Abstract
Huntington’s disease (HD) is a hereditary neurodegenerative disorder characterized by motor, cognitive and behavioural abnormalities. HD is caused by a mutation in the huntingtin gene which produces an enlarged chain of CAG triplets in this gene and an expanded chain of poliglutamines in the N terminal portion of the protein. HD is characterized by neuronal loss and atrophy of several brain nuclei, preferentially in the striatum.
The pathogenic mechanisms responsible for HD are partially unknown. Mutant huntingtin aggregates in insoluble filaments, changes its localization from the cytoplasm to the nucleus and changes the transcription of genes, inhibits mitochondrial function, activates caspases, block microtubules, interacts with Ca2 + channels and excitatory receptors and inhibits the production of neurotrophic factors.
There are abnormalities of the ubiquitin proteasomal system (UPS) in HD. In samples of human brain from patients with HD, it has been observed that there are intranuclear inclusions of huntingtin fragments which stain with antibodies against ubiquitine. These inclusions are present even before the presence of clinical deficits and their severity correlates with the size of the expansion. Proteasomal function, however, is preserved suggesting that the polyglutamine chains block the ubiquitylation pathway. In human fibroblasts from patients with HD, activation of autophagy compensates the deficits of the UPS .
Similarly, in experimental models of HD there are intraneuronal inclusions which appear before the clinical deficits and stimulation of the autophagy reduces the number of inclusions. Autophagy could compensate deficits related with blockade of the UPS in HD.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Huntington’s disease (HD) is an inherited neurodegenerative disorder, characterized by motor, cognitive and behavioral deficits [1–3], produced by the expansion of a CAG trinucleotide repeat in the huntingtin gene, which causes an enlarged poliglutamine chain at the N-terminal region of huntingtin [4, 5]. The expanded huntingtin mutant protein, which is ubiquitously expressed in the body and the brain, causes preferential neurodegeneration in distinct brain regions with a pattern that varies according to the size of the expansion. This is done in such a way that subjects with relatively small expansions show neuronal loss almost limited to the striatum while those with large expansion show widespread degeneration throughout the brain (striatum > thalamus > cerebral cortex > diencephalon > brain stem > cerebellum) and even with very large expansions into the muscle. In the striatum, the huntingtin protein accumulates and causes striatal atrophy [6] and the death of the medium and small size projecting spiny neurons [7, 8].
At the cellular levels, the most relevant finding observed in the neurons, which remain in different brain nuclei, is the presence of intranuclear inclusions, present in most neurons, that stain with antibodies anti-ubiquitin and anti-huntingtin. Since normal huntingtin is a cytoplasmic protein, the presence of immunoreactive intranuclear inclusion suggests a translocation of the mutant protein from cytoplasm to nucleus and a change of protein function.
Pathogenic Mechanisms
The mechanism of the disease is unknown though a number of molecular events are related to the abnormal protein (Fig. 5.1).
The most important mechanisms involved in the neurodegeneration of neurons in HD are the following:
-
a.
Aggregation of huntingtin in insoluble protofilaments
-
b.
Translocation to the nucleus and changes in transcription
-
c.
Inhibition of mitochondrial function
-
d.
Activation of caspases and interaction with other proteins
-
e.
Blockade of microtubules
-
f.
Interaction with neuronal excitability and regulation of Ca2+
-
g.
Deficits of neurotrophins and excessive inflammation
Mechanism of toxicity of mutant huntingtin [9]. Normal huntingtin is localized in the cytoplasm and regulates vesicle transport, including that of BDNF. It also regulates gene transcription. The mutation produces residues resistant to degradation, which are hyperphosphorylated and interfere with BDNF receptor signaling. Insoluble aggregates, activate caspases directly or via mitochondrial effects
Most of these mechanisms are interrelated and linked in multiple circular pathways. It is difficult at this time to pinpoint which of these pathogenic mechanisms is primary and which are secondary. Without this knowledge it is difficult to design neuroprotective studies based on rational backgrounds. In any event, abnormal processing of the mutant huntingtin or poliglutamine fragments derived from it could play a role in other cellular and molecular abnormalities observed en patients with Huntington’s disease. In order to discuss the data already available regarding the processing of abnormal proteins or protein segments in HD we shall divide our discussion in two parts, (a) data obtained in patients with HD and (b) data obtained in experimental models (Fig. 5.2).
Polar zippers of poliglutamines [10]
Protein Processing in Patients with HD
Shortly after it was discovered that the gene defect responsible for HD was an expansion of the variable sequence of CAG in the 5′-terminal region of the huntingtin gene [4], which translates into an enlarged polyglutamine chain in the N-terminal region of the huntingtin protein, it was found that in models of four diseases characterized by polyglutamine expansions these polyglutamines form β-sheets strongly held together by hydrogen bonds [10]. It was also suggested that “glutamine repeats may function as polar zippers, for example, by joining specific transcription factors bound to separate DNA segments. Their extension may cause disease either by increased, nonspecific affinity between such factors or by gradual precipitation of the affected proteins in neurons” [10].
Neuropathological studies performed in brains of patients with HD showed neuronal intranuclear inclusions and dystrophic neurites containing fragments of polyglutamine derived from the N-terminal region of the huntingtin protein [11]. The extent of huntingtin accumulation increased with the size of the polyglutamine expansions. The neuronal intranuclear inclusions and the dystrophic neurites contained ubiquitin, which suggest that mutant huntingtin was labeled for protesomal processing but resistant to digestion [11]. The authors considered that abnormal aggregation of huntingtin was one of the pathogenic mechanisms of the disease.
The types, location, numbers, forms, and composition of microscopic huntingtin aggregates in human brain have been studied with a fusion protein antibody against the first 256 amino acids which preferentially recognizes aggregated huntingtin [12]. Neuropil aggregates are much more common than nuclear aggregates and can be present in large numbers in presymptomatic mutation carriers before the onset of clinical symptoms. There are also many more aggregates in cortex than in striatum, where they are actually uncommon. Although the striatum is the most affected region in HD, only 1–4 % of striatal neurons in all grades of HD have nuclear aggregates. Neuropil aggregates, which have been identified by electron microscopy to occur in dendrites and dendritic spines, could play a role in the known dendritic pathology that occurs in HD. Aggregates increase in size in advanced grades, suggesting that they may persist in neurons that are more likely to survive. Ubiquitination is apparent in only a subset of aggregates, suggesting that ubiquitin-mediated proteolysis of aggregates may be late or variable [12]. The preferential localization of the intranuclear inclusions in cortical neurons in spite of a more severe cell loss in the striatum could be interpreted in different ways as (a) there are fewer inclusions in striatal neurons because there are fewer surviving striatal neurons and (b) there are more intranuclear inclusions in surviving cortical neurons because ubiquitination is, somehow, a way of neutralization of the toxic properties of huntingtin.
Cellular clearing of abnormal proteins could take place through the ubiquitine-proteasome pathway or through autophagy, a process which could be divided in three categories, macro-autophagy, micro-autophagy and chaperone mediated autophagy. In general, both pathways, ubiquitination + proteolysis and autophagy, are inter-related and when one of these processes is blocked there is a compensatory activation of the other in healthy cells [13]. Therefore, if ubiquitin related proteolysis is depressed in patients with HD, one could expect that autophagy is an important compensatory pathway for protein processing and that stimulation of autophagy could be a possible disease modifying therapeutic approach.
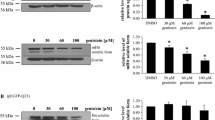
The role of autophagy in HD has been unveiled by recent studies which show that the age at onset of the disease could be modified by the V471A polymorphism of the ATG7 gene, a key factor in autophagy [14, 15]. We have further investigated the possible effects of manipulation of ubiquitination and autophagy in fibroblasts from patients with HD. These fibroblasts have increased levels of ubiquitilated proteins and higher levels of reactive oxygen species (ROS) , huntingtin and the autophagy marker LAMP2A. Baseline replication rates were higher in HD than in control fibroblasts but that was reverted after 12 passages. Epoxomicin, an inhibitor of the ubiquitin pathway, increases the activated caspase-3, HSP70, huntingtin, ubiquitinated proteins and ROS levels in fibroblasts from both HD and controls. Treatment with trehalose, an activator of autophagy, counteracts the effects of epoxomicin such as an increase in ROS, ubiquitinated proteins, huntingtin and activated caspase-3 levels and also increases the autophagy marker LC3 levels more in HD fibroblast than controls. These results suggest that activation of autophagy by trehalose could revert protein processing abnormalities observed in patients with HD (Fig. 5.3).
Epoxomicin increases ROS levels, which are reduced by trehalose in HD firboblasts. a 29, 79–dichlorofluorescin (DCF) immunocytochemistry (green) and total nuclei stained with bis-benzimide (blue) in control and HD fibroblasts and b percentage of DCF positive cells respect to the total number. (Scale bar = 20 mm). Values are expressed as the mean ± SD, n = 4 patients. Control cell number (mean per field), n = 4. HD cell number (mean per field), n = 4. The data of each patient was obtained using 4 replicates. Statistical analysis was performed by one-way ANOVA with repeated measures followed by Bonferroni multiple comparison test: *** = p < 0.001 vs solvent; + = p < 0.05, +++ = p < 0.001 HD vs controls; ΔΔΔ=p < 0.001 trehalose + epoxomicin vs epoxomicin [16]
Ubiquitination and Autophagy in Models of HD
Shortly after the discovery of the genetic mutation that causes HD several animal models of HD have been created in experimental animals from nematodes to mammals, most commonly mice and more recently pigs and sheep. These experimental models essentially confirm the same abnormalities of protein processing observed in patients with HD, and most importantly, the presence of neuronal intranuclaer inclusions in different brain areas, a correlation between the size of the expansion and density of inclusions, normal proteasomal function in vitro and alternative compensatory autophagic function. Both the UPS and autophagy pathway are important for processing of mutant huntingtin or its polyglutamine fragments since the inhibition of the two systems increases the the levels of huntingtin in PC12 cells [17].
Autophagic activity can be enhanced in Akt- and m-TOR-independent mechanisms by a number of small molecules including phenoxamine and its derivatives [18–20]. Autophagic enhancement in experimental models of HD reduces the number of inclusions [19].
Very interesting experiments have been published recently regarding the selective impairment of the striatum and its possible relation with autophagy [21]. These authors have shown that the striatal specific protein Rhes binds huntingtin and enhances its cytotoxicity, since Rhes deleted mice are protected in HD. In PC12 cells deletion of Rhes decreases autophagy and Rhes overexpression increases autophagy. These effects are independent of mTOR and opposite to the direction predicted by the known activation of mTOR by Rhes. Rhes robustly binds the autophagy regulator Beclin-1, decreasing its inhibitory interaction with Bcl-2, independent of JNK-1 signaling. Finally, co-expression of mutant huntingtin blocks Rhes-induced autophagy activation. Thus, the preferential striatal pathology and delayed onset of HD may reflect the striatal-selective expression and changes in autophagic activity of Rhes [21].
In mice and fly models of HD mTOR is bound to huntingtin aggregates [22].Treatment with rapamycin, which inhibits mTOR related kinase activity and increases autophagy, reduces the number of huntingtin aggregates [22].
Another important factor in the neuropathology is the cell-interactions during the disease and in the brain up to 90 % are glial cells supporting the survival and function of the neuronal cells. Glial pathology has also been found in HD models [23, 24] and in brains of patients with HD, analyzed post-mortem [25–27]. Mutant huntingtin is expressed also in glial cells and directly affects neuron pathology in HD [28], but comparing the amount of huntingtin inclusions, neurons accumulate much more than glial cells in mouse brains [29, 30]. Glial cells may be able to clear truncated proteins more efficiently than neurons, and astrocytes also produce neurotrophic factors and cytokines to regulate the forms and functions of neurons. Recent studies have shown that R6/1 mice implanted with a pump which delivers glial conditioned medium in one striatum have fewer intranuclear inclusions than littermates infused with artificial cerebro spinal fluid (Fig. 5.4) [31].
GCM treatment (30 days) diminishes the number of huntingtin inclusions in cortex and striatum. a Microphotographs of the right and left cortex with the specific antibody anti-HTT inclusions and b quantification of both cortex, left and right. c Microphotographs of the right and left striatum with the specific antibody anti-HTT inclusions and d quantification of both striatum, left and right. The values are expressed as the mean ± SEM (n = 6 mice in each experimental group). The statistical analysis was performed by student’s t test. * = p < 0.05, ** = p < 0.01 GCM vs control CSF groups [31]
This protective effect of glial conditioned medium on huntingtin pathology correlates with enhancement of autophagy [31]. The relative sparing of glia from huntingtin related pathology is, therefore, probably related to the production by glia of soluble factors, which diffuse into the glia conditioned medium and enhance authophagy and clearance of mutant huntingtin (Fig. 5.5).
GCM treatment for 15 days activates autophagy pathways. a Representative bands and western blot quantification of LC3 II/LC3 I ratio. The presence of LC3 in autophagosomes and the conversion of LC3 to the lower migrating form LC3-II are used as an indicator of autophagy. b Representative bands and quantification of Beclin-1 antibody, direct marker of the autophagosome formation. c Western blots representative bands and quantification of p-62 specific antibody, as a substrate of autophagy and d LAMP-2 quantification, corrected by b-actin. Values are from the hemibrains, expressed as mean ± SEM (n = 6 in each group). Statistical analysis performed by t student. *p,0.05 GCM vs control CSF group and + p,0.05 right vs left hemispheres. e Representative microphotographs of Lysosomal-associated membrane protein, LAMP-2 A antibody, present in lysosomes and endosomes, which implies autophagy activation. LAMP-2A staining (green) and nuclei (Hoescht, blue) immunofluorescence in striatum of R6/1 mice, with CSF and GCM infusion. (406 magnification, scale bar = 10 mm). f Quantification of number of cells with LAMP-2A positive vesicle distribution in the perinuclear region, around the nucleus cell. Numbers of cells counted in 6 random fields of the striatum at 206 magnification. Mean ± SEM (n = 6 in each group). Statistical analysis performed by student’s t test. *p,0.05, **p,0.01 GCM vs control CSF group; +p,0.05, +++p,0.001 right vs left hemispheres [31]
In summary, there is evidence of impairment of the UPS by mutant huntingtin. It is also clear that cells of patients with HD try to compensate the UPS dysfunction by enhancing autophagy. This strategy could be used therapeutically.
References
Bruyn GW, Went LN. Huntington’s chorea. In: Vinken BG, Vinken PJ, Klawans HL, editors. Handbook of clinical neurology. Amsterdam: Elsevier; 1986. pp. 267–97.
Huntington G. On chorea. In: Butler PSW, editor. The medical and surgical reporter: a weekly journal. 1872. pp. 317–21.
Ruiz PJ, et al. Huntington’s disease: a multidisciplinary study. Eur J Neurol. 1995;2(3):185–91.
Gusella JF, MacDonald ME. Hunting for Huntington’s disease. Mol Genet Med J Neurol. 1993;3:139–58.
Gusella JF, et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306(5940):234–8.
Vonsattel JP, Keller C, Cortes Ramirez EP. Huntington’s disease—neuropathology. Handb Clin Neurol. 2011;100:83–100.
Roze E, et al. Pathophysiology of Huntington’s disease: an update. Rev Neurol (Paris). 2008;164(12):977–94.
Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57(5):369–84.
Ross CA. Huntington’s disease: new paths to pathogenesis. Cell. 2004;118(1):4–7.
Perutz M. Polar zippers: their role in human disease. Protein Sci. 1994;3(10):1629–37.
DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–3.
Gutekunst CA, et al. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci. 1999;19(7):2522–34.
Li X, et al. Inhibiting the ubiquitin-proteasome system leads to preferential accumulation of toxic N-terminal mutant huntingtin fragments. Hum Mol Genet. 2010;19(12):2445–55.
Metzger S, et al. Age at onset in Huntington’s disease is modified by the autophagy pathway: implication of the V471A polymorphism in Atg7. Hum Genet. 2010;128(4):453–9.
Metzger S, et al. The V471A polymorphism in autophagy-related gene ATG7 modifies age at onset specifically in Italian Huntington disease patients. PLoS One. 2013;8(7):e68951.
Fernandez-Estevez MA, et al. Trehalose reverses cell malfunction in fibroblasts from normal and Huntington’s disease patients caused by proteosome inhibition. PLoS One. 2014;9(2):e90202.
Wu JC, et al. The regulation of N-terminal Huntingtin (Htt552) accumulation by Beclin1. Acta Pharmacol Sin. 2012;33(6):743–51.
Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3(6):331–8.
Tsvetkov AS, et al. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A. 2010;107(39):16982–7.
Williams A, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4(5):295–305.
Mealer RG, et al. Rhes, a striatal-selective protein implicated in Huntington disease, binds beclin-1 and activates autophagy. J Biol Chem. 2014;289(6):3547–54.
Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–95.
Reddy PH, et al. Behavioural abnormalities and selective neuronal loss in HD transgenic mice expressing mutated full-length HD cDNA. Nat Genet. 1998;20(2):198–202.
Yu ZX, et al. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. J Neurosci. 2003;23(6):2193–202.
Myers RH, et al. False-negative results with levodopa for early detection of Huntington’s disease. N Engl J Med. 1982;307(9):561–2.
Myers RH, et al. Homozygote for Huntington disease. Am J Hum Genet. 1989;45(4):615–8.
Sapp E, et al. Early and progressive accumulation of reactive microglia in the Huntington disease brain. J Neuropathol Exp Neurol. 2001;60(2):161–72.
Bradford J, et al. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J Biol Chem. 2010;285(14):10653–61.
Shin JY, et al. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol. 2005;171(6):1001–12.
Tydlacka S, et al. Differential activities of the ubiquitin-proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J Neurosci. 2008;28(49):13285–95.
Perucho J, et al. Striatal infusion of glial conditioned medium diminishes huntingtin pathology in r6/1 mice. PLoS One. 2013;8(9):e73120.
Acknowledgements
The authors thankfully acknowledge the support of CIBERNED and CAM grants (PI: Dr. MA Mena) and the help of Mrs. C. Marsden with the editing of the manuscript in English.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Mena, M., Perucho, J., Fernandez-Estevez, M., Yébenes, J. (2015). Autophagy Pathways in Huntington’s Disease. In: Fuentes, J. (eds) Toxicity and Autophagy in Neurodegenerative Disorders. Current Topics in Neurotoxicity, vol 9. Springer, Cham. https://doi.org/10.1007/978-3-319-13939-5_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-13939-5_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-13938-8
Online ISBN: 978-3-319-13939-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)