Abstract
In this paper we demonstrate a simulation framework that enables the direct and quantitative comparison of post-processing methods for diffusion weighted magnetic resonance (DW-MR) images. DW-MR datasets are employed in a range of techniques that enable estimates of local microstructure and global connectivity in the brain. These techniques require full alignment of images across the dataset, but this is rarely the case. Artefacts such as eddy-current (EC) distortion and motion lead to misalignment between images, which compromise the quality of the microstructural measures obtained from them. Numerous methods and software packages exist to correct these artefacts, some of which have become de-facto standards, but none have been subject to rigorous validation. The ultimate aim of these techniques is improved image alignment, yet in the literature this is assessed using either qualitative visual measures or quantitative surrogate metrics. Here we introduce a simulation framework that allows for the direct, quantitative assessment of techniques, enabling objective comparisons of existing and future methods. DW-MR datasets are generated using a process that is based on the physics of MRI acquisition, which allows for the salient features of the images and their artefacts to be reproduced. We demonstrate the application of this framework by testing one of the most commonly used methods for EC correction, registration of DWIs to b = 0, and reveal the systematic bias this introduces into corrected datasets.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Diffusion Tensor
- Eddy Current
- Image Space
- Simulation Framework
- Diffusion Weighted Magnetic Resonance Image
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Diffusion-weighted magnetic resonance (DW-MR) imaging is a powerful, non-invasive technique that allows us to probe the microstructure of biological tissue. The technique is well suited to the brain, and is used by clinicians and researchers studying its structure in health and disease.
A DW measurement is made by applying a diffusion-sensitising gradient in a particular direction across a sample, before acquiring an MR image. The image contains information on the diffusion of water in this direction. This diffusion is influenced by the underlying microstructure, and by acquiring a range of images with varying gradient strength and direction we can probe this structure. Many techniques, both model-based [1] and model-free [2] use the rich information provided by such datasets to characterise brain microstructure.
Unfortunately images acquired with DW-MRI are susceptible to a number of artefacts [3]. For example, \(B_0\) field inhomogeneities and the uneven magnetisation of the brain due to its magnetic susceptibility can lead to spatial displacements of several pixels. This prevents comparison between these images and others that do not contain these artefacts, such as T1- and T2-weighted images. Some artefacts lead to spatial offsets between the DW-MR images in a dataset, which compromises their anatomical correspondence and undermines the estimates of microstructure obtained from them. For example, motion can lead to rigid offsets between images, and eddy currents lead to a shear, scaling and translation of the image in the phase encoding (PE) direction that varies according to the amount of diffusion sensitisation used (typically summarised by the b-value) and the direction it is applied in.
Techniques for dealing with these artefacts can broadly be divided into those implemented at acquisition time, involving either some modification to the acquisition process or the collection of supplementary data, and post-processing methods implemented after acquisition time. Post-processing techniques are the most widely used, as they have several advantages: they can be applied retrospectively to already acquired data, a user can revert to the original data if the technique does not work as hoped, and they don’t require additional scan-time, which is often expensive.
The literature contains a vast body of post-processing techniques and software packages for correcting artefacts in DW-MRI [4–6]. Ideally their corrections would be validated by comparison to the ground truth, i.e. a map of the spatial deformations caused by the artefacts, but these cannot be obtained for real data. As a result the literature relies on either qualitative visual assessments of image alignment [7], or quantitative assessments of surrogate measures of alignment that have questionable validity, such as tract length [8], or reduced residuals from fits to microstructural models [9]. The lack of an objective ground truth means existing techniques cannot be systematically assessed, preventing end-users from making an informed choice. The development of new methods is also hindered, as any improvements over existing ones are difficult to demonstrate.
Simulation could provide us with a ground truth that would enable us to assess methods objectively, allowing researchers to make informed decisions when selecting post-processing methods. Simulation systems exist for MRI [10, 11] but there is nothing satisfactory for DW-MRI. Several systems are designed to simulate DWIs of white matter bundles [12] but these are unable to generate the full brain images required for the assessment of post-processing methods. Methods that do attempt to simulate full-brain DWIs exhibit at least one of two serious limitations. The first is the failure to model the full process of image acquisition [13], i.e. the recording of a signal in frequency space which is Fourier transformed to generate a spatial image, which precludes the inclusion of realistic artefacts. The second is the use of a heavily simplified model to create the DW contrast, which means the simulations do not capture some of the features of DWIs that makes their processing uniquely challenging, such as the variation of contrast with the direction of diffusion weighting [14].
In this work we introduce a simulation framework that allows for realistic DW-MR images to be generated, enabling the effectiveness of correction techniques to be assessed objectively and directly. The framework simulates the physics of MRI acquisition by solving Bloch’s and Maxwell’s equations, which ensures the images and their artefacts capture the key features of their real-world counterparts. The complexity of diffusion contrast is captured using a model-based approach. The framework is flexible, and allows for a range of artefacts to be modelled including EC, motion, B\(_0\) inhomogeneities and magnetic susceptibility. We demonstrate an application of this framework to EC artefacts, by providing a quantitative assessment of the most commonly used correction technique, registration of all DWIs to b = 0.
2 Methods
In this section we describe our simulation framework for producing realistic DW-MR images, and discuss its application to validating eddy-current correction schemes. An overview of the framework is discussed in Sect. 2.1, our implementation of it in Sect. 2.2 and its application in Sect. 2.3.
2.1 Simulation Framework
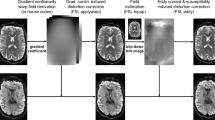
The framework (Fig. 1) combines a physics-based approach to the MR image acquisition process with a model-based representation of diffusion in order to simulate realistic DW-MR datasets. To provide a meaningful validation a simulation must capture the key characteristics of DW-MR images and their artefacts. Many of the artefacts are introduced during the acquisition of the MR signal in k-space, so it is necessary to reproduce this process for a faithful simulation.
The framework takes four main inputs. The first is a geometric object that specifies the proton density and location of white matter (WM), grey matter (GM) and cerebrospinal fluid (CSF) along with their T1 and T2 values. The second is a representation of diffusion-weighting. These two inputs are combined to produce a geometric object with its proton density reduced by an attenuation factor determined by the diffusion model. The third input is a pulse sequence, detailing the RF pulses and gradients. The fourth are details of any artefacts to be included, e.g. motion. The effects of eddy currents are included in the pulse sequence. The MR simulator takes the attenuated object, pulse sequence and artefacts and solves Bloch’s and Maxwell’s equations at each point in the object, summing the resultant signal in order to generate the k-space measurements. This is Fourier transformed to produce the output DWI.
2.2 Implementation
A full-brain segmentation was used as the geometric object input. It was created with T1- and T2-weighted images from a single subject from the WU-Minn HCP dataset [15], using FSL’s FAST [16]. Diffusion weighting was achieved using the diffusion tensor (DT) [17]. Although the DT can not faithfully represent complex WM anatomy, such as crossing fibres, empirically we find it adequate for capturing the main features that make the processing of DWIs particularly challenging: the contrast between WM, GM and CSF, as well as the variation of signal with both the direction and strength of diffusion weighting (see Fig. 3). To account for the departure from Gaussian diffusion at higher b-values, separate tensors were fit to the b = 1000 s mm\({}^{-2}\) and b = 2000 s mm\({}^{-2}\) DWIs in the HCP dataset using FSL’s DTIFIT. The tensor from the b = 1000 s mm\({}^{-2}\) shell was used to predict attenuation for simulated DWIs with \(b\le \) 1000 s mm\({}^{-2}\), and the tensor from the b = 2000 s mm\({}^{-2}\) shell was used for simulations with b-values above this.
Eddy currents were added to the pulse sequence using the method in [14], by superposing a sum of decaying exponentials on each gradient field:
where \(t_i\) corresponds to the time each diffusion gradient is turned on or off, \(\tau \) is the decay time, \(\varepsilon \) is a constant determining the relationship between the strength of eddy and diffusion gradients and a + or - is selected depending on whether the gradient is being turned on or off. We performed simulations with \(G^{\mathrm {diff}}\) = 40 m T m\(^{-1}\), and selected \(\varepsilon \,=\,0.006\) and \(\tau \) = 100 ms to represent typical values found in a clinical scanner [9].
We used POSSUM [11] to simulate the physics of MR acquisition. POSSUM allows for the creation of pulse sequences, signal generation and image reconstruction. By solving for the magnetization vectors over time at every voxel in the object it allows for effects such as spin history, motion during pulse readout and B\(_0\) inhomogeneities to be accounted for. By default, POSSUM simulates gradient-echo echo-planar imaging (EPI) sequences. To simulate the spin-echo EPI sequences typically used for DWI acquisition, we replace the default tissue-specific T\(_2\)* values with their corresponding T\(_2\) values.
2.3 Application to Eddy-Current Artefacts
To demonstrate the application of our framework we use it to assess one of the most routinely employed methods for EC correction, registration of all DWIs to b = 0, by comparing evaluated and ground truth displacement fields.
Comparison of spatial displacement fields is the most direct way to evaluate post-processing methods. For the case of EC distortions, we obtain a mapping from distorted to undistorted space from an analysis of how the influence of eddy currents on k-space translates into geometric distortions in image space. The relationship between k-space and image space is expressed as a Fourier transform:
where \(k_i =\int \bar{\gamma } G_i(t) \, \mathrm {d}t\), \(\bar{\gamma }\) is the gyromagnetic ratio, \(G_x\) and \(G_y\) are the imaging gradients applied, \(S(k_x,k_y)\) is the MR signal in k-space, and \(\rho (x,y)\) is the image in real space. In the presence of eddy currents, our imaging gradients are modified by additional gradients \(\left( G_x^E,G_y^E,G_z^E\right) \) and a spatially invariant term \(\epsilon _0\), causing our phase term to become modified. Assuming the phase-encoding (PE) direction is aligned with the y-axis, our signal equation becomes [9]:
where
Neglecting the decay of the EC field, which we have found has a negligible effect on our obtained displacement fields, the ratios \(\frac{{\varDelta }k_i}{k_y}\) are constants. As a result the effects of EC induced gradients is to introduce two shears and a scaling which correspond to displacements along the PE direction in image space, as shown in Fig. 2. For any given slice z is constant, so we observe these deformations as a shear, scaling and translation of the image. The simulation framework allows us to find the EC induced offsets \(\frac{{\varDelta }k_i}{k_y}\) and thus obtain the transformations they lead to in image space; this gives us a ground truth displacement field that we can compare to the output of each EC correction tool.
The top row shows the k-space offsets that occur when EC induced gradients are added to the x-, y- and z-gradient channels, and their corresponding image space shifts are shown in the bottom row. K-space displacements are obtained from the simulation framework, and the image space displacement fields calculated according to the analytical relationship described in Sect. 2.3.
3 Experiments and Results
In this section we explain our experiment design and show results for our validation of the simulation framework (Sect. 3.1) and its application to assessing registration to b = 0 for EC correction (Sect. 3.2).
3.1 Validation of the Framework
We first assess how well the simulated images capture the most important characteristics of real images. POSSUM has been shown to provide realistic MR simulation without diffusion weighting [11, 18], so here we focus on assessing the simulation of diffusion weighting. In the case of DW-MR the key characteristic is the variation in contrast as the strength and direction of diffusion weighting changes. To test this we compared a real and a simulated, artefact-free dataset with identical parameters: a 3 T scanner with three shells, b = 300/700/2000 s mm\({}^{-2}\), 8/32/64 directions with 1/4/8 b = 0 images, TR/TE = 3000/109 ms. Figure 3a compares the changes in contrast with varying b-value. Figure 3b compares changes in contrast with varying direction of b-vector.
3.2 Application to Eddy-Current Artefacts
To assess the effectiveness of registration to b = 0 as a method for eddy correction we applied three techniques to a simulated dataset. The first is FSL’s eddy_correct, which performs registration of each DWI to b = 0. To provide some comparison we also used a method designed to circumvent registration to b = 0, proposed by Zhuang et al. [8]. Zhuang’s technique assumes eddy-distortions are a function of the applied diffusion gradients and obtains this relationship by only registering DWIs with similar contrast. It predicts a shear, scaling and translation parameter for every slice of each volume. Zhuang’s technique is tailored to ECs, and uses a constrained, 3 degree of freedom (DOF) registration algorithm, whilst eddy_correct uses a full 12 DOF. To control for this, our third method registers each DWI to b = 0 using the same constrained, 3 DOF registration algorithm used by Zhuang.
The simulated dataset consisted of two shells, b = 700/2000 s mm\({}^{-2}\), 32/64 directions with 4/8 b = 0 images, TR/TE = 3000/90 ms, 78\(\times \)110\(\times \)60 with isotropic voxel size 1.85 mm. Rician noise was added to give SNR = 20. EC gradients were added to the pulse sequences according to the model in Sect. 2.2. No other artefacts were added.
The three correction methods were also applied to a real dataset, to see if the findings from simulation manifest themselves in real data. The dataset was acquired on a 3T Siemens scanner with similar parameters to the simulated dataset: b = 700/2000 s mm\({}^{-2}\), 32/64 directions with 4/8 b = 0 images, TR/TE = 7500/103 ms, 96\(\times \)96\(\times \)55 with isotropic voxel size 2.5 mm. To test the effectiveness of correction an outline was drawn around the b = 0 image for the dataset, which is not affected by EC distortions, and then superposed on the corrected DWIs. This allows for a visual assessment of correction.
Figure 4 reports the mean error in the displacement fields predicted by the three eddy correction methods. Figure 5 shows the spatial distribution of these errors for a typical slice from one gradient of the b = 2000 s mm\({}^{-2}\) shell. Figure 6 shows the results for correction on a real dataset.
Mean error in displacement field across the brain. The ground truth displacement was calculated from k-space shifts as described in Sect. 2.3. The first 32 volumes have b = 700 s mm\({}^{-2}\), the remaining 64 b = 2000 s mm\({}^{-2}\).
Correction applied to real data. Anterior portion of the brain shown. Red outlines were drawn on the b = 0 images. Note these outline are different for the b = 700 and b = 2000 images as the CSF in the b = 0 has been cut out in the b = 2000 image. This is because the CSF rim is attenuated fully in the b = 2000 data and this needs to be reflected in the outline for it to be a useful guide for testing alignment (Color figure online).
4 Discussion and Conclusions
We have presented a simulation framework that allows for DW-MR datasets to be generated for the purposes of testing, providing an objective and quantitative means of assessing post-processing techniques. Our framework improves on previous attempts at simulation in two key areas. Firstly, our simulated images are able to provide a much more realistic representation of the contrast differences found across DW-MR datasets. This is demonstrated in Fig. 3a, which shows that the signal attenuation with increasing b-value is well matched to the attenuation found in real data, and Fig. 3b, which shows that variations in contrast with varying b-vector are captured, particularly noticeable in the white matter. Previous simulations have used a single representative mean diffusivity or diffusion tensor for each tissue type to provide diffusion-weighting [13, 14], which leads to vastly oversimplified contrast which will bias assessments of correction schemes. The second improvement comes from our modelling of the MR acquisition process. Some attempts at simulations have modelled artefacts by applying geometric transforms in the spatial domain [13]. These distortions are not fully realistic, and furthermore without simulating the full image generation process certain artefacts cannot be introduced, such as those caused by motion during the read-out phase. Our approach extends both the range and realism of artefacts that can be simulated.
The simulation framework allowed us to quantitatively assess registration to b = 0 as a method for correcting eddy currents. Figure 4 shows that registration to b = 0 performs poorly for both b = 700 and b = 2000 shells. In the b = 700 shell the displacement fields obtained from eddy_correct are off by 0.5-1 pixels, and this rises to 1–2.5 pixels for the b = 2000 images. By contrast Zhuang’s scheme, which explicitly avoids registration to b = 0, is able to provide correction with mean errors of less than 0.5 pixel for both shells. This performance cannot be attributed to Zhuang’s method explicitly modelling the two shears and one scaling that ECs gives rise to because results from the constrained, 3 DOF registration to b = 0 were poor, particularly for the b = 2000 shell where it performed worse than eddy_correct. It is notable that the constrained registration performed slightly better than eddy_correct for the b = 700 shell and slightly worse for the b = 2000 shell. This is likely because the registration used by eddy_correct is better optimised which allows it to cope better with the decreased SNR in the b = 2000 shell. Figure 5 shows how these errors are spatially distributed in the brain. As expected, these errors are largest at the edges of the brain, where the scalings have the largest effect.
The results from simulation corroborate with our findings on real data. Figure 6 shows that data corrected by the two methods for registration to b = 0 overscaled the images, causing them to overlap with the b = 0 outlines. This effect is particularly noticeable for the b = 2000 shell, where overlaps of more than two pixels are clear, in agreement with our findings on simulated data.
These results are important in the context of techniques that use DW-MR data. Data is most commonly acquired at b = 1000, and our results indicate we can expect errors of more than 0.5 pixels in such images if they are corrected using registration to b = 0. These are enough to cause anatomical misalignment in regions of partial volume, such as the boundaries between grey matter and CSF which will compromise any information on microstructure obtained from such data. Our results demonstrate this effect will be even more severe for data acquired at b = 2000, which is becoming more common with the increasing popularity of high angular resolution (HARDI) techniques.
Future work will focus on further development of the framework. A more realistic model of diffusion could be used, such as NODDI [1], to prevent the need to fit multiple diffusion tensors. Currently a single T1 and T2 value is used for each tissue type, which could be replaced by spatial maps which will allow for variations within tissue types. The inclusion of spatially varying EC gradients will allow for higher order effects to be modelled, allowing us to model artefacts at much higher b-values.
Using the simulation framework, we can quantitatively assess the effectiveness of artefact correction schemes. In demonstrating the framework’s application to ECs, we have shown that one of the most commonly used correction techniques introduces a systematic error that is significant enough to undermine any analysis performed on data corrected using this scheme. The framework is flexible and allows for simulation of the full range of artefacts found in DW-MR, including motion, susceptibility and B\(_0\) inhomogeneities. We hope that this framework will become a key aspect of the validation of any post-processing schemes, which will allow users to make decisions on their choice of processing techniques that are informed by objective, quantitative evidence.
References
Zhang, H., Schneider, T., Wheeler-Kingshott, C.A., Alexander, D.C.: NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage 61(4), 1000–1016 (2012)
Tournier, J., Calamante, F., Gadian, D.G., Connelly, A., et al.: Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage 23(3), 1176–1185 (2004)
Le Bihan, D., Poupon, C., Amadon, A., Lethimonnier, F.: Artifacts and pitfalls in diffusion MRI. J. Magn. Reson. Imaging 24(3), 478–488 (2006)
Oguz, I., Farzinfar, M., Matsui, F., Budin, F., Liu, Z., Gerig, G., Johnson, H.J., Styner, M.: DTIPrep: quality control of diffusion-weighted images. Front. neuroinformatics. 8, 1–11 (2014)
Jenkinson, M., Smith, S.: A global optimisation method for robust affine registration of brain images. Med. Image Anal. 5(2), 143–156 (2001)
Andersson, J.L.R., Skare, S., Ashburner, J.: How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20(2), 870–888 (2003)
Mangin, J.-F., Poupon, C., Clark, C., Le Bihan, D., Bloch, I.: Distortion correction and robust tensor estimation for MR diffusion imaging. Med. Image Anal. 6(3), 191–198 (2002)
Zhuang, J., LU, Z.-L., Vidal, C.B., Damasio, H.: Correction of eddy current distortions in high angular resolution diffusion imaging. J. Magn. Reson. Imaging 37(6), 1460–1467 (2013)
Jezzard, P., Barnett, A.S., Pierpaoli, C.: Characterization of and correction for eddy current artifacts in echo planar diffusion imaging. Magn. Reson. Med. 39(5), 801–812 (1998)
Kwan, R.K.-S., Evans, A.C., Pike, G.B.: MRI simulation-based evaluation of image-processing and classification methods. IEEE Trans. Med. Imaging 18(11), 1085–1097 (1999)
Drobnjak, I., Gavaghan, D., Süli, E., Pitt-Francis, J., Jenkinson, M.: Development of a functional magnetic resonance imaging simulator for modeling realistic rigid-body motion artifacts. Magn. Reson. Med. 56(2), 364–380 (2006)
Neher, P.F., Laun, F.B., Stieltjes, B., Maier-Hein, K.H.: Fiberfox: Facilitating the creation of realistic white matter software phantoms. Magn. Reson. Med. 72(5), 1460–1470 (2013)
Bastin, M.E.: Correction of eddy current induced artefacts in MR diffusion iterative cross-correlation. Magn. Reson. Imaging 17(7), 1011–1024 (1998)
Nunes, R.G., Drobnjak, I., Clare, S., Jezzard, P., Jenkinson, M.: Performance of single spin-echo and doubly refocused diffusion-weighted sequences in the presence of eddy current fields with multiple components. Magn. Reson. Imaging 29(5), 659–667 (2011)
Van Essen, D.C., Ugurbil, K., et al.: The human connectome project: a data acquisition perspective. Neuroimage 62(4), 2222–2231 (2012)
Zhang, Y., Brady, M., Smith, S.: Segmentation of brain MR images through a hidden markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20(1), 45–57 (2001)
Basser, P.J., Mattiello, J., LeBihan, D.: Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. Imaging 103, 247–254 (1994)
Drobnjak, I., Pell, G.S., Jenkinson, M.: Simulating the effects of time-varying magnetic fields with a realistic simulated scanner. Magn. Reson. Imaging 28(7), 1014–1021 (2010)
Acknowledgements
MG is supported by the EPSRC (EP/L504889/1). MG and HZ are supported by the Royal Society International Exchange Scheme with China. HZ is additionally supported by the EPSRC (EP/L022680/1) and the MRC (MR/L011530/1). ID is supported by the Leverhulme Trust.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Graham, M.S., Drobnjak, I., Zhang, H. (2015). A Simulation Framework for Quantitative Validation of Artefact Correction in Diffusion MRI. In: Ourselin, S., Alexander, D., Westin, CF., Cardoso, M. (eds) Information Processing in Medical Imaging. IPMI 2015. Lecture Notes in Computer Science(), vol 9123. Springer, Cham. https://doi.org/10.1007/978-3-319-19992-4_50
Download citation
DOI: https://doi.org/10.1007/978-3-319-19992-4_50
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-19991-7
Online ISBN: 978-3-319-19992-4
eBook Packages: Computer ScienceComputer Science (R0)