Abstract
Mathematical modeling has the potential to assist radiofrequency ablation (RFA) of tumors as it enables prediction of the extent of ablation. However, the accuracy of the simulation is challenged by the material properties since they are patient-specific, temperature and space dependent. In this paper, we present a framework for patient-specific radiofrequency ablation modeling of multiple lesions in the case of metastatic diseases. The proposed forward model is based upon a computational model of heat diffusion, cellular necrosis and blood flow through vessels and liver which relies on patient images. We estimate the most sensitive material parameters, those need to be personalized from the available clinical imaging and data. The selected parameters are then estimated using inverse modeling such that the point-to-mesh distance between the computed necrotic area and observed lesions is minimized. Based on the personalized parameters, the ablation of the remaining lesions are predicted. The framework is applied to a dataset of seven lesions from three patients including pre- and post-operative CT images. In each case, the parameters were estimated on one tumor and RFA is simulated on the other tumor(s) using these personalized parameters, assuming the parameters to be spatially invariant within the same patient. Results showed significantly good correlation between predicted and actual ablation extent (average point-to-mesh errors of 4.03 mm).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In radiofrequency ablation (RFA), a probe is placed within the malignant tissue with electrodes at its tip to create heat, thus causing coagulative necrosis at temperatures above \(50\,^{\circ }\mathrm{{C}}\). In order to prevent recurrence, the procedure needs to generate a necrosis that covers completely the tumor, which relies on optimal heat duration and probe placement. The results of RFA are significantly improved by the experience of the clinicians [1]: for the same physician, survival rate of treated patients increased twofolds over a four years period. This learning curve is partly due to the difficult assessment of the cooling effect of the large vessels, porous circulation and blood coagulation, which results in suboptimal ablation and local recurrences in up to 60 % of the cases [2]. To improve the planning of the procedure, computerized simulations have been devised to better predict the extent of the necrosis and eventually modify the probe position or the duration of heating for example. A patient-specific tool showing the extent of ablation given the probe position, the heat duration and patient images will potentially be beneficial in providing a personalized treatment planning and guidance, as it could improve the current clinical outcome. Studies [3–5] have investigated the finite element method (FEM) to simulate the heat transfer on generic human anatomies and eventually optimize the placement of the probes [6]. Simulations with animal-specific [7] or patient-specific anatomies were also recently considered [8] with the inclusion of cooling effects computed from simulated hepatic venous flow and hepatic parenchymal flow. However, nominal tissue parameters were employed in these studies with values often based on ex vivo experiments on animal tissue sometimes with a large varying range between published studies. Because tissue properties are patient-specific and can depend on the current state of the tissue, a proper estimation of those parameters is needed but has been often overlooked. In this paper, we present a computational model of heat transfer and cellular death during RFA which is based on patient-specific tissue parameters and anatomies estimated from CT (Sect. 2). Our framework is adapted to situation where no temperature map is available. We rely on the Lattice-Boltzmann Method (LBM) to compute not only heat diffusion, cellular necrosis as in [8] but also blood and parenchyma flow in the liver tissue. This latter method is based on a computational fluid Dynamics (CFD) solver which incorporates a porous part to deal with the liver parenchyma. This framework is particularly efficient for the personalization as it provides a fast solver and naturally accounts for the flow transition between veins and parenchyma. The model is then personalized based on the first ablation. This information is used to plan subsequent ablation(s) of the same or additional lesions to treat. This can be validated in case of the ablation of multiple tumors inside the liver, assuming that the parameters are spatially invariant within the same patient. In Sect. 3, heat conductivity and porosity were selected as the most sensitive parameters for predicting the necrosis extent. After their estimation on patient specific data, we demonstrate improved prediction accuracy. Sect. 4 concludes the paper.
2 Mathematical Model of RFA Simulation
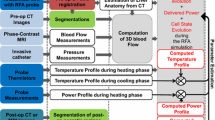
The simulation of heat transfer inside the liver depends on the patient-specific anatomy (Sect. 2.1) and on the blood flow inside the main vessels and the parenchyma considered as a porous medium (Sect. 2.2). The different steps of our method are illustrated in Fig. 1.
2.1 Patient-Specific Liver Anatomy
Volumetric binary images of the parenchyma, tumors, hepatic veins, vena cava, portal vein and the hepatic artery when visible are generated from a semi-automatic segmentation [9] of preoperative CT images. A multi-label mask image is created to identify the different structures of this detailed anatomical model of patient’s liver and circulation tree. To define the computational domain, a level set representation of the liver, without tumor and vessels is computed. From the vessel masks, a porosity map is created to identify the porous parenchyma and the vessels. Because non-visible, the walls of the vessels are extrapolated using 26 connectivities dilatation of the vessels masks. The vessel extremities, which do not have walls are manually identified.
2.2 Hepatic Blood Flow Computation
Model Description. The blood in the main vessels and in the parenchyma are combined in the generalized 3D incompressible Navier-Stokes equation for fluid flow in porous media, thus considerably easing the definition of boundary conditions between vessels and parenchyma, improving on [8]. More precisely, writing \(\mathbf {v}\) as the blood velocity and \(p\) the pressure inside the liver, we solve:
The added force \(\mathbf {F}\) represents the total body force due to the presence of a porous medium [10]. F depends on the porosity coefficient \(\epsilon \) (fraction of blood volume over the total volume) whose default values are 1 in the CT-visible vessels, 0.1 in the porous parenchyma [7], and 0.04 in the vessel walls to model an impermeable medium. Experiments have been performed to obtain a sufficiently small porosity (0.04) to avoid flowing through the vessel wall. At the border of the liver, no flux boundary conditions are used whereas Dirichlet boundary conditions are applied at the inlets of portal vein and vena cava and at the outlet of the vena cava: the portal vein and vena cava inflow, \(\varphi _p\) and \(\varphi _{vc_{in}}\) are fixed as well as the vena cava outlet pressure \(p_0\) (see Fig. 2 for more details). This method makes the boundary conditions simple to treat: no boundary conditions are fixed on the extremities of the vessels inside the parenchyma thanks to the use of the porosity map. This framework mainly avoids the occurrence of shear stress on the vessel walls due to their much lower value of porosity. Figure 2 illustrates flows calculated in a patient-specific geometry.
Implementation using LBM. Equation 1 is solved using Lattice Boltzmann Method (LBM) for fast computation on general purpose graphics processing units (GPU). LBM has been developed for CFD and is now a well-established discretization method. In RFA, it has been validated in [8] through a comparison with an analytical solution, for a similar accuracy as FEM. An isotropic Cartesian grid with 19-connectivity topology and Neumann boundary conditions is employed. A Multiple-Relaxation-Time (MRT) model is used for increased stability [11]. At position \(\mathbf {x}\) for the edge \(\mathbf {e}_i\), the governing equation is: \(\mathbf {f}(\mathbf {x}+\mathbf {e_i} \varDelta x,t+\varDelta t) = \mathbf {f}(\mathbf {x},t)+ \mathbf {A} [\mathbf {f}^{eq} (\mathbf {x},t)-\mathbf {f}(\mathbf {x},t)]+\varDelta t \mathbf {g}(\mathbf {x},t).\) In this equation, \(\mathbf {f}(\mathbf {x})={\{f_i(\mathbf {x})\}}_{i=1..19}\) is the vector of distribution function with \(f_i(\mathbf {x})\) being the probability of finding a particle travelling along the edge \(\mathbf {e}_i\) of the node \(\mathbf {x}\) at a given time; \(c= \varDelta x/\varDelta t\); \(c_s^2 = 1/4\); \(\varDelta x\) is the spacing; \(f^{eq}_i(\mathbf {x},t)=\omega _i\rho [1+\frac{\mathbf {e_i} . \mathbf {v}}{c c_s^2}]\); \(g_i(\mathbf {x},t)=\omega _i\rho \frac{\mathbf {e_i} . \mathbf {F}}{c_s^2}\), \(\varvec{\omega } = {\{\omega _i\}}_{i=1..19} \) is the vector of weighting factors and \(\mathbf {A}\) the MRT matrix. The fluid mass density and velocity are computed from the LBM distributions as \(\rho = \sum ^{19}_{i=1} f_i(\mathbf {x},t)\) and \(\rho \mathbf {v} = \sum ^{19}_{i=1} \mathbf {e_i} f_i(\mathbf {x},t)+\frac{\varDelta t}{2} \rho \mathbf {F}\) and are updated at every node of the grid for every timestep \(\varDelta t\).
2.3 Model of Heat Transfer and Cellular Necrosis in Liver Tissue
This model describes how the heat flows from the probe through the liver while accounting for the cooling effect of the main vessels and parenchyma. Its main parameters are then optimized to match the observed extent of the necrosis. Assuming blood vessels and surrounding tissue isolated from each other, the temperature is computed by solving the diffusion equation:
everywhere in the domain, to which we add either the cooling term \(H (T_{b0}-T)/(1-\epsilon )\) when a point belongs to a large vessel, where blood velocity is high, (Pennes model [12]) or \(-\epsilon \rho _b c_b \mathbf {v}\cdot \nabla T/(1-\epsilon )\) when it belongs to the parenchyma, where tissue is dominating (WK model [13]). \(T\), \(Q\), \(\mathbf {v}\) and \(T_{b0}\) stand for temperature, source term, blood velocity and the mean temperature (assumed constant) of the blood in large vessels. A weakly coupling model is considered: the blood flow has an influence on the temperature distribution through the WK model but the temperature does not affect the blood flow (coagulation is not considered here), which allows us to speed up the calculations since the blood flow distribution is computed only once, at the beginning of the simulation. The discretization of the bioheat equation also relies on LBM using a no-flux boundary condition on the liver boundary defined as a level-set function. Tissue necrosis is computed using a 3-state model based on the simulated temperatures [14] using Eq. 3, where \(k_{f}, k_{b}\) are the damage and recovery rate respectively.
2.4 Parameter Estimation
As parameters in the heat transfer and cellular necrosis equation are customarily taken from the literature, we aim to personalize them given the observed extent of the necrotic region measured post-operatively as temperature maps are not readily available. Most of the parameters are defined as constant whereas the heat capacity \(c_t\) and conductivity \(dt=\bar{dt}*(1+0.00161)*(T-310) W(m K)^{-1}\) are temperature dependent and therefore spatially distributed [8]. Using DAKOTAFootnote 1, we first perform a sensitivity analysis to know which parameters mostly influence the volume and the point-to-mesh error [15] of the computed necrosis area. Then, we optimize the most sensitive ones: the heat conductivity and the porosity, as to minimize the average point-to-mesh error between computed and observed necrotic region. To this end, we use a gradient-free optimization method, the Constrained Optimization BY Linear Approximations (COBYLA), which required only a few numbers of forward simulations.
3 Experiments and Results
All experiments were executed on a Windows 7 desktop machine (Intel Xeon, 2.80 GHz, 45 GB RAM, 24 CPUs) with a Nvidia Quadro 6000 1.7 GB (448 CUDA cores).
3.1 Sensitivity Analysis
We want to know the sensitivity of four uncertain parameters of our model: \(\bar{dt}\), H, \(\bar{k_f}\), \(\epsilon \) on the volume of the computed necrosis but also on the point-to-mesh error between the computed necrosis area and the one computed with the nominal parameters from the literature. To this end, a synthetic case has been setup to speed-up the process (Fig. 3). The range of parameters values used [3] are reported in Table 1. These parameters of interest were modelled with a uniformly distributed uncertainty, and the sensitivity analysis was performed using variance based decomposition to compute the global sensitivity indices (so-called Sobol indices). \(\bar{dt}\) has the largest total effect (0.58) as compared with \(\bar{k_f}\), H and \(\epsilon \) (0.16, 0.15, 0.43) on the volume of the lesion, whereas \(\epsilon \) and \(\bar{dt}\) have the same larger total effect (0.37) as compared with \(\bar{k_f}\) and H (0.16, 0.35) on the point-to-mesh error with respect to lesion obtained with nominal values. As it is not reasonable to try to estimate all of these four parameters at once, we decided to estimate only two of them: we chose \(\bar{d_t}\) for its effect on the volume, as the nominal value of \(\bar{k_f}\) [14] seemed accurate and \(\epsilon \). H was chosen large enough to maintain a constant temperature of \(37\,^{\circ }\mathrm{C}\) in the CT-visible vessels. We fixed the other parameters to nominal values for the personalization on patient data.
3.2 Verification of the Optimization Framework
In order to confirm the accuracy of the optimization framework, we considered a synthetic case on a regular cuboid domain where all the phenomena occurring during RFA were present (diffusion, reaction and advection) and with nominal parameters of tissue properties. First we simulated a necrotic area with generic parameters by emulating the clinical RFA protocol: during 7 min, we heated at \(105\,^{\circ }\mathrm{C}\), the simulation continued for 3 more minutes without heating so that each cell reached a steady state. Then, the main parameters: \(\bar{d_t}\) and \(\epsilon \) are estimated by minimizing the mean of the point-to-mesh error between the computed necrotic region and the one created at the first step. We managed to obtain the estimated parameters with 6.1 % of error on \(\bar{d_t}\) and 2.1 \(\%\) on \(\epsilon \) in 32 min after 36 iterations with a mean of the point-to-mesh error of \(10^{-3}\) mm.
3.3 Evaluation on Patient Data
We evaluated our model on 3 patients, with 7 ablations (several tumors ablated for each patient) for whom pre- and post-operative CT images were available. Clinical RFA protocol was simulated: the probe was deployed within the tumor and cells in a diameter defined pre-operatively around the center of the tumor probe tip were heated at \(105\,^{\circ }\mathrm{C}\) during 7 min or 2 times 7 min. The diameter and heat duration were iteratively increased according to the size of the tumor. The simulation continued for 3 more minutes without any heat source so that each cell reached a steady state. The parameters were considered spatially invariant within the same patient. Nevertheless, the cell death model locally changes the properties of the tissue, and different parameters are related to different location inside the liver (\(H\) and \(\epsilon \) are related to large vessels, and parenchyma respectively), but we consider that they have a constant value. The parameters are estimated on one tumor by reducing the error with the ground-truth. Then, we computed the cell death area of the other tumor(s) of this patient with the personalized parameters. The computed cell death area compared qualitatively well with the observed post-operative necrosis zone for tumor located at different place inside the liver, close to large vessels, on the border (Fig. 4, the predicted lesion was manually segmented by an expert and registered to preoperative image). Quantitatively, average point-to-mesh errors (Table 2) were within tolerance in clinical routine for the four tumors estimated with the personalization of the main biological parameters, as the probes can be deployed in steps of 1 cm. The estimated heat conductivities were lower than the nominal value (0.512 W(m K)\(^{-1}\)), whereas the porosity was very close (0.1). Other experiments on patient 1 showed also a significant improvement of the correlation between predicted and actual ablation extent compared to the prediction using only nominal parameters (average point-to-mesh errors of 4.44 mm vs 4.98 mm, average Dice score of 72.0 % vs 68.5 %). Patient 2 presents a quite large Dice difference between the two cases. It might be due to segmentation or registration issues, and potentially to model limitations (assumptions, etc.), which will be further investigated in a pre-clinical setup. The main parameters are first considered with nominal values taken from the literature and then optimized to match the observed extent of the necrosis. Current errors can be explained by segmentation and registration processes but also by the limited number (2) of personalized parameters.
4 Discussion and Conclusion
The personalization of the sensitive tissue parameters allows to have a better estimation of the necrosis and to predict the outcome of RFA in case of multiple tumors inside the liver. As our framework totally rely on LBM, no advanced meshing techniques are required. All the computations are directly done from patient images: heat propagation and cell death modeling as well as the heat sink effect of blood vessels and porous circulation in the liver. The coupled computation of the porous and hepatic flow eliminates the difficulties in the setting of boundary conditions in the parenchyma, and the occurence of shear stress on the vessels wall is avoided as the Pennes Model is used in the big vessels where the flow is not accounted for. The current method needs several tumors for validation and is worth using only when no temperature maps are available, but it could be easily translated into clinical settings. Adaptation for RFA under image-guidance is considered: RFA procedure is usually done in several steps (increase in probe diameters for example). An intra-operative image is acquired at the end of the first step and used to personalize key parameters, providing a powerful guidance tool. No post-operative images are required. A necessary step before deploying this method in clinical settings is a pre-clinical validation with extensive data on larger populations to evaluate the computational model of RFA and to consider potential safety issue of the proposed application. Even if promising results are achieved with the use of patient-specific parameters, the impact of possible biases in the post- to the pre-operative image registration like the impact of the average simulation of the probe need to be investigated as well as the sensitivity of the results with respect to segmentation.
Notes
- 1.
http://dakota.sandia.gov - multilevel framework for sensitivity analysis.
References
Hildebrand, P., Leibecke, T., Kleemann, M., Mirow, L., et al.: Influence of operator experience in radiofrequency ablation of malignant liver tumours on treatment outcome. Eur. J. Surg. Oncol. (EJSO) 32, 430–434 (2006)
Kim, Y.S., Rhim, H., Cho, O.K., Koh, B.H., Kim, Y.: Intrahepatic recurrence after percutaneous radiofrequency ablation of hepatocellular carcinoma: analysis of the pattern and risk factors. Eur. J. Radiol. 59, 432–441 (2006)
Altrogge, I., Preusser, T., Kroger, T., Haase, S., Patz, T., Kirby, R.M.: Sensitivity analysis for the optimization of radiofrequency ablation in the presence of material parameter uncertainty. Int. J. Uncertain. Quantification 2(3), 295–321 (2012)
Chen, X., Saidel, G.M.: Mathematical modeling of thermal ablation in tissue surrounding a large vessel. J. Biomech. 131, 011001 (2009)
Jiang, Y., Mulier, S., Chong, W., Diel Rambo, M., et al.: Formulation of 3D finite elements for hepatic radiofrequency ablation. IJMIC 9, 225–235 (2010)
Kröger, T., Pätz, T., Altrogge, I., Schenk, A., et al.: Fast estimation of the vascular cooling in RFA based on numerical simulation. Open Biomed. Eng. J. 4, 16–26 (2010)
Payne, S., Flanagan, R., Pollari, M., Alhonnoro, T., et al.: Image-based multi-scale modelling and validation of radio-frequency ablation in liver tumours. Philos. T. Roy. Soc. A 369, 4233–4254 (2011)
Audigier, C., Mansi, T., Delingette, H., Rapaka, S., Mihalef, V., Sharma, P., Carnegie, D., Boctor, E., Choti, M., Kamen, A., Comaniciu, D., Ayache, N.: Lattice Boltzmann method for fast patient-specific simulation of liver tumor ablation from CT images. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013, Part III. LNCS, vol. 8151, pp. 323–330. Springer, Heidelberg (2013)
Criminisi, A., Sharp, T., Blake, A.: GeoS: geodesic image segmentation. In: Forsyth, D., Torr, P., Zisserman, A. (eds.) ECCV 2008, Part I. LNCS, vol. 5302, pp. 99–112. Springer, Heidelberg (2008)
Guo, Z., Zhao, T.: Lattice-boltzmann model for incompressible flows through porous media. Phys. Rev. E 66, 036304 (2002)
Pan, C., Luo, L.S., Miller, C.T.: An evaluation of lattice boltzmann schemes for porous medium flow simulation. Comput. Fluids 35, 898–909 (2006)
Pennes, H.H.: Analysis of tissue and arterial blood temperatures in the resting human forearm. J. Appl. Physiol. 85, 5–34 (1998)
Klinger, H.: Heat transfer in perfused biological tissue I: general theory. B. Math. Biol. 36, 403–415 (1974)
O’Neill, D., Peng, T., Stiegler, P., Mayrhauser, U., et al.: A three-state mathematical model of hyperthermic cell death. Ann. Biomed. Eng. 39, 570–579 (2011)
Zheng, Y., Barbu, A., Georgescu, B., Scheuering, M., Comaniciu, D.: Fast automatic heart chamber segmentation from 3d CT data using marginal space learning and steerable features. In: IEEE 11th International Conference on Computer Vision, 2007, ICCV 2007, pp. 1–8. IEEE (2007)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this paper
Cite this paper
Audigier, C. et al. (2014). Parameter Estimation for Personalization of Liver Tumor Radiofrequency Ablation. In: Yoshida, H., Näppi, J., Saini, S. (eds) Abdominal Imaging. Computational and Clinical Applications. ABD-MICCAI 2014. Lecture Notes in Computer Science(), vol 8676. Springer, Cham. https://doi.org/10.1007/978-3-319-13692-9_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-13692-9_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-13691-2
Online ISBN: 978-3-319-13692-9
eBook Packages: Computer ScienceComputer Science (R0)







