demonstrated as a marked reduction in its emission intensity. The degree of FRET was used to estimate a distance of 38 Å between the bound H33342 and the NBD-Cl bound at the catalytic site of P-gp (Qu and Sharom 2002). H33342 binds to the “H-site” of P-gp (Shapiro and Ling 1997b) (Sect. “Fluorescent P-gp Substrates and the Kinetics of Transport”), and the FRET data also indicates that this site is found 10–14 Å below the external surface of the membrane, within the cytoplasmic leaflet (Qu and Sharom 2002).
A FRET strategy was also used to identify a broad location for the “R-site” of drug binding on P-gp (Shapiro and Ling 1998). The ligand for the “R-site” was LDS-751 and it displayed a marked increase in the fluorescence intensity on binding to P-gp, indicative of a hydrophobic environment. LDS-751 was chosen as the “R-site” ligand since it is a FRET partner for the intrinsic tryptophan residues and it caused a 40 % reduction in the fluorescence of the latter. FRET was proposed to occur with the cytosolic, solvent exposed tryptophan residues and an approximate location for the “R-site” was also placed in the cytoplasmic leaflet of the membrane. That the location of both the “R- and S-sites” is found in the cytoplasmic leaflet of the bilayer was used to support a role for P-gp as a flippase that translocates lipids between leaflets of the bilayer.
Although not a FRET interaction, there was also a spectral interaction between H33342 and LDS-751 upon binding to P-gp, which again demonstrates that the protein is capable of binding more than one substrate or modulator simultaneously. This observation is in agreement with the photo-affinity and mutagenesis data presented in previous sections. It also highlights the utility of biochemical approaches and how molecular understanding can be reached with a combination of approaches. Moreover, the investigations with fluorescence spectroscopy have provided not only information on the location of binding sites, but the properties and characteristics of them.
Can Structural Information Locate Drug-Binding Sites?
The preceding sections have detailed how our understanding of the nature of drug binding to P-gp, and the location of this binding, has improved in the last three decades. The availability of structural data would boost our understanding further and reconcile some of the contradictory observations from biochemical and biophysical studies. In order to reveal the sites of drug binding, this structural data will need to be obtained in the presence of bound substrate. Furthermore, provision of data to fully describe the mechanism of drug translocation will require structural information in multiple conformations such as nucleotide-free, nucleotide-bound, post-hydrolytic and post-dissociation of drug. In order to reveal the chemical nature of drug interaction with the protein (i.e. hydrogen bonding versus π–π stacking) we will need structural data at high resolution; near or below the 2 Å level.
Overview: Structure of P-gp
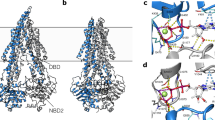
The availability of purification procedures that provided high yields of fully functional P-gp begun to surface in the mid-1990s, although they used classical multiple step chromatographic methods (Liu and Sharom 1996; Callaghan et al. 1997). The use of affinity tags (i.e. polyhistidine) from the late 1990s improved the quality and purity of the protein, and the ease of its purification (Lerner-Marmarosh et al. 1999; Taylor et al. 2001; Loo and Clarke 1995b). The first attempts at obtaining structural data for P-gp also begun in earnest soon after these milestones. Initial attempts used electron microscopy (EM) with single particle analysis (SPA) and eventually progressed to 2-D crystals in a membrane environment. It was not until 2009 that the first structure of P-gp (mouse) using an X-ray crystallography approach was published (Aller et al. 2009), following considerable effort by multiple teams in the field. The structure (3.8 Å resolution) continues to generate debate amongst researchers and has now undergone rounds of refinement and reinterpretation. Another structure (Jin et al. 2012), of a P-gp homologue in C. elegans, has been obtained to higher resolution (3.4 Å), however it was in an identical conformation and generated in the absence of bound substrate/modulator. Progress has been steady and the information has illuminated our understanding of drug binding to the protein. However, like most scientific endeavours, it has raised several new questions.
The First Structure of P-gp: Electron Microscopy
The first structure of any full-length ABC protein was obtained in 1997 (Rosenberg et al. 1997) using electron microscopy-single-particle analysis (EM-SPA) of hamster P-gp purified from drug selected CHRB30 cells (Callaghan et al. 1997). The structure was obtained to 2.5 nm (25 Å) resolution and the SPA technique was done with P-gp in detergent micelles. The resolution of EM-SPA will not provide high resolution for a protein of this size and was intended merely to generate a preliminary view of the gross organisation and dimensions of P-gp. The protein displayed a cylindrical geometry with a diameter of 10 nm and a height of 8 nm. Given that the average height of a bilayer is ~4 nm, the authors posited that half of the structure in the “vertical” plane was found within the membrane. Based on the dimensions of other proteins and the size of membrane spanning helices, it appeared that the TMD of P-gp was somewhat loosely packed. Perhaps the standout feature, and at the time the most controversial one, was the central pore of diameter ~5 nm. Based on the staining properties of the particles it was suggested that the pore was aqueous. Lectin-gold labelling of the extensively glycosylated hamster P-gp isoform was used to orientate the particles and thereby identify the NBDs. The central “aqueous” pore of the P-gp particles was suggested to be open to the extracellular environment. The final observation from this structure was that the TMDs also contained a discontinuity (or gap) within the plane of the membrane that may provide access for substrates to the central cavity. The latter point was intentionally speculative, but it is in agreement with the photo-affinity/MALDI-TOF data proposing that P-gp contains gates for substrates (Ecker et al. 2002).
Gradual Evolution of the EM-Derived Structure of P-glycoprotein
The structural efforts with EM evolved to higher resolution over the next few years and the next data was obtained for 2-D crystals of P-gp, reconstituted into membranes (Rosenberg et al. 2001). The key feature of this manuscript was the presentation of data for P-gp trapped in multiple conformations; namely AMP-PNP bound (i.e. nucleotide bound) and ADP.Vi trapped (i.e. post-hydrolytic). Comparison of the TMD regions revealed extensive conformational transitions during passage between the three catalytic states. This observation would be the spur for numerous pharmacology studies on the effects of events at the NBDs on drug binding. Moreover, it provided direct evidence that events at the NBDs instigate the major conformational transitions essential to the drug translocation process. A subsequent publication obtained resolution improvements and focussed on comparison between the nucleotide-free and -bound conformations (Rosenberg et al. 2003). The investigation provided confirmation of the central pore with dimensions of 5–6 nm in diameter and 5 nm depth. The central pore was open to the extracellular space and closure at the cytoplasmic face was produced by the NBDs. Significant structural transformation occurs in this region in the presence of nucleotide, and the TM helices rearrange to support a “helix rotation” motion. This results in the opening of the internal cavity along the length of the membrane, which may enable the entry of hydrophobic drugs to the putative drug-binding pocket.
The final instalment of EM-based structures for P-gp was resolved to approximately 8 Å for protein trapped in the nucleotide-bound state (Rosenberg et al. 2005). The structure was obtained from cryo-EM of 2-D crystals of P-gp to provide further resolution of the overall topography and packing of the transmembrane helices. The structure displayed a pseudo-2-fold symmetry within two blocks of six TM helices, showing the presence of the 12 putative membrane spanning helices and some evidence of inter-domain cross-over facilitated by the sixth helix in each domain. With the integration of available biochemical data, the authors speculated the location of numerous TM helices. However, due to the low resolution, definite identifications could not be assigned. Although only limited predictions of the location of helices were possible from the structure, the comparison between conformations of P-gp provided crucial information on the molecular mechanism of drug translocation.
Crystal Structure of a Nucleotide-Bound P-gp Homologue; Sav1866
As is often the case in structural biology, structures are obtained from some unlikely sources and often for uncharacterised proteins. In keeping with this theme, the first full-length structure generated using X-ray crystallography for an ABC export pump was obtained for the S. aureus exporter Sav1866 (Dawson and Locher 2006). Little was known about this pump prior to the structure and ensuing biochemical studies confirmed it was a multidrug efflux pump. The structure was obtained to 3.0 Å and contained 12 transmembrane helices, six from each of the monomers. Unlike the import proteins, there was considerable cross-over of TM helices between the two monomers in the structure. The structure was obtained in the absence of any bound substrate, however successful crystallisation was obtained in the presence of ADP. Consequently, the two NBDs were arranged in close apposition in the standard “sandwich dimer” configuration of the Walker A/B and signature motifs. This post-hydrolytic structure also contained a central aqueous cavity in an externally facing configuration.
The same research team published a subsequent structure for Sav1866 (3.4 Å) determined in the presence of AMP-PNP, a non-hydrolysable analogue of ATP (Dawson and Locher 2007). The latter structure was obtained to investigate the physiological relevance of the earlier structure, as in the ADP-bound state the dimerised half-transporter, adopted an outward-facing conformation (Dawson and Locher 2006). The apposition and arrangement of the NBDs was similar between the two structures and the structure obtained in the presence of AMP-PNP also showed an outward-facing conformation with the central cavity open to the extracellular surface of the membrane. The outward-facing configuration subsequent to nucleotide binding or hydrolysis is substantiated by considerable evidence with ABC proteins. However, the lack of difference between the AMP-PNP and ADP-bound states is more puzzling given the reported differences with P-gp (Rosenberg et al. 2001; Martin et al. 2001). It may transpire that only following dissociation of ADP will the protein adopt an inward-facing conformation, exposing a central cavity (binding site) to scan the inner leaflet of the lipid bilayer and bind substrates with high affinity (Higgins and Linton 2004; Smith et al. 2002). That bound ADP was sufficient for the transporter to adopt the outward-facing conformation was interpreted to suggest that the detergent solution, (in which the protein was obtained), shifted the conformational equilibrium of the protein and may not reflect in vivo conditions. The crystal structure provided a molecular basis for Sav1866 mechanism as it clearly showed a coupling between the ATP-bound state and drug release as a result of the outward conformation.
X-Ray Crystal Structure of Mouse P-gp
In 2009 the first X-ray crystal-based structure of a mammalian ABC transporter, namely mouse P-gp, was resolved to 3.8–4.4 Å (Aller et al. 2009). Mouse P-gp has 87 % sequence identity to human P-gp and from a pharmacological perspective is indistinguishable from the human isoform (Taylor et al. 1999). Three distinct structures were solved in the publication and included one in the absence of nucleotide/drug, and two structures in the absence of nucleotide, but in the presence of two cyclic hexapeptide stereoisomers. The presence of bound drug was greeted with enthusiasm for its potential to provide a molecular basis for substrate promiscuity of the transporter; however, the resolution remains short of atomic level detail.
The P-gp structure, obtained in the absence of both nucleotide and substrate or inhibitor, showed an inward-facing conformation resolved to 3.8 Å (PDB 3G5U). The model was generated using multiwavelength anomalous dispersion mapping. A striking feature, and one that has engendered considerable debate (Gottesman et al. 2009; Li et al. 2014), is the physical separation of the two NBDs by a distance of 30 Å. It remains unclear if this is the “native” configuration of empty P-gp, or whether it is an artefact of crystallisation. The latter is likely given that protein is in detergent micelles (i.e. relaxed) and the crystal unit–unit contact is conferred by the NBDs. Due to the large separation of NBDs, the two blocks of TMs were observed to adopt a conformation forming a large internal cavity. The presence of a central cavity is in agreement with previous structural and biochemical data, albeit with different dimensions. The characteristic “domain swapping” of helices was consistent with that first documented for Sav1866. Moreover, the central cavity is open to the cytoplasmic face of the membrane, but closed to the extracellular surface by TM helices. The volume of the pocket within the central cavity was estimated at 6000 Å3. This is an extraordinary dimension that renders it capable of facilitating the simultaneous binding of drugs at different regions within the cavity. This is evident by the molecular surface area of established P-gp substrates; vinblastine 154 Å2, paclitaxel 221 Å2, and doxorubicin 206 Å2. It was also suggested that two pairs of TM helices may form putative portals in the inner leaflet of the membrane to enable substrate passage from the lipid bilayer into the central pore. Furthermore, solvent accessible amino-acid residues lining the internal cavity were investigated and showed the majority of the residues contributing to binding of the cyclic peptides are hydrophobic and aromatic (58 of the 73 solvent accessible residues). Within the drug-binding pocket, it was found that hydrophobic and aromatic residues are present at the upper (extracellular) region, whereas polar and charged residues are more frequent in the lower or cytoplasmic region. This supports data obtained for the topology of TM6/12, both of which contribute to cavity lining (Crowley et al. 2010b; Rothnie et al. 2004; Storm et al. 2008).
Unfortunately, it appears that the mouse P-gp could not co-crystallise in the presence of any of the well-characterised arrays of substrates and modulators. The authors did have success in co-crystallising P-gp with two stereoisomeric cyclic peptide inhibitors, cyclic-tris-(R)-valineselenazole (QZ59-RRR) and cyclic-tris-(S)-valineselenazole (QZ59-SSS). This was the first report using these two uncharacterised compounds that appear to act as inhibitors, or modulators, of P-gp. It is unclear if they are substrates for transport and whether their binding overlaps with any of the established substrates/modulators. Co-crystallised P-gp adopted the expected inward-facing conformation and the stereoisomers surprisingly appeared to bind at distinct sites. QZ59-RRR was observed to bind in the “centre of the transporter” and may interact with TMs 1, 5, 6, 7, 11 and 12. Conversely, QZ59-SSS simultaneously binds at two distinct sites at the “upper” and “lower” regions of the central cavity facilitated by TMs 1, 2, 6, 7, 11 and 12. A portion of QZ59-SSS at the upper site was observed in a disordered state and there is no available explanation for this. At the resolution obtained for the structure, assigning directly interacting residues or the points of interaction between peptide and drug remains speculative. There was high structural similarity of crystallised P-gp in the absence of inhibitor, co-crystallised with QZ59-RRR or co-crystallised with two molecules of QZ59-SSS. This is in conflict with the proposed induced fit mechanism for substrate transport by P-gp, which would cause conformational changes upon drug binding. Biochemical data will be essential to ascertain stoichiometry of binding and to elucidate which residues directly interact with the cyclic peptides and more established substrates and inhibitors.
In 2013, three crystal structures of mouse P-gp were published (Ward et al. 2013) (see Sect. “Crystal Structure of a P-gp Homologue from C. elegans”) in a follow-up study, and in response to the structure obtained from C. elegans (Jin et al. 2012). All the structures were in the inward-facing conformation and obtained in the absence of nucleotide. Two of the structures were crystallised in the absence of inhibitors, however one was obtained in the presence of a nanobody bound to the C-terminal side of NDB1, preventing the dimerisation of the NBDs and thereby inhibiting ATP hydrolysis of the protein. The three structures presented different separation distances between the NBDs, which was suggested as proof of conformational flexibility of the inward-facing conformation of P-gp. In particular, two crystal structures (in the absence of inhibitor or drug) diffracted to 3.8 and 4.0 Å, corresponding to NBD separations of 31 and 36 Å, respectively; which is different to the original structure by the consortium. The final structure was co-crystallisation with a nanobody inhibitor Nb592 to a resolution of 4.1 Å and an NBD separation of 30 Å. It was suggested that the observed conformational changes to the presence of a “TM-hinge region” formed by extracellular loops between the TM helices. The conformational flexibility of the inward-facing conformation, most drastically noticed in the cytosolic domains, is therefore a consequence of small movement at this pivot point. Another highly plausible interpretation is that the use of detergent micelles enables considerable conformational freedom compared to the more rigid environment of a phospholipid bilayer. In fact, the literature is replete with biochemical studies demonstrating the difference in activity and functional properties of P-gp between lipid- and detergent-based milieu.
Crystal Structure of a P-gp Homologue from C. Elegans
Shortly following the publication of the mouse P-gp crystal structure, the X-ray crystallographic derived structure of a P-gp homologue from Caenorhabditis elegans was resolved to 3.4 Å (PDB 4KSB) (Jin et al. 2012). The P-gp structure from C. elegans was obtained in the absence of nucleotide or drug, and with the transporter in an inward-facing conformation. The substrate profile and amino-acid sequence vary considerably between C. elegans and mammalian P-gp, with only 46 % identity to the murine isoform. However, the authors argue that that structural information from the C. elegans isoform can provide mechanistic insight into the function of the protein.
The C. elegans version of the structure of P-gp displayed separation between the NDBs of 53 Å, which is considerably larger than that observed in the crystal structures of mouse P-gp (Aller et al. 2009; Ward et al. 2013). Another difference was that the structure contained only one portal formed by the TMs and a discontinuity in the helical structure of TM10/12, which line the portal. The greater preponderance of disordered loops in this region was thought to increase the number of contact points, or interactions, between drugs and amino-acid residue.
The interactions between TMDs and NBDs are observed in this structure with four intracellular helices (IH1–IH4), providing a domain interface that may be involved in communication. To further support the significance of this interface, the sequence identity between amino-acid residues in this region for C. elegans and human P-gp was high. Additionally, a highly conserved collection of amino acids in IH1, IH4 and NBD1 were proposed to form a salt bridge at the TMD–NBD1 interface. A similar structure was observed in the TMD–NBD2 interface, with conserved residues in IH2, IH3 and NDB2.
The crystal structure of C. elegans P-gp, and of the murine isoforms, has provided a wealth of information on the functional organisation of the transporter, tantalising glimpses into the drug-binding domain/sites and testable hypotheses regarding the inter-domain communication that transduces drug binding to provision of energy.
Bioinformatics Approaches to Reveal Drug-Binding Sites
The generation of a high-resolution protein structure should not be considered the closure of a research field; it is more often the instigator of considerable further investigation. The structures of helices, sheets and loops, that we view, are actually models built from the primary data—i.e. X-ray diffraction patterns. Advances in bioinformatics, in particular protein molecular modelling, enable rigorous interpretation of the structures, their stability and facilitates their refinement. Currently, we do not have structural data of sufficient resolution to unequivocally determine which residues are involved in mediating drug–P-gp contact. Molecular modelling, drug docking and further biochemical data, will build on the solid foundation of structural data to provide this. The following sections describe how bioinformatics and biophysical approaches have begun to unravel the mysteries of drug binding to P-gp.
Human P-gp Homology Modelling
Following the release of Sav1866 X-ray structure in 2006, O’Mara and Tieleman investigated the suitability of this template for homology modelling. Homology models of P-gp were generated to represent different stages in the transport cycle; namely, closed and open (O’Mara and Tieleman 2007). The ADP-bound Sav1866 structure was used to render the closed state (i.e. nucleotide bound). To produce a model in the absence of nucleotide (i.e. open), insights were used from the importer protein BtuCD and the NBDs from the maltose importer (i.e. MalK subunits). The open state model of P-gp was verified for orientation and energy minimisation. Significant conformational changes were observed in the TMDs that altered the geometry and residue distribution of the central cavity. The authors reported a rotation of the TM helices to re-orientate polar residues lining the central cavity in the closed state to inter-helical regions, thereby exposing hydrophobic residues. This supports previous data of the closed state representing a low affinity extrusion pore that becomes a high affinity binding site for recognition and transport of substrates (Dawson and Locher 2006).
Another study, by Ravna et al. also investigated the use of Sav1866 as a template for the structure of P-gp (Ravna et al. 2007). This model strongly supported biochemical data that attributed specific TM helices to drug binding since the orientation and nature of the residues lining the central cavity would support the interaction of hydrophobic substrates. The authors reported that although the sequence identity between Sav1866 and P-gp was only 31 %, the conserved secondary structures and overall geometry between the two proteins suggest that the Sav1866 may be a realistic template for the structure.
These fledgling attempts at providing molecular models of P-gp structure were used in a number of publications to facilitate reconciliation of biochemical data from a structural perspective (Crowley et al. 2009, 2010b; Storm et al. 2007, 2008). This strategy was vital in not only interpretation of results, but it provided a focus or direction for subsequent mutagenesis studies, and therein lies the true value of the technique. Subsequently, a molecular model of human P-gp was generated from the murine structure and the effects of ATP and Mg2+ binding were investigated (O’Mara and Mark 2014). The inclusion of Mg·ATP was associated with a highly stable structure of P-gp and confirmed the asymmetry of NBD organisation. The model was subsequently used with electron paramagnetic resonance spectroscopy to demonstrate alternating access of TM6/12 during the translocation process (van Wonderen et al. 2014). Recently, molecular dynamic simulations have been used with the model to propose sites for binding and translocation of morphine and nicardipine (Subramanian et al. 2015).
Refinement of the Murine Crystal Structure of P-gp
Figure 3 presents a comparison of the NBD separation in mouse (3G5U, 4KSC and 4M1 M) and C. elegans (4F4C) structures with the distances between the Cα atoms of the last resolved residue in the C-terminus of both NBD1 and NBD2 shown. Clearly, the separation between these catalytic sites is highly variable between the four P-gp structures and the incongruity of this value may highlight the significant conformational flexibility of the protein.
Separation of nucleotide-binding domains for mouse (3G5U, 4KSB and 4M1M) and C. elegans (4F4C) P-gp crystal structures. The distance between the final residue in the C-terminus of each NBD was measured and denoted by grey residues connected by a grey line (where visible). For mouse P-gp structures the separation was measured between residues 626 and 1271, while the distance of separation for C. elegans was measured between residues 659 and 1250. Mouse P-gp 3G5U (purple) shows a separation of 14.90 Å while other mouse structures 4KSB (blue) and 4M1M (green) show a separation of 31.06 and 16.56 Å, respectively. The distance measured between the NBDs in the C. elegans structure, 4F4C (orange) was 34.27 Å
Comparison of the structure from murine and C. elegans P-gp revealed considerable differences, most notably possible registry shifts. Consequently, in 2014, Li et al. published a refined model (4M1M) of the murine P-gp structure (Li et al. 2014) as an update to the original structure (Aller et al. 2009). The refined structure was generated using single-wavelength anomalous dispersion (SAD) phasing of the original mouse Pgp structural data. The initial dataset was produced using multi-wavelength anomalous dispersion (MAD) phasing, which has largely been superseded by SAD (Li et al. 2014). To refine the original model, SAD phasing was conducted on a previously collected dataset free of radiation damage. Registry shift corrections were made to the TM helices forming the central cavity, intracellular helices and elbow helices, amongst other corrections. The refinements resulted in a significant increase in the percentage of residues present in the favourable Ramachandran region (95 % in the refined model compared to 56 % in the original structure). The SAD-phased map showed improvement in many criteria and revealed features of the protein that were not resolved in the original model. Finally, the figure of merit (FOM) value for the refined structure improved from a MAD score classified as unacceptable to a SAD score that is.
The refined structural model for murine P-gp identified nine aromatic residues within the TMDs that are also present in human P-gp, but not in the C. elegans isoform (Jin et al. 2012; Li et al. 2014). The authors speculated that evolutionary differences in residue composition are indicative of different functions and account for the significant substrate promiscuity observed in mammalian P-gp isoforms. Significant changes in the position of TM4 were observed in the refined structure, and this helix is implicated in formation of the portal for drug entry. The improved murine structure is a superior template to generate new homology models in order to investigate the molecular basis for polyspecific drug recognition. The continued travails with the structure of P-gp provide considerable evidence for stringent and rigorous assessment of data, moreover, investigators should ensure equal quantities of care and of course, patience, in the fact that structural biology is a long journey.
Portals and Drug Accessibility to the Central Cavity
Structural data has consistently shown the presence of portal(s) in the TMD formed by pairs of TM helices. The topography of these pairs is such that substrates present in the lipid bilayer can enter the binding pocket through the interface between the two TMDs. Extensive biochemical data has suggested gates are formed by the interaction of one TM helix in one TMD with another TM helix in the second TMD. The X-ray structures of mouse P-gp have shown the presence of two portals formed by TMs 4 and 6 in TMD1 and TMs 10 and 12 in TMD2 (Aller et al. 2009; Ward et al. 2013). Only one portal capable of acting as a drug entry site was observed in the C. elegans structure, with the other portal sterically hindered by an N-terminal helical hairpin (Jin et al. 2012).
Do these portals merely provide an open entry point to the central cavity for drugs? Are the portals simply sites of interactions for modulators to regulate the translocation process? If the portals represent an initial binding site, then why do pharmacological studies on drug binding display a single association step? Structural investigations with P-gp have revealed information regarding the location of binding sites, the presence of a central cavity, and the identification of putative substrate portals. However, many fundamental questions regarding the drug-binding site remains, and several new questions posed in this paragraph have emerged. Only through an iterative process that combines biochemical, structural and computational studies will we finally reveal this enigmatic process.
Describing the Nature of Drug-Binding Sites
In addition to identifying their location, the nature and biophysical properties of the drug-binding sites have been studied extensively. Describing the nature of the drug–protein interactions will also shed light on the pharmacophoric features essential for high affinity binding, and reveal differences between substrates and modulators. Such essential fundamental information may also translate into improved design of anticancer drugs to avoid the clutches of P-gp, or inhibitors to halt its effect on chemotherapy efficacy. Finally, understanding how the binding sites alter their interaction with drug in multiple conformations of the protein will facilitate a molecular description of multidrug recognition and translocation.
Substrate Recognition
From the earliest investigations, the ability of P-gp to interact with an astonishing number of compounds was a “standout feature”. P-gp is one member of the triad of multidrug efflux pumps that prevent cytotoxic drug accumulation in cancer and regulate pharmacokinetic profiles in healthy individuals. However, the three pumps have distinct substrate profiles, albeit with some inevitable overlap. Clearly, there must be chemical–physical features of a drug that render it as a substrate or modulator of P-gp; in other words, the elusive pharmacophore. The most widely observed features are the hydrophobicity of substrates, a planar aromatic ring system and a cationic nitrogen moiety. A number of investigations have attempted to generate a detailed list of pharmacophoric features (Stouch and Gudmundsson 2002; Montanari and Ecker 2015; Ueda et al. 1997; Demel et al. 2008; Orlowski and Garrigos 1999); although to date, a widely accepted and definitive set is unavailable.
Hydrogen bond formation has oft-been considered a staple requirement for high affinity binding and to maintain selective drug interaction with enzymes, receptors and transporters. With this established dogma, Anna Seelig analysed a large number of compounds known to interact with P-gp and characterised the number and location of functional groups capable of participating in hydrogen bonding (Seelig 1998). The author described three chemical units based on a fixed spatial separation of electron donor groups, and possessing any one of these features would classify a molecule as a P-gp substrate. The three chemical units were categorised as possessing a spatial separation of: (1) 4.6 ± 0.6 Å between two electron donor groups, (2) 2.5 ± 0.3 Å between two electron donor groups, or, (3) 4.6 ± 0.6 Å between three electron donor groups. Binding strength (i.e. affinity) of the compound, positively increased with the number of hydrogen donor features it exhibited. A review of structure––activity relationships (SAR) (Stouch and Gudmundsson 2002) concluded that the reliance on hydrophobicity may be an over-simplification of the drug–P-gp interaction and may simply reflect the degree of membrane intercalation. Of course, this property is a key element of the hydrophobic vacuum cleaner models of P-gp (Homolya et al. 1993; Raviv et al. 1990), which demonstrate that drugs are “extracted” from the lipid milieu. This review also suggested that the examination of hydrogen bond donor/acceptor moieties on substrates gave the most successful determinants of binding. Moreover, they stated that the presence of multiple binding sites would cloud the issue and experimental discrimination between them was an essential step; unfortunately, this has not yet been possible to achieve experimentally. Another article (Raub, 2005) also indicated that the experimental tools were not yet available to discriminate between sites and that considerably greater sophistication in SAR investigations was required. In particular, strategies to improve our understanding of the structural determinants for P-gp binding/transport will require “…reducing molecular size, replacing electronegative atoms, blocking or masking H-bond donors with N-alkylation or bulky flanking groups, introducing constrained conformation, or by promoting intramolecular hydrogen bonds…”. These are indeed important investigative strategies. However, the current pessimism by funding councils and the pharmaceutical industry towards generating P-gp inhibitors may render this goal unattainable.
Due to the vast number of structurally different compounds P-gp can recognise, it is widely accepted that the protein–drug interactions are intricate and complex. A general pharmacophore model of P-gp drugs was hypothesised from a genetic algorithm similarity program (GASP) and relates to the verapamil-binding site. GASP analysis was used to examine a highly diverse set of compounds from a number of drug classifications, to derive a general pharmacophore for P-gp substrates (Pajeva and Wiese 2002). Multiple pharmacophore elements were proposed including: two hydrophobic regions, three hydrogen bond acceptors and one hydrogen bond donor. Interaction points for individual drugs were analysed and compared with the wide variations in specific interactions attributed to different binding modes for the compounds within the defined verapamil site. Furthermore, the authors found the binding affinity of P-gp substrates and inhibitors was influenced by the number of pharmacophore points simultaneously involved in the interaction. The verapamil-binding site contains multiple points for hydrophobic and hydrogen bond interactions, and different drugs interact with a specific subset of these points.
Another study that focussed on understanding how P-gp recognises such chemically and structurally distinct compounds, utilised a combination of biochemical (primarily ATPase assays with multiple drugs) and molecular modelling techniques (Garrigues et al. 2002). The investigation revealed that, for the compounds tested, there were two regions for drug interaction on P-gp. On one site, drugs including cyclosporin A, verapamil, actinomycin D and peptides would compete for binding. The alternate site was exclusively for vinblastine; although it must be remembered that drugs not tested in this investigation may interact at this site. This study also concluded that the binding region of P-gp contained multiple individual sites located near to each other. Moreover, the binding affinity of drugs will depend on the distribution of recognition elements such as hydrogen bond donors and hydrophobic moieties in the site. The presence of more than one site capable of drug recognition in the overall binding region provided P-gp with its characteristic promiscuity.
Another investigation, prior to the publication of the X-ray structure of murine P-gp, generated a molecular model to examine drug binding (Vandevuer et al. 2006). The model was developed using data from a wide range of sources including electron microscopy, cysteine-disulphide cross-linking and mutagenesis data. Molecular docking programs were used to assess the binding of selected drugs, with energetics of the interaction used to assign relative potencies. Seven drugs were docked to the cavity within the centre of the protein and the nature of their interactions with the polypeptide was analysed. A number of different types of interactions between drug and P-gp were identified, which included hydrogen bonding, π–π stacking, and cation–π interactions. The findings were general support for the pharmacophore model described by Pajeva and Wiese (Pajeva and Wiese 2002). The ligands were found to bind at different positions within the cavity and multiple binding locations were observed for a single ligand. Taken together, this study provided evidence of multiple drug-binding sites and information describing the drug–protein interactions at these locations.
Do Modulators and Substrates Bind at Distinct Sites on P-gp?
The descriptions presented thus far indicate that P-gp is likely to have a large binding domain, which is likely to reside within the central cavity. A number of helices from the TMDs line the cavity and it has been proposed that substrates may enter this cavity directly from the lipid milieu through defined portals. A number of compounds have been identified that inhibit the transport of anticancer drugs by P-gp and thereby restore the efficacy of chemotherapy. Often these “inhibitors” are classified as competitive, despite the lack of formal data to indicate binding at the identical site to substrate. In addition, many “inhibitors” of the transport of anticancer drugs are themselves substrates for transport. A classical inhibitor would prevent the transport of a true substrate, but not be translocated across the membrane per se. The interaction of two compounds that are both transported would be revealed as an apparent inhibitory effect on the compound whose transport was being measured. As will be shown in subsequent sections, there is a vast amount of data to indicate that P-gp allosteric binding sites can influence binding and transport of drugs. This intricate and complex network of interactions necessitates clarification of nomenclature. The term modulator will be used to indicate a compound that interferes with the transport process for a substrate. Moreover, the modulator must bind to P-gp at a site distinct from the substrate in question; i.e. at an allosteric site.
Photo-affinity labelling approaches demonstrated distinct binding sites for the P-gp substrate [125I]-IAAP and the modulator (±) cis(Z)flupentixol (Dey et al. 1997, 1999; Maki et al. 2003). [125I]-IAAP was shown to photo-label P-gp at two sites on the protein and that the presence of cis(Z)flupentixol was able to enhance the binding. The enhancement of photo-labelling (and presumably binding) is clear evidence for a positive allosteric interaction. In addition, cis(Z)flupentixol displayed a differential ability to alter the photo-labelling of [125I]-IAAP at the N- and C-terminal sites. Also, ATP-bound and vanadate-trapped P-gp displays considerably reduced photo-labelling by [125I]-IAAP and the high affinity binding site is only “regenerated” following dissociation of nucleotide (Maki and Dey 2006). However, cis(Z)flupentixol binding to P-gp in the ATP or vanadate-trapped configuration (without nucleotide dissociation) is also able to “regenerate” [125I]-IAAP binding, suggestive of an allosteric site that is essentially unaffected by events at the NBDs (Maki and Dey 2006). cis(Z) flupentixol treatment of P-gp generated proteolytic cleavage patterns that were identical to those obtained following trapping of the protein with ADP-vanadate or ATP-γ-S (Ghosh et al. 2006). It appears that either binding site for cis(Z)flupentixol may be located in TM12, since mutation of the phenylalanine residue to alanine (F983A) affects the tryptic digestion pattern observed in the presence of modulator. Alternatively, the residue may be involved in the conformational changes accompanying cis(Z)flupentixol binding. The data using this strategy clearly demonstrates alternate binding sites for a modulator and substrate; in addition, binding at the two sites confers distinct conformational transitions in P-gp.
One of the X-ray structures of mouse P-gp (Aller et al. 2009) was obtained in the presence of two enantiomeric modulators that showed inhibitors bound within the central cavity. This has led to efforts aimed at localising the H- and R-sites. One investigation focussed on the ability of the two enantiomers (QZ59-RRR and QZ59-SSS) to modulate substrate transport from the H-site (Hoechst 33342) and the R-site (Daunorubicin) (Martinez et al. 2014). The SSS-enantiomer was found to alter transport of both the H- and R-site substrates in a competitive manner. The RRR-enantiomer also interacts with transport of Hoechst 33342, but with a lower potency than the SSS-enantiomer. In addition, the RRR-enantiomer inhibits daunorubicin transport in a non-competitive manner. The data may demonstrate that the QZ59-SSS binding overlaps with both the H-site and the R-site. In addition, the binding of QZ59-RRR may occur at the H-site, but may allosterically affect the R-site. It is worth noting that the competitive and non-competitive interactions were measured exclusively with transport, and not binding assays. Binding is the first step of transport and the unequivocal attribution of the location of QZ59-SSS or QZ59-RRR binding requires alternative assay systems. Nonetheless, the data reveal that modulators and transported substrates can bind at distinct sites on P-gp; however, some sites may be competent for both transport and allosteric modulation.
Complex Interactions Between Substrates and Modulators on P-gp
The data summarised in Sect. “Do Modulators and Substrates Bind at Distinct Sites on P-gp?” (and elsewhere) clearly demonstrate that the nature and location of drug binding to P-gp is a complex one and involves an intricate network of sites on the protein. This was further illustrated by an investigation of the effects of 34 P-gp interacting drugs (i.e. substrates and modulators) on the characteristics of ATP hydrolysis (Litman et al. 1997). They then examined the interactions between these drugs by studying them in pairs; for example, using verapamil and a series of transported substrates. This ambitious study led to the identification of at least four major types of drug interactions on, or with, P-gp. These four types comprise classical competitive and non-competitive inhibition, allosteric inhibition and co-operative stimulation of ATP hydrolysis.
Another series of manuscripts also examined the effects of pairs of compounds on the ATPase activity of P-gp in order to deduce the nature of drug binding (Orlowski and Garrigos 1999; Pascaud et al. 1998; Garrigos et al. 1997). For example, Pascaud et al. measured the alteration of P-gp ATPase activity in the presence of vinblastine, azidopine, verapamil and five dihydropyridine modulators; nicardipine, nimodipine, nitrendipine and nifedipine (Pascaud et al. 1998). ATPase activity was stimulated by verapamil and nicardipine but not by vinblastine. Amongst the dihydropyridines, the potency to stimulate activity was greatest for the more hydrophobic compounds. The authors painstakingly measured and assessed the nature of interaction between the drugs. They generated a “sticky paper model” for the drug binding to P-gp based on the ATPase assay effects. The binding “domain” consisted of a series of individual sites for drugs; one class of site was specific for transported substrates, and others for modulators. The modulator sites affected ATPase activity of a substrate independently, while others worked in tandem. Some sites were exclusive to a class of compounds, whilst others displayed broad specificity to a number of drugs.
Collectively, the enzymatic assay results described in this section were in agreement with the concept of a multiple-site binding model for drugs. Moreover, the sites are involved in a complex allosteric network of either positive or negative heterotropic interactions. Interpretation of the data is complex due to the use of a functional assay. ATPase activity consists of drug/nucleotide association, instigation of conformational change, ATP catalysis, nucleotide/drug dissociation and resetting. Drug interactions may affect one or more of these steps in the process.
Classical Receptor-Drug Analysis of Multidrug Binding to P-gp
As alluded to in the previous section, it is difficult to assign binding sites for drugs based on assays that involve multistep functional activity. Direct binding of drugs to proteins can be measured using radioligand-binding assays. Simple equilibrium assays provide affinities of ligands (KD), binding capacity (BMAX), the inhibition constant for modulators (KI) and the nature of interaction between two drugs readily demonstrated by the relative effects on KD/BMAX using classic Schild plots (Larazeno and Birdsall 1993; Kenakin 1997). The use of kinetic assays to determine the association and dissociation rate constants will provide definitive proof of allosteric interactions as defined by the law-of-mass action.
Several teams used the radioligand approach to examine the binding of drugs to P-gp, both to discover novel “inhibitors” of the protein, and to reveal information on the binding interaction (Ferry et al. 1992, 1995; Martin et al. 1997). The dissociation constant (KD) was measured for several substrates of P-gp and revealed considerable differences in the affinities of ligands to bind to P-gp. The presence of distinct binding affinities argues against one of the assumptions of the hydrophobic vacuum cleaner model, which suggested that interaction is purely based on non-specific adsorption to a hydrophobic interface. Assays measuring the heterologous displacement of radiolabelled substrates were used to estimate the potency (IC50) of displacing compound to bind to P-gp. However, the IC50 value is only a relative measure of affinity that may be converted to an inhibition constant (i.e. KI) using the Cheng–Prusoff equation (Larazeno and Birdsall 1993). This transformation is only applicable in cases where the displacing compound and radioligand share a competitive interaction. Consequently, the dissociation rate constant was also measured for the radioligand and the ability of displacing compound to affect this rate was determined. Several dihydropyridines, Hoechst 33342 and Tariquidar (to name a few), were able to increase the rate of [3H]-vinblastine dissociation from P-gp (Ferry et al. 1995; Martin et al. 1997; Pascaud et al. 1998). According to the law-of-mass action, this can only occur through an allosteric effect; thereby, confirming the presence of multiple drug-binding sites.
Another approach to discriminate between competitive and non-competitive interaction involves the use of Langmuir Adsorption (Saturation) Isotherms to analyse radioligand-binding assays (Kenakin 2004). Saturation isotherms were generated for [3H]-vinblastine in the presence of increasing concentrations of other substrates or modulators (Martin et al. 2000). An alteration in the BMAX for [3H]-vinblastine was observed in the presence of the substrates Rhodamine123, Hoechst 33342 and the modulators XR9051, XR9576 (Tariquidar) and GF120918 (Elacridar). The reduced BMAX, in the absence of any alteration in the apparent KD for [3H]-vinblastine, was indicative of a non-competitive interaction. The non-competitive interaction was confirmed as a negative allosteric heterotropic one following demonstration of an elevated dissociation rate constant (k off). Based on the data with [3H]-vinblastine, it appears that other transported substrates bind at distinct sites on P-gp. Similarly, the modulators also interact at a site distinct from that of [3H]-vinblastine. Is there any overlap, or does the binding site on P-gp contain a distinct interaction “zone” for all compounds?
To address this question, similar analyses were undertaken using a tritiated version of the most potent P-gp modulator, XR9576 (i.e. Tariquidar) (Martin et al. 2000). In the presence of vinblastine, the BMAX for [3H]-XR9576 was unaffected, however there was a dose-dependent shift to the right in its saturation binding curves. The data was analysed using a Schild plot, which was characterised by a slope significantly different to 1.0, which is indicative of a non-competitive interaction. This is in full agreement with data obtained for [3H]-vinblastine in the presence of XR9576. Hoechst 33342 also produced a shift in the saturation binding curves of [3H]-XR9576 to the right. However, in the case of Hoechst 33342, the slope was not significantly different to 1.0 and revealed a competitive binding interaction with [3H]-XR9576. Whole cell accumulation assays indicated that [3H]-XR9576 was not a substrate for transport by P-gp and the unlabelled compound also inhibited the ATPase activity of purified protein (Martin et al. 1999). The binding data revealed that XR9576 and the transported substrate, vinblastine, bound at distinct sites on P-gp. However, since XR9576 interacts with P-gp at the same location as the transported substrate Hoechst 33342, their shared site is indeed capable of transport. Why is [3H]-XR9576 not transported by P-gp? The rate of [3H]-XR9576 dissociation from P-gp was considerably slower than that observed for vinblastine or paclitaxel. The high affinity binding of [3H]-XR9576, in particular the slow k off rate, may prevent efficient transport across the bilayer since rapid dissociation from the external facing binding site is essential for transport. This suggests that it is the nature of the ligand and its interaction with the binding site that determine whether transport across the bilayer occurs.
The data in the preceding paragraph demonstrate that the modulator XR9576 and the substrate Hoechst 33342 bind at the same site. Is this a generic property of the binding sites; namely that they are capable of interacting with either class of compounds? This would confirm that the nature of the ligand, rather than the site properties, dictates whether transport occurs. To address this, the interaction of a number of other modulators with [3H]-XR9576 was investigated.
In particular, the effects of nicardipine, XR9051 and GF120918 (Elacridar) on the saturation isotherms and the dissociation rate of [3H]-XR9576 were measured. Both nicardipine and XR9051 increased the apparent KD of [3H]-XR9576 without any effect on the BMAX, which is suggestive of competition. XR9051 did not alter the dissociation rate of [3H]-XR9576, thereby confirming a competitive interaction; which is not unexpected given the structural similarity of the two ligands. In contrast, nicardipine caused an increase in the dissociation rate, indicating a non-competitive allosteric interaction. Consequently, nicardipine must bind at a site distinct from the binding site shared by XR9576, XR9051 and Hoechst 33342. GF120918 caused a dose-dependent decrease in the BMAX of [3H]-XR9576 and [3H]-vinblastine without a change in the KD, indicating that GF120918 does not bind to the same sites as either. Another study had demonstrated a non-competitive interaction between GF120918 and, which suggests the presence of a fourth site on P-gp.
Collectively, the evidence collected from these investigations was used to classify at least four distinct drug-binding sites on P-gp. This provided crucial support not only for the existence of multiple binding sites on P-gp but also insight into the complex ways in which the binding sites communicate with each other. The data also demonstrated that substrates and modulators can bind at equivalent sites. It is therefore the nature of interaction at the site that demonstrates whether a drug will be transported.
Summary and Perspectives
Over more than three decades, scientists have attempted to localise drug-binding sites and to provide detail on the molecular interactions with P-gp. These attempts have involved biochemistry, pharmacology, structural and bioinformatics approaches. The purpose of this article was to summarise the numerous sources of data for this key question in the ABC transporter field. We have moved on considerably since the early hydrophobic vacuum cleaner and drug-flippase models. However, drugs do appear to enter the binding domain directly from the lipid milieu and do so via portals or gates formed at the interface between the two TMDs. The binding domain is located in the central cavity of P-gp, which is lined by multiple helices. The precise dimensions of this cavity remain the source of considerable debate within the structural biology field. The binding domain contains multiple sites for interaction with drugs and the drug–protein interactions involve a combination of hydrogen bonding and π–π stacking, particularly with phenylalanine residues. Initial binding of a drug causes local conformational changes leading to a substrate-induced fit that generates high affinity. The individual sites are proximal and each appears capable of interacting with transported substrates and modulators. Moreover, it is the molecular interaction between drug and site that determines whether translocation of drug across the membrane occurs.
The primary objective remains to unequivocally determine the residues involved in the interaction with drugs at the individual sites within the domain.
References
Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323:1718–1722
Beck WT, Cirtain MC, Glover CJ, Felsted RL, Safa AR (1988) Effects of indole alkaloids on multidrug resistance and labeling of P-glycoprotein by a photoaffinity analog of vinblastine. Biochem Biophys Res Commun 153:959–966
Bruggemann EP, Germann UA, Gottesman MM, Pastan I (1989) Two different regions of P-glycoprotein [corrected] are photoaffinity-labeled by azidopine. J Biol Chem 264:15483–15488
Bruggemann EP, Currier SJ, Gottesman MM, Pastan I (1992) Characterization of the azidopine and vinblastine binding site of P-glycoprotein. J Biol Chem 267:21020–21026
Callaghan R, Berridge G, Ferry DR, Higgins CF (1997) The functional purification of P-glycoprotein is dependent on maintenance of a lipid-protein interface. Biochim Biophys Acta 1328:109–124
Chang G (2003) Structure of MsbA from Vibrio cholera: a multidrug resistance ABC transporter homolog in a closed conformation. J Mol Biol 330:419–430
Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB (1986) Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 47:381–389
Chiba P, Burghofer S, Richter E, Tell B, Moser A, Ecker G (1995) Synthesis, pharmacologic activity, and structure-activity relationships of a series of propafenone-related modulators of multidrug resistance. J Med Chem 38:2789–2793
Crowley E, Callaghan R (2010) Multidrug efflux pumps: drug binding—gates or cavity? FEBS J 277:530–539
Crowley E, O’Mara ML, Reynolds C, Tieleman DP, Storm J, Kerr ID, Callaghan R (2009) Transmembrane helix 12 modulates progression of the ATP catalytic cycle in ABCB1. Biochemistry 48:6249–6258
Crowley E, O’mara ML, Kerr ID, Callaghan R (2010a) Transmembrane Helix 12 plays a pivotal role in coupling energy provision and drug binding in ABCB1. FEBS J 277(19):3974–3985
Crowley E, O’Mara ML, Kerr ID, Callaghan R (2010b) Transmembrane helix 12 plays a pivotal role in coupling energy provision and drug binding in ABCB1. FEBS J 277:3974–3985
Dano K (1973) Active outward transport of daunomycin in resistant Ehrlich ascites tumor cells. Biochim Biophys Acta 323:466–483
Dawson RJ, Locher KP (2006) Structure of a bacterial multidrug ABC transporter. Nature 443:180–185
Dawson RJ, Locher KP (2007) Structure of the multidrug ABC transporter Sav 1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett 581:935–938
Demel MA, Schwaha R, Kramer O, Ettmayer P, Haaksma EE, Ecker GF (2008) In silico prediction of substrate properties for ABC-multidrug transporters. Expert Opin Drug Metab Toxicol 4:1167–1180
Dey S, Ramachandra M, Pastan I, Gottesman MM, Ambudkar SV (1997) Evidence for two nonidentical drug-interaction sites in the human P-glycoprotein. Proc Natl Acad Sci USA 94:10594–10599
Dey S, Hafkemeyer P, Pastan I, Gottesman MM (1999) A single amino acid residue contributes to distinct mechanisms of inhibition of the human multidrug transporter by stereoisomers of the dopamine receptor antagonist flupentixol. Biochemistry 38:6630–6639
Ecker G, Chiba P, Hitzler M, Schmid D, Visser K, Cordes HP, Csollei J, Seydel JK, Schaper K-J (1996) Structure-activity relationship studies on benzofuran analogs of propefenone-type modulators of tumor cell multidrug resistance. J Med Chem 39:4767–4774
Ecker GF, Csaszar E, Kopp S, Plagens B, Holzer W, Ernst W, Chiba P (2002) Identification of ligand-binding regions of P-glycoprotein by activated-pharmacophore photoaffinity labeling and matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Mol Pharmacol 61:637–648
Ferry DR, Kampf K, Goll A, Glossmann H (1985) Subunit composition of skeletal muscle transverse tubule calcium channels evaluated with the 1,4-dihydropyridine photoaffinity probe [3H]azidopine. EMBO J 4:1933–1940
Ferry DR, Russell MA, Cullen MH (1992) P-glycoprotein possesses a 1,4-dihydropyridine selective drug acceptor site which is allosterically coupled to a vinca alkaloid selective binding site. Biochem Biophys Res Commun 188:440–445
Ferry DR, Malkhandi JP, Russell MA, Kerr DJ (1995) Allosteric regulation of [3H]vinblastine binding to P-glycoprotein of MCF-7 Adr cells by dexniguldipine. Biochem Pharmacol 49:1851–1861
Frillingos S, Sahin-Toth M, Wu J, Kaback HR (1998) Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J 12:1281–1299
Garrigos M, Mir LM, Orlowski S (1997) Competitive and non-competitive inhibition of the multidrug-resistance-associated P-glycoprotein ATPase. Further experimental evidence for a multisite model. Eur J Biochem 244:664–673
Garrigues A, Loiseau N, Delaforge M, Ferte J, Garrigos M, Andre F, Orlowski S (2002) Characterization of two pharmacophores on the multidrug transporter P-glycoprotein. Mol Pharmacol 62:1288–1298
Ghosh P, Moitra K, Maki N, Dey S (2006) Allosteric modulation of the human P-glycoprotein involves conformational changes mimicking catalytic transition intermediates. Arch Biochem Biophys 450:100–112
Glossmann H, Ferry DR, Striessnig J, Goll A, Moosburger K (1987) Resolving the structure of the Ca2+ channel by photoaffinity labeling. TIPS 8:95–100
Gottesman MM, Ambudkar SV, Xia D (2009) Structure of a multidrug transporter. Nat Biotechnol 27:546–547
Greenberger LM (1993) Major photoaffinity drug labeling sites for iodoaryl azidoprazosin in P-glycoprotein are within, or immediately C-terminal to, transmembrane domains 6 and 12. J Biol Chem 268:11417–11425
Greenberger LM, Yang C-PH, Gindin E, Horwitz SB (1990) Photoaffinity probes for the a1-adrenergic receptor and the calcium channel bind to a common domain in P-glycoprotein. J Biol Chem 265:4394–4401
Gribar JJ, Ramachandra M, Hrycyna CA, Dey S, Ambudkar SV (2000) Functional characterization of glycosylation-deficient human P-glycoprotein using a vaccinia virus expression system. J Membr Biol 173:203–214
Hafkemeyer P, Dey S, Ambudkar SV, Hrycyna CA, Pastan I, Gottesman MM (1998) Contribution to substrate specificity and transport of nonconserved residues in transmembrane domain 12 of human P-glycoprotein. Biochemistry 37:16400–16409
Higgins CF (1992) ABC transporters; from microorganisms to man. Annu Rev Cell Biol 8:67–113
Higgins CF, Linton KJ (2004) The ATP switch model for ABC transporters. Nat Struct Mol Biol 11:918–926
Homolya L, Hollo Z, Germann UA, Pastan I, Gottesman MM, Sarkadi B (1993) Fluorescent cellular indicators are extruded by the multidrug resistance protein. J Biol Chem 268:21493–21496
Isenberg B, Thole H, Tummler B, Demmer A (2001) Identification and localization of three photobinding sites of iodoarylazidoprazosin in hamster P-glycoprotein. Eur J Biochem 268:2629–2634
Jin MS, Oldham ML, Zhang Q, Chen J (2012) Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490:566–569
Kenakin TP (1997) Pharmacologic analysis of drug-receptor interaction. Lippincott-Raven, Philadelphia
Kenakin T (2004) Principles: receptor theory in pharmacology. Trends Pharmacol Sci 25:186–192
Lankelma J, Spoelstra EC, Dekker H, Broxterman HJ (1990) Evidence for daunomycin efflux from multidrug resistant 2780AD human ovarian carcinoma cells against a concentration gradient. Bioch Biophys Acta 1055:217–222
Larazeno S, Birdsall NJM (1993) Estimation of competitive antagonist affinity from functional inhibition curves using the Gaddum, Schild and Cheng-Prusoff equations. Br J Pharmacol 109:1110–1119
Lerner-Marmarosh N, Gimi K, Urbatsch IL, Gros P, Senior AE (1999) Large scale purification of detergent-soluble P-glycoprotein from Pichia pastoris cells and characterization of nucleotide binding properties of wild-type, Walker A, and Walker B mutant proteins. J Biol Chem 274:34711–34718
Li J, Jaimes KF, Aller SG (2014) Refined structures of mouse P-glycoprotein. Protein Sci 23:34–46
Ling V, Thompson LH (1974) Reduced permeability in CHO cells as a mechanism of resistance to colchicine. J Cell Physiol 83:103–116
Ling V, Kartner N, Sudo T, Siminovitch L, Riordan JR (1983) Multidrug-resistance phenotype in Chinese hamster ovary cells. Cancer Treat Rep 67:869–874
Litman T, Zeuthen T, Skovsgaard T, Stein WD (1997) Competitive, non-competitive and cooperative interactions between substrates of P-glycoprotein as measured by its ATPase activity. Biochim Biophys Acta 1361:169–176
Liu R, Sharom FJ (1996) Site-directed fluorescence labeling of P-glycoprotein on cysteine residues in the nucleotide binding domains. Biochemistry 35:11865–11873
Liu R, Sharom FJ (1997) Fluorescence studies on the nucleotide binding domains of the P-glycoprotein multidrug transporter. Biochemistry 36:2836–2843
Liu R, Siemiarczuk A, Sharom FJ (2000) Intrinsic fluorescence of the P-glycoprotein multidrug transporter: sensitivity of tryptophan residues to binding of drugs and nucleotides. Biochemistry 39:14927–14938
Loo TW, Clarke DM (1993) Functional consequences of phenylalanine mutations in the predicted transmembrane domain of P-glycoprotein. J Biol Chem 268:19965–19972
Loo TW, Clarke DM (1995a) Membrane topology of a cysteine-less mutant of human P-glycoprotein. J Biol Chem 270:843–848
Loo TW, Clarke DM (1995b) Rapid purification of human P-glycoprotein mutants expressed transiently in HEK 293 cells by nickel-chelate chromatography and characterization of their drug stimulated ATPase activities. J Biol Chem 270:21449–21452
Loo TW, Clarke DM (1997) Identification of residues in the drug-binding site of human P-glycoprotein using a thiol-reactive substrate. J Biol Chem 272:31945–31948
Loo TW, Clarke DM (1999) Identification of residues in the drug-binding domain of human P-glycoprotein. Analysis of transmembrane segment 11 by cysteine-scanning mutagenesis and inhibition by dibromobimane. J Biol Chem 274:35388–35392
Loo TW, Clarke DM (2000) Identification of residues within the drug-binding domain of the human multidrug resistance P-glycoprotein by cysteine-scanning mutagenesis and reaction with dibromobimane. J Biol Chem 275:39272–39278
Loo TW, Clarke DM (2001) Defining the drug-binding site in the human multidrug resistance P-glycoprotein using a methanethiosulfonate analog of verapamil, MTS-verapamil. J Biol Chem 276:14972–14979
Loo TW, Clarke DM (2002) Location of the rhodamine-binding site in the human multidrug resistance P-glycoprotein. J Biol Chem 277:44332–44338
Loo TW, Clarke DM (2005) Do drug substrates enter the common drug-binding pocket of P-glycoprotein through “gates”? Biochem Biophys Res Commun 329:419–422
Loo TW, Bartlett MC, Clarke DM (2003a) Permanent Activation of the Human P-glycoprotein by Covalent Modification of a Residue in the Drug-binding Site. J Biol Chem 278:20449–20452
Loo TW, Bartlett MC, Clarke DM (2003b) Simultaneous binding of two different drugs in the binding pocket of the human multidrug resistance P-glycoprotein. J Biol Chem 278:39706–39710
Loo TW, Bartlett MC, Clarke DM (2003c) Substrate-induced conformational changes in the transmembrane segments of human P-glycoprotein. Direct evidence for the substrate-induced fit mechanism for drug binding. J Biol Chem 278:13603–13606
Loo TW, Bartlett MC, Clarke DM (2006a) Transmembrane segment 1 of human P-glycoprotein contributes to the drug-binding pocket. Biochem J 396:537–545
Loo TW, Bartlett MC, Clarke DM (2006b) Transmembrane segment 7 of human P-glycoprotein forms part of the drug-binding pocket. Biochem J 399:351–359
Lugo MR, Sharom FJ (2005) Interaction of LDS-751 and Rhodamine 123 with P-Glycoprotein: evidence for simultaneous binding of both drugs. Biochemistry 44:100
Lugo MR, Sharom FJ (2009) Interaction of LDS-751 with the drug-binding site of P-glycoprotein: a Trp fluorescence steady-state and lifetime study. Arch Biochem Biophys 492:17–28
Maki N, Dey S (2006) Biochemical and pharmacological properties of an allosteric modulator site of the human P-glycoprotein (ABCB1). Biochem Pharmacol 72:145–155
Maki N, Hafkemeyer P, Dey S (2003) Allosteric modulation of human P-glycoprotein. Inhibition of transport by preventing substrate translocation and dissociation. J Biol Chem 278:18132–18139
Martin C, Berridge G, Higgins CF, Callaghan R (1997) The multi-drug resistance reversal agent SR33557 and modulation of vinca alkaloid binding to P-glycoprotein by an allosteric interaction. Br J Pharmacol 122:765–771
Martin C, Berridge G, Mistry P, Higgins C, Charlton P, Callaghan R (1999) The molecular interaction of the high affinity reversal agent XR9576 with P-glycoprotein. Br J Pharmacol 128:403–411
Martin C, Berridge G, Higgins CF, Mistry P, Charlton P, Callaghan R (2000) Communication between multiple drug binding sites on P-glycoprotein. Mol Pharmacol 58:624–632
Martin C, Higgins CF, Callaghan R (2001) The vinblastine binding site adopts high- and low-affinity conformations during a transport cycle of P-glycoprotein. Biochemistry 40:15733–15742
Martinez L, Arnaud O, Henin E, Tao H, Chaptal V, Doshi R, Andrieu T, Dussurgey S, Tod M, di Pietro A, Zhang Q, Chang G, Falson P (2014) Understanding polyspecificity within the substrate-binding cavity of the human multidrug resistance P-glycoprotein. FEBS J 281:673–682
Montanari F, Ecker GF (2015) Prediction of drug-ABC-transporter interaction—recent advances and future challenges. Adv Drug Deliv Rev
Naito M, Yusa K, Tsuruo T (1989) Steroid hormones inhibit binding of Vinca alkaloid to multidrug resistance related P-glycoprotein. Biochem Biophys Res Commun 158:1066–1071
O’Mara ML, Mark AE (2014) Structural characterization of two metastable ATP-bound states of P-glycoprotein. PLoS ONE 9:e91916
O’Mara ML, Tieleman DP (2007) P-glycoprotein models of the apo and ATP-bound states based on homology with Sav 1866 and MalK. FEBS Lett 581:4217–4222
Orlowski S, Garrigos M (1999) Multiple recognition of various amphiphilic molecules by the multidrug resistance P-glycoprotein: molecular mechanisms and pharmacological consequences coming from functional interactions between various drugs. Anticancer Res 19:3109–3123
Pajeva IK, Wiese M (2002) Pharmacophore model of drugs involved in P-glycoprotein multidrug resistance: explanation of structural variety (hypothesis). J Med Chem 45:5671–5686
Pascaud C, Garrigos M, Orlowski S (1998) Multidrug resistance transporter P-glycoprotein has distinct but interacting binding sites for cytotoxic drugs and reversing agents. Biochem J 333(Pt 2):351–358
Pawagi AB, Wang J, Silverman M, Reithmeier RA, Deber CM (1994) Transmembrane aromatic amino acid distribution in P-glycoprotein. A functional role in broad substrate specificity. J Mol Biol 235:554–564
Pleban K (2004) P-glycoprotein substrate binding domains are located at the transmembrane domain/transmembrane domain interfaces: a combined photoaffinity labeling-protein homology modeling approach. Mol Pharmacol 67:365–374
Qu Q, Sharom FJ (2001) FRET analysis indicates that the two ATPase active sites of the P-glycoprotein multidrug transporter are closely associated. Biochemistry 40:1413–1422
Qu Q, Sharom FJ (2002) Proximity of bound Hoechst 33342 to the ATPase catalytic sites places the drug binding site of P-glycoprotein within the cytoplasmic membrane leaflet. Biochemistry 41:4744–4752
Raub TJ (2005) P-glycoprotein recognition of subtrates and circumvention through rational drug design. Mol Pharm 3(1):3–25
Raviv Y, Pollard HB, Bruggemann EP, Pastan I, Gottesman MM (1990) Photosensitized labeling of a functional multidrug transporter in living drug-resistant tumor cells. J Biol Chem 265:3975–3980
Ravna AW, Sylte I, Sager G (2007) Molecular model of the outward facing state of the human P-glycoprotein (ABCB1), and comparison to a model of the human MRP5 (ABCC5). Theor Biol Med Model 4:33
Riordan JR, Ling V (1985) Genetic and biochemical characterization of multidrug resistance. Pharmacol Ther 28:51–75
Rosenberg MF, Callaghan R, Ford RC, Higgins CF (1997) Structure of the multidrug resistance P-glycoprotein to 2.5 nm resolution determined by electron microscopy and image analysis. J Biol Chem 272:10685–10694
Rosenberg MF, Velarde G, Ford RC, Martin C, Berridge G, Kerr ID, Callaghan R, Schmidlin A, Wooding C, Linton KJ, Higgins CF (2001) Repacking of the transmembrane domains of P-glycoprotein during the transport ATPase cycle. EMBO J 20:5615–5625
Rosenberg MF, Kamis AB, Callaghan R, Higgins CF, Ford RC (2003) Three-dimensional structures of the mammalian multidrug resistance P-glycoprotein demonstrate major conformational changes in the transmembrane domains upon nucleotide binding. J Biol Chem 278:8294–8299
Rosenberg MF, Callaghan R, Modok S, Higgins CF, Ford RC (2004) 3-D structure of P-glycoprotein: the transmembrane regions adopt an asymmetric configuration in the nucleotide-bound state. J Biol Chem M410296200
Rosenberg MF, Callaghan R, Modok S, Higgins CF, Ford RC (2005) Three-dimensional structure of P-glycoprotein: the transmembrane regions adopt an asymmetric configuration in the nucleotide-bound state. J Biol Chem 280:2857–2862
Rothnie A, Storm J, Campbell J, Linton KJ, Kerr ID, Callaghan R (2004) The topography of transmembrane segment six is altered during the catalytic cycle of P-glycoprotein. J Biol Chem 279:34913–34921
Safa AR (1988) Photoaffinity labeling of the multidrug-resistance-related P-glycoprotein with photoactive analogs of verapamil. Proc Natl Acad Sci USA 85:7187–7191
Safa AR (1999) Photoaffinity analogs for multidrug resistance-related transporters and their use in identifying chemosensitizers. Drug Resist Updat 2:371–381
Safa AR (2004) Identification and characterization of the binding sites of P-glycoprotein for multidrug resistance-related drugs and modulators. Curr Med Chem Anticancer Agents 4:1–17
Safa AR, Mehta ND, Agresti M (1989) Photoaffinity labeling of P-glycoprotein in multidrug resistant cells with photoactive analogs of colchicine. Biochem Biophys Res Commun 162:1402–1408
Seelig A (1998) A general pattern for substrate recognition by P-glycoprotein. Eur J Biochem 251:252–261
Shapiro AB, Ling V (1995) Reconstitution of drug transport by purified P-glycoprotein. J Biol Chem 270:16167–16175
Shapiro AB, Ling V (1997a) Effect of quercetin on Hoechst 33342 transport by purified and reconstituted P-glycoprotein. Biochem Pharmacol 53:587–596
Shapiro AB, Ling V (1997b) Positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. Eur J Biochem 250:130–137
Shapiro AB, Ling V (1998) Transport of LDS-751 from the cytoplasmic leaflet of the plasma membrane by the rhodamine-123-selective site of P-glycoprotein. Eur J Biochem 254:181–188
Shapiro AB, Fox K, Lam P, Ling V (1999) Stimulation of P-glycoprotein-mediated drug transport by prazosin and progesterone. Evidence for a third drug-binding site. Eur J Biochem 259:841–850
Sharom FJ (1997) The P-glycoprotein efflux pump: how does it transport drugs? J Membr Biol 160:161–175
Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF (2002) ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell 10:139–149
Sonveaux N, Shapiro AB, Goormaghtigh E, Ling V, Ruysschaert JM (1996) Secondary and tertiary structure changes of reconstituted P-glycoprotein. A Fourier transform attenuated total reflection infrared spectroscopy analysis. J Biol Chem 271:24617–24624
Sonveaux N, Vigano C, Shapiro AB, Ling V, Ruysschaert JM (1999) Ligand-mediated tertiary structure changes of reconstituted P-glycoprotein. A tryptophan fluorescence quenching analysis. J Biol Chem 274:17649–17654
Spoelstra EC, Westerhoff HV, Pinedo HM, Dekker H, Lankelma J (1994) The multidrug-resistance-reverser verapamil interferes with cellular P-glycoprotein-mediated pumping of daunorubicin as a non-competitive substrate. Eur J Biochem 221:363–373
Storm J, O’Mara ML, Crowley EH, Peall J, Tieleman DP, Kerr ID, Callaghan R (2007) Residue G346 in transmembrane segment six is involved in inter-domain communication in P-glycoprotein. Biochemistry 46:9899–9910
Storm J, Modok S, O’Mara ML, Tieleman DP, Kerr ID, Callaghan R (2008) Cytosolic region of TM6 in P-glycoprotein: topographical analysis and functional perturbation by site directed labeling. Biochemistry 47:3615–3624
Stouch TR, Gudmundsson O (2002) Progress in understanding the structure-activity relationships of P-glycoprotein. Adv Drug Deliv Rev 54:315–328
Subramanian N, Condic-jurkic K, Mark AE, O’mara ML (2015) Identification of possible binding sites for Morphine and Nicardipine on the multidrug transporter P-glycoprotein using umbrella sampling techniques. J Chem Inf Model
Taylor JC, Ferry DR, Higgins CF, Callaghan R (1999) The equilibrium and kinetic drug binding properties of the mouse P-gp1a and P-gp1b P-glycoproteins are similar. Br J Cancer 81:783–789
Taylor AM, Storm J, Soceneantu L, Linton KJ, Gabriel M, Martin C, Woodhouse J, Blott E, Higgins CF, Callaghan R (2001) Detailed characterization of cysteine-less P-glycoprotein reveals subtle pharmacological differences in function from wild-type protein. Br J Pharmacol 134:1609–1618
Ueda K, Okamura N, Hirai M, Tanigawara Y, Saeki T, Kioka N, Komano T, Hori R (1992) Human P-glycoprotein transports cortisol, aldosterone and dexamethasone but not progesterone. J Biol Chem 267:24248–24252
Ueda K, Taguchi Y, Morishima M (1997) How does P-glycoprotein recognize its substrates? Semin Cancer Biol 8:151–159
van Wonderen JH, McMahon RM, O’Mara ML, McDevitt CA, Thomson AJ, Kerr ID, Macmillan F, Callaghan R (2014) The central cavity of ABCB1 undergoes alternating access during ATP hydrolysis. FEBS J 281:2190–2201
Vandevuer S, van Bambeke F, Tulkens PM, Prevost M (2006) Predicting the three-dimensional structure of human P-glycoprotein in absence of ATP by computational techniques embodying crosslinking data: insight into the mechanism of ligand migration and binding sites. Proteins 63:466–478
Ward AB, Szewczyk P, Grimard V, Lee CW, Martinez L, Doshi R, Caya A, Villaluz M, Pardon E, Cregger C, Swartz DJ, Falson PG, Urbatsch IL, Govaerts C, Steyaert J, Chang G (2013) Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc Natl Acad Sci USA 110:13386–13391
Wu Q, Bounaud PY, Kuduk SD, Yang CP, Ojima I, Horwitz SB, Orr GA (1998) Identification of the domains of photoincorporation of the 3′- and 7-benzophenone analogues of taxol in the carboxyl-terminal half of murine mdr1b P-glycoprotein. Biochemistry 37:11272–11279
Zhang X, Collins KI, Greenberger LM (1995) Functional evidence that transmembrane 12 and the loop between transmembrane 11 and 12 form part of the drug-binding domain in P-glycoprotein encoded by MDR1. J Biol Chem 270:5441–5448
Acknowledgements
The authors would like to acknowledge funding from Worldwide Cancer Research (#12-0008) and the Wellcome Trust (#WT094392MA).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Mittra, R., Coyle, E.M., Callaghan, R. (2016). Just How and Where Does P-glycoprotein Bind All Those Drugs?. In: George, A. (eds) ABC Transporters - 40 Years on. Springer, Cham. https://doi.org/10.1007/978-3-319-23476-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-23476-2_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-23475-5
Online ISBN: 978-3-319-23476-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)