Abstract
Brain lesions, especially White Matter Lesions, are not only associated with cardiac and vascular disease, but also with normal aging. Quantitative analysis of WMLs in large clinical trials is becoming more and more important. Based on intensity features and tissues’ prior, we develop a computer-assisted WMLs segmentation method, with the expectation that our approach would segment WMLs in Magnetic Resonance Imaging (MRI) sequences without user intervention. We first train a SVM nonlinear classifier to classify the MRI data voxel-by-voxel. In detail, the attribute vector is constructed by the intensity features extracted from Multi-channel MRI sequences, i.e. fluid attenuation inversion recovery (FLAIR), T1-weighted, T2-weighted, proton density-weighted (PD), and the tissues’ prior provided by partial volume estimate (PVE) images in native space. Based on the prior that the lesions almost exist in white matter, we then present an algorithm to eliminate the false-positive labels. Subsequent further segmentation through Region-Scalable Fitting (RSF) evolution on FLAIR sequence is employed to effectively segment precise lesions boundary and detect missing lesions. Compared with the manual segmentation results from an experienced neuroradiologist, experimental results for real images show desirable performances and high accuracy of the proposed method.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
White Matter lesions (WMLs) are small groups of dead cells that clump together in the white matter of the brain. The human brain is made of both gray and white matter. Information is typically stored and archived in the gray area, but the white parts play an important role when it comes to shuttling signals back and forth and retrieving information from one place and bringing it to the next. Lesions can slow or stop this process. Alzheimer’s disease, Multiple Sclerosis, and dementia are three of the most common ailments connected with lesions, but the list is usually quite long. Segmenting the lesions from Magnetic Resonance (MR) image is the prerequisite step for performing various quantitative analyses of the disease. Since brain lesion patterns are quite heterogeneous, for their shapes are deformable, their location may vary widely with different subjects, the segmentation of lesions is a challenge.
Leemput et al. [1] proposed to set up a multivariate Gaussian model by a digital brain atlas that contains information about the normal tissue signal distribution, and use it to detect lesions as outliers. Lao et al. [2] employ the previously segmented WMLs annotated by neuroradiologists to train a SVM classifier to classify new scans and reduce false-positive by utilizing anatomical knowledge and measures of distance from the training set. Freifeld et al. [3] perform healthy tissue segmentation using a probabilistic model, termed constrained-GMM, while lesions are simultaneously identified as outlier Gaussian components.
For different medical imaging mechanisms, there are different appearances for one subject. In general, the normal tissues are more distinguishable from each other in T1-weighted than other sequences, so many researchers utilize T1-weighted image to segment the normal tissues. But on the other hand, the lesions in the T1-weighted lesions appear hypointense or isointense relative to GM, which makes the lesions segmentation difficult. In the T2-weighted and PD-weighted, although these lesions appear hyperintense, it is hard for us to distinguish lesions for their intensities’ overlap with CSF. In the FLAIR images, the intensities of lesions are hyperintense and more distinguishable than others, which makes a lot of related studies mainly utilize the FLAIR images to segment lesions [4, 5]. But due to the effects of pulsatile fluid flow and partial volume, there exhibit higher intensities in some edge regions of the GM and CSF, which might introduce artificial anatomical errors in the final segmentation results and reduce the accuracy rate. Based on the above basis, combining more than one modality of the MR protocols, i.e. multi-channel images, has the benefits of increasing the intensity feature space to produce a better discrimination between brain tissues and reducing the uncertainty to increase the accuracy of the segmentation [2].
Therefore, we would rather integrate information from multi-channel for WMLs segmentation than just using a single-channel image. In this paper, we formulate the WMLs segmentation problem as a WMLs classification problem at first. After that, we treat the problem as a segmentation problem, using the classification results as the initial region of interest (ROI) for active contour to improve the final detection.
2 Methods
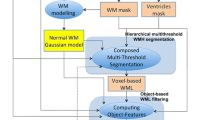
For quantitative analysis and comparison, we have tested our method over 45 subjects. Mean age of these subjects was 62 (mean: 62.2, SD: 5.9, range: 54–77, median: 61). All 45 participants’ exams consisted of transaxial T1-w, T2-w, PD and FLAIR scans. All scans except T1-w were performed with a 3 mm slice thickness, no slice gap, a 240 \(\times \) 240 mm FOV and a 256 \(\times \) 256 scan matrix. T1-w scans were performed with a 1.5 mm slice thickness, same slice gap, FOV and scan matrix. We use the manual segmentation results from an experienced neuroradiologist as the reference for assessments of our method performance. Figure 1 shows the detailed framework of our computer-assisted WMLs segmentation.
2.1 Image Pre-processing
To compensate for differences owing to subject movement between scans, all sequences of the same individual need be co-registered. As FLAIR sequence contains the most distinctive lesion-healthy tissue differentiation for segmentation of white matter lesions [6], FLAIR image space of each subject serves as the reference space which all other corresponding sequences (T1, T2 and PD) are co-registered to. A rigid transformation based on mutual-information [7] was applied for co-registration of multi-channel images. To remove the non-brain tissue for restricting our analysis on the brain tissue only, we employed a deformable model based skull stripping algorithm [8] to the co-registered T1-weighted images to generate an initial brain tissue mask, which will be used to skull-strip the other modality images. To correct the intensity inhomogeneity, we use the N3 method developed by [9].
2.2 The Voxel-Level Feature Extraction
The problem of image segmentation is first regarded as a classification task, and the goal of segmentation is to assign a label to individual voxel or a region. So, it is very important to extract the effective voxel-level image feature.
Intensity Normalization. Intensity is one of the most dominant and distinguishable low-level visual features in describing image, and has been employed for segmentation. We extract the intensity of multi-channel MR images for the attribute vector. As for the acquisitions of different sequences differing from one another, the magnitudes of the intensities were unequal in different sequences. We adopt the following steps to normalize the intensities, \(I^{FLAIR}\), \(I^{T1}\), \(I^{T2}\) and \(I^{PD}\) individual and construct the attribute vector.
-
1.
For a subject, find all the non-background voxel’s locations in FLAIR image space, i.e. \(\varOmega \), and make them as domain \(\varOmega _{non-background}\).
-
2.
Computing the average intensity, \(\mu ^{FLAIR}\), \(\mu ^{T1}\), \(\mu ^{T2}\) and \(\mu ^{PD}\) in domain, respectively.
-
3.
Divide the original intensity \(I^*\) by the corresponding average intensity \(\mu ^*\), and get the normalized intensity \({norI}^*\).
-
4.
We compose the 4 different normalized intensities \({norI}^*\) of each voxel as the corresponding first 4 attribute vector components in the domain \(\varOmega _{non-background}\).
There is one note that has to be mentioned. Although the co-registration algorithm that we have implemented has been shown high accuracy of registration, there will always be inevitable misregistration in the processing. Smoothing has been shown to ameliorate the effects of registration error [10], so we employ a mean filtering for its easy implementation to all sequences. To assist the extraction of voxel-level image feature, we implement the same normalized operations to the filtered data. Then we construct the attribute vector components of each voxel by composing the 8 normalized intensities.
Computing the PVE-Label. A single voxel in a medical image may contain several tissue types as a result of the finite resolution of the imaging devices. This case is known as partial volume effect (PVE). Tohka [11] presented a method integrated in VBM8 for the accurate, robust, and efficient estimation of partial volume model parameters, which is crucial to the accurate estimation of tissue volumes, etc.
As the normal tissues are more distinguishable from each other in T1-weighted than other sequences, we adopt this method primarily utilizing the intensity values of co-registered T1-weighted image to generate a PVE lable image volume in native space for exact tissue classification. Each voxel in PVE lable image volume is labeled with a number ranging from 1 to 3 in accord with the voxel intensity in co-registered T1-weighted image. The integers (1, 2, 3) represent CSF, GM and WM, respectively, while values between those integers mean the partial volume effect. This PVE lable image provides the 9th attribute vector component of the voxel.
2.3 Voxel-Wise Segmentation of WML by SVM
In machine learning, support vector machines (SVM) introduced by Cortes and Vapnik [12] are supervised learning models with associated learning algorithms that analyze data and recognize patterns, used for classification and regression analysis. In this paper, we mainly concentrate on the detection of WM lesions, so we classify the voxels of the MR images into two classes: lesion (assigned with label 1) and non-lesion (assigned with label 0).
All the data are prepared by the above proceeding steps. 2 subjects which are empirically chosen form the training set whose samples are made of all the lesion voxels and their surrounding normal tissue voxels. The training set consists of 43960 voxel samples (19137 voxel for lesion, others for non-lesion). These training samples are provided to SVM to generate a nonlinear classifier for classifying the remaining 43 subjects voxel-by-voxel. Every non-background voxel will be assigned by a label, lesion or normal tissues. We call the results as initial predicted labels, since false positive labels will be eliminated by the methods proposed next.
2.4 Elimination of False-Positive Labels
Most of the false-positive labels are in the appearance of very small regions or even disperse points for the noise or the intensity inhomogeneity and the misregistration between the multiple modalities. Although we have adopted inhomogeneity correction and the average filtering to remit the adverse effect of the unavoidable factors, there are still some unexpected results.
Elimination Based on the Prior. The movement of the CSF and partial volume effect give rise to the intensity overlap between CSF and partial GM area and WMLs, which will result in the some false-positive segmentation, so we need to take steps to eliminate the false-positive.
Based on the prior that WMLs are almost existing in WM regions [13] and around the ventricle, we construct the corresponding template of each subject by the segmented WM and ventricle from the PVE-label image. The later is achieved by the morphology operation on the tissue classification. Then we eliminate the false-positive label by quantifying the fusion between the initial predicted label and the template. In detail, when a predicted lesion area’s fusion degree is lower than a certain degree, this area will be considered as false-positive label and then be deleted. The experimental results show that this method of false-positive elimination makes a remarkable improvement on the accuracy of segmentation. This result will be named second eliminated label.
Further Segmentation by the Region-Scalable Fitting. Intensity inhomogeneities are inevitable phenomenon in MR images and may cause considerable difficulties in WMLs segmentation. Region-Scalable Fitting (RSF) model [14], a region-based active contour model, can deal with intensity inhomogeneity through drawing upon intensity information in local regions at a controllable scale to guide the motion of the contour.
As the brain images consist of several regions, the RSF model which only segment image into two regions (object and background) can’t be used directly in whole image. We construct the ROI only containing the potential lesion by all the voxels with value of PVE-label \( \ge \) 1.5. Since brain images are complex, and lesion volumes are relatively small, it is impractical to use the standard initial contour (small circles) as in [19, 20] and others. Hence, we use the segmentation of the lesions from the SVM classifier as an initialization for the curve, which makes it possible to apply active contour techniques for lesion delineation [3].
As mentioned above, the WMLs have the most distinctive performance of lesion-healthy tissue differentiation in FLAIR sequence [6], so we apply the RSF model only on the FLAIR sequence for further segmentation. In this way, the lesion is the object and pixels around it are the background. As a result, the WMLs’ boundary can be detected more precisely than the second eliminated lesions, and some missed lesion area can be detected automatically with the evolution of active contour. This further segmentation performs an effective detection of lesions and we call this result as final segmented labels.
3 Experiments and Results
3.1 Validation Methods
We evaluate our approach by comparing automatically segmented lesions to the ground truth for the 43 remaining testing subjects. We compute the true-positive rate (TPR), true-negative rate (TNR), the precision and the Dice similarity coefficient (DSC) of our approach: \(TPR=\frac{TP}{TP+FN}\), \(TNR=\frac{TN}{TN+FP}\), \(Precision(PI)=\frac{TP}{TP+FP}\) and \(DSC=\frac{2TP}{2TP+FP+FN}\), where TP and FP are the number of true- and false-positive voxels, and TN and FN are the number of true- and false-negative voxels, respectively.
3.2 Results
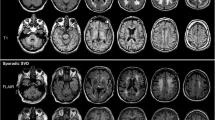
Our method was examined on ACCORD-MIND MRI dataset [15]. We compare our algorithm with the WMLs segmentation algorithm developed by Lao et al. [2], Lesion Segmentation Toolbox (LST) [16], LesionTOADS [17], Zhan et al. [18]. Figures 2 and 3 show the automatic segmentation versus manual lesion segmentation in different sizes of lesion burden. Figure 4 shows the outstanding performance of our approach in dealing with each subject. This is due to our introduction of active contour model for delineating the lesion boundaries precisely in the presence of intensity inhomogeneities.
In Table 1 we summarize the statistic data (mean) of the above 4 evaluation index in these 6 results: Lao et al., LST, LesionTOADS, Zhan et al., SVM + Elimination (second eliminated labels) and SVM + Elimination + RSF evolution (final segmented labels). From this table, we can conclude that the elimination step and the further segmentation step of RSF evolution perform effectively improvement in WM lesion segmentation.
4 Conclusion
We have developed and evaluated an computer-assisted algorithm for automated segmentation of White Matter lesions, based on integrating intensity feature information from multiple-channel MR sequences and the tissues’ prior provided by PVE images to train a nonlinear classifier by SVM for the initially effective segmentation of lesions. The experimental results have shown the robustness and accuracy of the classification. We then eliminate the false-positive labels, which is caused by pulsatile fluid flow and partial volume, based on the prior that the lesions almost exist in white matter. This strategy not only improve the accuracy of the first SVM classification, but also can provide an precisely initial ROI to the active contour evolution for delineating the lesion boundaries. The RSF evolution provides results that agree highly with human experts’ segmentation.
References
Van Leemput, K., Maes, F., Vandermeulen, D., Colchester, A., Suetens, P.: Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Trans. Med. Imag. 20, 677–688 (2001)
Lao, Z., Shen, D., Liu, D., Jawad, A.F., Melhem, E.R., Launer, L.J., Bryan, R.N., Davatzikos, C.: Computer-assisted segmentation of white matter lesions in 3D MR images using support vector machine. Acad. Radiol. 15, 300–313 (2008)
Freifeld, O., Greenspan, H., Goldberger, J.: Lesion detection in noisy MR brain images using constrained GMM and active contours. In: 4th IEEE International Symposium Biomedical Imaging, pp. 596–599. IEEE Press, From Nano to Macro (2007)
Khayati, R., Vafadust, M., Towhidkhah, F., Nabavi, S.M.: Fully automatic segmentation of multiple sclerosis lesions in brain MR FLAIR images using adaptive mixtures method and markov random field model. Comput. Biol. Med. 38, 379–390 (2008)
Souplet, J.C., Lebrun, C., Ayache, N., Malandain, G.: An automatic segmentation of T2-FLAIR multiple sclerosis lesions. In: Grand Challenge Work.: Multiple Sclerosis Lesion Segmentation Challenge, pp. 1–11 (2008)
Liu, J., Smith, C.D., Chebrolu, H.: Automatic multiple sclerosis detection based on integrated square estimation. In: 2009 IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, pp. 31–38 (2009)
Maes, F., Collignon, A., Vandermeulen, D., Marchal, G., Suetens, P.: Multimodality image registration by maximization of mutual information. IEEE Trans. Med. Imag. 16, 187–198 (1997)
Smith, S.M.: Fast robust automated brain extraction. Hum. Brain Mapp. 17, 143–155 (2002)
Sled, J.G., Zijdenbos, A.P., Evans, A.C.: A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imag. 17, 87–97 (1998)
Shen, D., Liu, D., Liu, H., Clasen, L., Giedd, J., Davatzikos, C.: Automated morphometric study of brain variation in XXY males. Neuroimage 23, 648–653 (2004)
Tohka, J., Zijdenbos, A., Evans, A.: Fast and robust parameter estimation for statistical partial volume models in brain MRI. Neuroimage 23, 84–97 (2004)
Cortes, C., Vapnik, V.: Support-vector networks. Mach. Learn. 20, 273–297 (1995)
Van Leemput, K., Maes, F., Bello, F., Vandermeulen, D., Colchester, A.C.F., Suetens, P.: Automated segmentation of MS lesions from multi-channel MR images. In: Taylor, C., Colchester, A. (eds.) MICCAI 1999. LNCS, vol. 1679, pp. 11–21. Springer, Heidelberg (1999)
Li, C., Kao, C.Y., Gore, J.C., Ding, Z.: Minimization of region-scalable fitting energy for image segmentation. IEEE Trans. Imag. Process. 17, 1940–1949 (2008)
Action to control cardiovascular risk in diabetes memory in diabetes. http://www.accordtrial.org
Schmidt, P., Gaser, C., Arsic, M., Buck, D.: An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 59, 3774–3783 (2012)
Shiee, N., Bazin, P.L., Ozturk, A., Reich, D.S., Calabresi, P.A., Pham, D.L.: A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage 49, 1524–1535 (2010)
Zhan, T., Zhan, Y., Liu, Z., Xiao, L., Wei, Z.: Automatic method for white matter lesion segmentation based on T1-fluid-attenuated inversion recovery images. IET Computer Vision (2015)
Rousson, M., Deriche, R.: A variational framework for active and adaptative segmentation of vector valued images. In: Proceedings Workshop on Motion and Video Computing, pp. 56–61. IEEE Press (2002)
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Trans. Imag. Process. 10, 266–277 (2001)
Acknowledgments
This work was supported in part by the Graduate Student Research Innovation Project of Jiangsu Province, the NUST Graduate Student Research Innovation Project, the Research Fund for the Doctoral Program of Higher Education of China (RFDP) (No. 20133219110029), the Fundamental Research Funds for the Central Universities (No. 30915012204), the Key Research Project for the Science and Technology of Education Department Henan Province (No. 15A520056) and also sponsored by the Natural Science Foundation of China under Grant No. 61171165 and 11431015.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Yu, R., Xiao, L., Wei, Z., Fei, X. (2015). Automatic Segmentation of White Matter Lesions Using SVM and RSF Model in Multi-channel MRI. In: Zhang, YJ. (eds) Image and Graphics. ICIG 2015. Lecture Notes in Computer Science(), vol 9217. Springer, Cham. https://doi.org/10.1007/978-3-319-21978-3_57
Download citation
DOI: https://doi.org/10.1007/978-3-319-21978-3_57
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-21977-6
Online ISBN: 978-3-319-21978-3
eBook Packages: Computer ScienceComputer Science (R0)