Abstract
The open-ended microbial diagnostic approaches such as the complete or partial sequencing of the 16S ribosomal gene by Sanger sequencing or by pyrosequencing have provided new insights into the diversity of the oral microbiota. These techniques have recently been used to evaluate the microbiota associated with osseointegrated implants and these results have expanded the knowledge on the diversity of the microbial communities associated with peri-implantitis. Taken together, the results of these studies suggest that the diversity of the microbial community of peri-implantitis and periodontitis might not be as similar as previously thought. Although certain known periodontal pathogens may also be associated with the etiology of peri-implantitis, apparently there were many differences between these two clinical conditions, involving distinct microorganisms. Further investigations on the diversity of peri-implant microbiota would be essential in order to define effective preventive and therapeutic strategies for peri-implantitis. It is also important to standardize laboratory protocols to make the results of the open-ended diagnostic techniques based on PCR amplification more comparable throughout the different research groups.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
5.1 Introduction
Peri-implant diseases are characterized by the presence of an inflammatory process that affects the peri-implant tissues under loading (Mombelli and Lang 1998; Heydenrijk et al. 2002). The signs of this infection vary from a mild inflammatory process of the peri-implant mucosa, including bleeding on probing and suppuration, to clinical attachment and bone loss (Heitz-Mayfield 2008; Zitzmann and Berglundh 2008). Similarly to periodontal diseases, peri-implant diseases result from a disruption in host-compatible/pathogenic microorganisms that may lead to two specific clinical situations: peri-implant mucositis, which is a lesion restricted to the peri-implant soft tissue, and therefore reversible; and peri-implantitis, which affects the soft tissue and the bone tissue adjacent to the osseointegrated dental implant (Mombeli 1999; Zitzmann and Berglundh 2008) (Fig. 5.1). Recent evidence has indicated that peri-implant mucositis may affect 63.4 % of subjects and 30.7 % of implants, and peri-implantitis 18.8 % of subjects and 9.6 % of implants (Atieh et al. 2013)
Microbiological studies have shown that the biofilm associated with implant failures differs substantially from that of healthy implants (Mombelli and Mericske-Stern 1990; Sanz et al. 1990; Leonhardt et al. 1999; Hultin et al. 2002; Quirynen et al. 2006; Renvert et al. 2007; Shibli et al. 2008). In humans, the subgingival biofilm around dental implants with clinically healthy marginal peri-implant tissues have demonstrated a microbiota with high proportions of coccoid cells, low proportions of anaerobic and Gram-negative species and a low prevalence of periodontal pathogens (Mombelli et al. 1987; Lee et al. 1999; Renvert et al. 2007; Shibli et al. 2008). In contrast, a peri-implantitis pocket seems to harbor a microbiota similar to that found in periodontitis, with high levels and proportions of Porphyromonas gingivalis, Prevotella intermedia, Prevotella nigrescens, Tannerella forsythia and Aggregatibacter actinomycetemcomitans (Becker et al. 1990; Mombelli 1993; Hultin et al. 2002; Leonhardt et al. 2003a, b; Quirynen et al. 2006; Shibli et al. 2008; Kumar et al. 2012). Therefore, it is assumed that the same pattern of colonization that occurs in healthy periodontal tissues or in periodontal disease may also occur around the subgingival surface of dental implants. This is a relevant piece of information and has direct clinical implications since the treatments proposed for peri-implant diseases have been based on this microbial similarity.
The introduction of open-ended microbial diagnostic approaches in the early 2000s, such as complete or partial sequencing of the 16S ribosomal gene by Sanger sequencing or by pyrosequencing have provided new insights into the diversity of the oral microbiota associated to periodontal health and disease (Paster et al. 2001; Faveri et al. 2008; Shchipkova et al. 2010). These techniques have recently been used to evaluate the microbiota associated with osseointegrated implants and these results have expanded the knowledge on the composition of these microbial communities.
This chapter presents a current overview of the composition of the biofilms associated with peri-implantitis, with focus on current knowledge about the diversity of these biofilms, based on the results of the studies that have used cutting-edge open-ended approaches. In addition, a brief discussion regarding the strengths and weaknesses of these new diagnostic techniques is also presented. This body of information might help to understand the shifts occurring in the composition of peri-implant biofilm structure that may lead to the development of peri-implantitis.
5.2 Microbial Profile in Peri-Implantitis
Several authors have used different microbiological techniques to study the role of bacterial biofilm in peri-implant diseases in humans (Pontoriero et al. 1994; Lee et al. 1999; Hultin et al. 2002; Renvert et al. 2007; Persson and Renvert 2013) and in animal models (Eke et al. 1998; Tillmanns et al. 1998; Shibli et al. 2003; Charalampakis et al. 2014). Most of these studies have demonstrated a clear relationship between some specific bacterial species and peri-implant mucositis or peri-implantitis (Hultin et al. 2002; Renvert et al. 2007; Persson and Renvert 2013). Indeed, after the implant surface has been exposed to the oral cavity a complex subgingival microbiota is established in a ‘pristine’ peri-implant pocket within 1–2 weeks, and apparently, the stability of the biofilm community is reached after 3 months (Quirynen et al. 2006).
Early studies characterizing the microbiota of healthy implants by dark field microscopy, described coccoid bacteria as the main morphotype, with a low proportion of spirochetes, fusiforms and motile and curved rods (Sanz et al. 1990; Mombelli and Mericske-Sterm 1990; Silverstein et al. 1994). These results were corroborated by culture techniques that described high levels of Gram-positive facultative cocci, Actinomyces and Veillonella spp., low total anaerobic rods and a low frequency of detection of Fusobacterium spp. and “Black-pigmented Bacteroides” (Leonhardt et al. 1999; Hultin et al. 2002). Therefore, these studies indicated that the microbiota colonizing clinically healthy implants was quite similar to that associated with healthy periodontal sites in periodontally healthy subjects.
Several studies have also compared the microbiota of healthy and diseased implants. Mombelli et al. (1987) described that the microbiota of peri-implantitis sites presented much higher levels of motile rods, spirochetes and fusiforms than that of healthy implants. In another study, subgingival biofilm samples taken from implants with peri-implantitis in 37 subjects, and from healthy implants in another 51 subjects were compared using culture methods (Leonhardt et al. 2003a). The authors analyzed the prevalence of P. intermedia, A. actinomycetemcomitans, P. gingivalis and Staphylococcus ssp. In the group not affected by peri-implantitis, P. intermedia was detected in 26 % of subjects and P. gingivalis in 2 %, as opposed to 66 and 31 % of the subjects presenting peri-implantitis. The prevalence of A. actinomycetemcomitans was low in both groups, around 3 %. In addition, Staphylococcus spp. were only found in peri-implantitis, in 17 % of the subjects.
Checkerboard DNA-DNA hybridization has been also used to examine the microbial profiles of supra- and subgingival biofilms in subjects with and without peri-implantitis (Shibli et al. 2008). Higher mean counts of P. gingivalis, Treponema denticola and T. forsythia (red complex species) were found in supra- and subgingival biofilms of subjects with peri-implantitis. In addition, the proportions of the pathogens from the red complex were elevated, while host-compatible beneficial microorganisms, such as the Actinomyces species, were reduced in diseased compared with healthy implants (Fig. 5.2). The microbiological profiles of supra- and subgingival environments did not differ substantially within healthy or diseased implants. Persson and Renvert (2013) analyzed the levels of 78 bacterial species from 166 implants with peri-implantitis and 47 healthy implants. Nineteen bacterial species were found at higher counts in peri-implantitis, including A. actinomycetemcomitans, Campylobacter gracilis, Campylobacter rectus, Campylobacter showae, Helicobacter pylori, Haemophilus influenzae, P. gingivalis, Staphylococcus aureus, Staphylococcus anaerobius, Streptococcus intermedius, Streptococcus mitis, T. forsythia, T. denticola and Treponema socranskii. The authors suggested that a cluster of seven bacterial species could be associated with peri-implantitis. The total bacterial load for these seven species (T. forsythia, P. gingivalis, T. socranskii, S. aureus, S. anaerobius, S. intermedius, and S. mitis) was approximately four times higher in implants with peri-implantitis (6.5 × 105) than in the healthy ones (1.8 × 105).
Bar stacks of the mean proportions of each microbial complex in supra-and subgingival plaque samples taken from 22 subjects with a healthy implant and 22 subjects with peri-implantitis. The percentage of DNA probe counts for each species was determined at each site and then across subjects in each group. Species in the complexes were summed and the proportions of which each complex were comprised were determined. The colors represent the different complexes described by Socransky et al. (1998). The grey color represents species that do not fall into any complex. The significance of differences in mean proportions was sought using the Mann–Whitney U-test (Data adapted from Shibli et al. 2008)
Overall, the available data on the composition of subgingival biofilm associated with peri-implantitis indicate elevated levels of certain bacterial species previously associated with periodontitis. However, other microorganisms not commonly implicated as etiological agents of periodontal diseases have also been detected in samples from peri-implantitis lesions, such as Enterobacter aerogenes, Enterobacter cloace, Escherichia coli, H. pylori, Peptostreptococcus micra, Pseudomonas spp, Candida spp, S. aureus and Staphylococcus epidermidis (Leonhardt et al. 1999; Mombelli and Décaillet 2011; Persson et al. 2010; Persson and Renvert 2013; Rams et al. 2013; Heitz-Mayfield and Lang 2010). Furthermore, in vitro studies have demonstrated that S. aureus has an affinity for titanium surfaces (Harris et al. 2007; Hudetz et al. 2008), which might indicate a specific role of this species in the etiology of peri-implantitis.
In summary, the overall results of the studies using culture and targeted molecular diagnostic techniques suggest that peri-implantitis is associated with a specific mixed microbiota that presents several similarities with the microbial profile associated with periodontal infections, as well as some other microorganisms not commonly associated with the etiology of periodontitis, such as Staphylococcus spp. and Enterobacteriaceae.
5.3 The Role of Open-Ended Molecular Diagnostic Tests in the Study of Oral Biofilm Diversity
During the last decade, a great progress has been made as regards the application of novel molecular microbiological methods in the studies of the oral microbiota. The cutting-edge open-ended molecular techniques allow for genome mapping of the entire microbial spectrum in a sample, and provide comprehensive characterization of both the cultivable and not-yet-cultivable microbiota associated with periodontal health and disease. These techniques allow an overview of the microbial communities as a whole, which represents an important advantage over culture and even over other molecular targeted methods, such as specific-specific polymerase chain reaction (PCR), DNA probes and microarrays (Hiyari and Bennett 2011; Wade 2011). The large body of information derived from these sequencing techniques has revealed new species that could act as pathogens in several oral infections (Paster et al. 2001; Faveri et al. 2008; Kumar et al. 2012), including peri-implantitis (Kayanagi et al. 2010, 2013; da Silva et al. 2014; Kumar et al. 2012). To date, the microbial diversity of peri-implantitis has been investigated using PCR-Denaturing Gradient Gel Electrophoresis (DGGE)-Sanger sequencing, PCR-cloning-Sanger sequencing and Next generation sequencing technologies, more specifically, pyrosequencing.
From 2001 to 2010, Sanger sequencing was the most widely used DNA sequencing method for studying the microbial diversity of the oral biofilm (Paster et al. 2001; Kumar et al. 2006; Faveri et al. 2008; Shchipkovaet al. 2010). Several studies published in the 1990s indicated that sequencing of the small ribosomal subunit gene (16S rDNA) could be useful for microbial identification (Weisburg et al. 1991; Green and Giannelli 1994; Cilia et al. 1996). This gene is the most common molecular marker used for identification and classification of prokaryotes due to its essential function, ubiquity and evolutionary properties (Case et al. 2007). It allows the detection and identification of a microorganism at a species level, or below, which is a crucial step while trying to understand etiology and treatment of human infections. Therefore, the construction and analysis of ribosomal gene libraries is a very important tool for studying microbial ecology.
Sanger sequencing is considered a chain-terminator method because DNA fragments of varying lengths are synthesized by incorporating nucleotides and dideoxy terminators (deoxyribonucleotide triphosphates [dNTPs] and dideoxynucleotide triphosphates [ddNTPs], respectively). Random incorporation of the ddNTPs causes chain termination that produces DNA fragments of every possible length. In a more recent modification, each ddNTP (A, C, T, or G) carries a unique fluorescent molecule, so that the extension products are both terminated and labeled with the appropriate fluorophore (Sanger et al. 1977; França et al. 2002). Terminated products must be purified from unincorporated ddNTPs, and the fragments are subsequently separated by size using capillary electrophoresis, in which the terminal nucleotide of each fragment is detected by fluorescence at wavelengths unique to each of the terminators (Prober et al. 1987). The read lengths for Sanger sequencing have increased in length, and 500–800 base reads can now be achieved routinely (França et al. 2002).
Although a large body of phylogenetic data for microbial identification has been generated via Sanger sequencing, new sequencing technologies that offer a series of additional benefits have emerged recently. One of these new sequencing technologies, pyrosequencing, is faster and more cost-effective than Sanger sequencing (Rastogi et al. 2013; Harrington et al. 2013) and allows thousands to hundreds of thousands of sequence reads to be generated in a single run (Harrington et al. 2013). For this method, specific genetic targets, such as hypervariable regions within bacterial 16S rDNAgenes may be amplified by PCR and subjected to DNA pyrosequencing. Sequencing by synthesis occurs through a DNA polymerase-driven generation of inorganic pyrophosphate, with the formation of ATP and ATP-dependent conversion of luciferin into oxyluciferin. The generation of oxyluciferin causes the emission of light pulses, and the amplitude of each signal is directly related to the presence of one or more nucleosides (Petrosino et al. 2009). Pyrosequencing is fundamentally different from Sanger sequencing in that bioluminescence results from strand elongation in real time, whereas, with Sanger sequencing, fluorescence is detected as a separate step after chain termination (Harrington et al. 2013). At present, pyrosequencing technology produces the longest reads of the next-generation sequencing platforms at approximately 700 pb (Harrington et al. 2013).
Sanger sequencing and pyrosequencing are powerful methods for evaluating oral biodiversity; however, DNA extraction and PCR amplification have been reported to be potential sources of biases associated with these techniques (Diaz et al. 2012; Abusleme et al. 2014). The understanding of possible limitations, intrinsic bias and inherent variability of the different diagnostic methods is crucial for the proper evaluation and interpretation of the results of the various studies. Diaz et al. (2012) evaluated the possible bias of DNA isolation and PCR amplification of 454-sequencing of 16S rDNA gene (Fig. 5.3). The authors used three different laboratory-created samples (mocks) of seven bacterial species (Streptococcus oralis, Streptococcus mutans, Lactobacillus casei, Actinomyces oris, Fusobacterium nucleatum, P. gingivalis and Veillonella sp.). Mock 1 contained equal numbers of 16S rDNA molecules, mock 2 equal numbers of cells and mock 3 unequal numbers of cells of these seven bacterial oral species. In theory, no difference in the number of reads of these species would be expected in mock 1, since they comprised equal amounts of genomic DNA for each species. On the other hand, mocks 2 and 3 could potentially be affected either by some bias of the PCR or sequencing processes or the cell lysis procedures. However, mock 1 did not show the estimated results, as F. nucleatum produced a higher number of reads and A. oris and L. casei a lower number of reads than expected. In addition to being under-represented in mock 1, A. oris and L. casei were also under-represented in mocks 2 and 3, which could be due to some PCR bias. Both S. mutans and P. gingivalis were shown in lower abundance than expected only in mocks 2 and 3, suggesting that these species were less effectively lysed. Other research groups have also observed some of these biases associated with the Sanger or pyro-sequencing techniques (de Lillo et al. 2004; Abusleme et al. 2014).
Pie charts of the accuracy of 16S ribosomal RNA (rRNA) amplification followed by 454-pyrosequencing in estimating abundance of species. Mock 1 contained equal numbers of 16S rRNA molecules, mock 2 equal numbers of cells and mock 3 unequal numbers of cells of these seven bacterial oral species. Expected numbers of sequence reads for mocks 2 and 3 were normalized according to the number of 16S rRNA copies in the genome of each organism (Data adapted from Diaz et al. 2013)
The results of the above-mentioned studies suggest that although pyrosequencing is a powerful technique for investigating the oral microbial diversity, the abundance of species is subject to empirical bias introduced through the methods used for DNA isolation and amplification. Investigators should be aware of these limitations in order to minimize technical errors by accounting for them while designing the studies and evaluating their data.
5.4 A Current Overview on the Microbial Diversity of Peri-Implantitis
Kayanagi et al. (2010) were the first to explore the microbial diversity of the subgingival biofilm around dental implants with different clinical conditions, by 16s rDNA PCR-cloning-Sanger sequencing. The authors selected three subjects that presented at least one healthy implant and one with peri-implantitis, as well as teeth with periodontitis. A total of 112 different taxa were identified from 335 sequenced clones sequenced. Among these taxa, 46 % (51 phylotypes) were not-yet cultivable and 20 % (22 phylotypes) were novel. The number of species detected in the subgingival biofilm of peri-implantitis, periodontitis and periodontally healthy sites was 77, 57 and 12, respectively. Some bacterial phyla, such as Chloroflexi, Tenericutes and Synergistetes were only detected at peri-implantitis sites, together with some species belonging to the Firmicutes phyla, such as Parvimonas micra, Peptostreptococcus stomatis, Pseudoramibacter alactolyticus and Solobacterium moorei. Interestingly, some bacterial species that have previously been associated with peri-implantitis, such as P. gingivalis and A. actinomycetemcomitans were found in low proportions in this study. However, due to the small sample size this study was unable to establish any type of association between taxa detected and clinical status. More recently, these authors (Kayanagi et al. 2013) continued to explore the microbial diversity of sites with periodontitis and peri-implantitis by adding three new subjects to the previously conducted study (Kayanagi et al. 2010). After screening 799 clones, a total of 333 species were identified, 63 % were not-yet cultivable and 23 % were novel. One hundred and ninety two species were detected in peri-implantitis and 142 in periodontitis. The most abundant phyla in both clinical groups were Firmicutes and Bacteroidetes, while Chloroflexi and Deferribacteres were only detected in peri-implantitis. Dialister spp., Eubacterium spp. and Porphyromonas spp. were more prevalent at peri-implantitis than periodontitis sites. According to the previous publication (Kayanagi et al. 2010), P. micra, P. stomatis, P. alactolyticus and S. moorei were limited to peri-implantitis sites and the most abundant species found among all samples was F. nucleatum. Interestingly, the authors described that the microbial composition of peri-implantitis was more diverse than that of periodontitis.
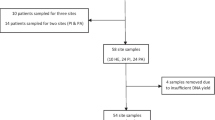
Recently, our research group used cloning and Sanger sequencing (da Silva et al. 2014) to investigate the microbial diversity of healthy implants placed in a group of subjects who had no diseased implants (Control group; n = 10) with that of healthy and diseased implants from another group of subjects (Test group; n = 10). The phylogenetic identity of 1387 16S rRNA gene clones was determined. Uncultivated phylotypes accounted for an average of 32.1 and 35.8 % of the taxa recovered from healthy implants in the Control and Test groups, respectively, and of 41.2 % from peri-implantitis. Higher proportions of some recognized periodontal pathogens from the orange complex (Socransky et al. 1998), such as F. nucleatum, P. micra, P. intermedia and C. gracilis were found in peri-implantitis sites (Table 5.1). Moreover, these sites presented significantly higher percentages of clones from de genera Desulfobulbus, Dialister, Filifactor, Fusobacterium, Mitsuokella and Porphyromonas in comparison with healthy implants. The biofilm associated with peri-implantitis harbored more pathogenic bacterial species from the orange complex, and other “unusual” putative periodontal pathogens, such as Filifactor alocis, Dialister invisus and Mitsuokella sp. HOT 131 in comparison with the healthy implants. Al-Radha et al. (2012) also found higher proportions of some species from the orange complex, such as Fusobacterium spp. and Prevotella ssp. in peri-implantitis sites using PCR-DGGE followed by Sanger sequencing. The authors also described that these species were more prevalent in the early stages of disease, whilst an increased diversity of species was present during the more advanced stages of disease.
Studies using these culture-independent techniques have also suggested that the Archaea domain might be associated with some oral infections, including periodontitis (Lepp et al. 2004; Matarazzo et al. 2011) and endodontic infection (Vianna et al. 2006). Therefore, it has been hypothesized that this domain could also have some association with the etiology of peri-implantitis. In 2011, we (Faveri et al. 2011) studied the prevalence and levels or Archaea in a group of 50 subjects presenting only healthy implants (Control, n = 25) or both healthy implants and peri-implantitis (Test, n = 25). In the peri-implantitis group, Archaea were detected in 48 %, 16 % and 8 % of diseased implants, healthy implants and teeth, respectively. Implants with peri-implantitis presented a significantly higher prevalence of Archaea in comparison with healthy implants and natural teeth. Methanobrevibacter oralis was the most prevalent phylotype and was detected in all Archaea positive samples, representing 92 % of the clones identified in the Control group, and 95.3 % of those identified in the Test group. The results of this study suggested an increased prevalence of Archaea in peri-implantitis sites, mainly the species M. oralis, in comparison with the healthy implants. Although these data do not necessarily denote that the Archaea domain has a direct function in tissue destruction, they suggest a possible role of this domain in the etiology of peri-implantitis. One possibility is that species from the Archaea domain may alter the ecosystem to a more anaerobic environment, which in turn would stimulate the further growth of strict anaerobes species, represented not only by methanogens but also by the members of the red complex, T. forsythia, T. denticola and P. gingivalis, as well as species of the orange complex.
Some authors have used the pyrosequencing technology to study the structure of the bacterial community associated with peri-implant health and disease (Kumar et al. 2012). Kumar et al. (2012) allocated 40 subjects in 4 clinical groups of 10 subjects each, as follows: peri-implantitis, healthy implants, chronic periodontitis, and periodontal health. The sequences represented 370s-OTUs and 84 genus level OTUs that were catalogued into the phyla Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, Spirochaetes, Candidatus Saccharibacteria (Syn. Candidate division TM7), Candidate division Sulphur River 1 (OP11) and Synergistes. Uncultivated phylotypes accounted for an average of 52.6 % 44.6 %, 77.8 % and 48.4 % of the taxa isolated from biofilms associated with periodontal health, periodontitis, healthy implants and peri-implantitis, respectively. The genera Anaerococcus, Anaerovorax, Anaerofilum, Exiguobacterium and Burkholderia were detected only in the peri-implantitis biofilm. A higher degree of similarity was observed between healthy and diseased implants than between healthy and diseased teeth. In addition, the biofilm associated with peri-implantitis showed statistically significant lower richness than healthy implants or diseased teeth, using the Shannon index to compare the microbial diversity. These data are somehow contradictory to those reported in the aforementioned studies (Kayanagi et al. 2010, 2013; da Silva et al. 2014). For the first time, authors also reported that there was greater abundance of Gram-negative anaerobes in the biofilm collected from healthy implants than that from peri-implantitis or periodontitis, as opposed to findings reported by other studies (Kayanagi et al. 2010, 2013).
More recently, Dabdoud et al. (2013) also used 16S rDNA pyrosequencing to explore the degree of congruence between adjacent peri-implant and periodontal microbiota in health and disease. The authors collected subgingival and peri-implant biofilm samples from 81 partially edentulous subjects with periodontal and peri-implant health and disease. Overall, the data revealed the presence of 12 phyla, 110 genera and 523 species. The predominant microorganisms were aerobes, evenly distributed between Gram-positive (194) and Gram-negative (148) species, followed by Gram-positive (47) or Gram-negative (99) anaerobes, and microorganisms that have not previously been identified (34). Staphylococcus and Treponema genera were statistically significantly associated with implant infection, but not with periodontal infection. Sixty percent of subjects shared fewer than 50 % of all species between their periodontal and peri-implant biofilms. In addition, 85 % of subjects shared fewer than 8 % of the most abundant species between tooth and implant. Also, the red complex pathogens were found in the peri-implantitis biofilm in only 37 % of the cases. Although these data corroborate the results of previous studies suggesting that certain periodontal pathogens may be present in both diseased teeth and implants (Mombelli et al. 1995; Rutar et al. 2001; Tabanella et al. 2009), the majority of the species, especially the most abundant types, showed distinct differences between periodontitis and peri-implantitis.
5.5 Concluding Remarks
The studies on the composition of the biofilm associated with peri-implantitis started in the late 1980s and from the beginning, the main focus of these studies has been the search for already known periodontal pathogens. A considerable amount of data from studies using culture and molecular targeted techniques supported the notion that most of the periodontal pathogens were also found in higher levels and proportions in peri-implantitis. Thus, at the end of the 2000s it was widely accepted that there was a great similarity between the composition of the subgingival biofilms of peri-implantitis and periodontitis. In the last few years, the use of cutting-edge open-ended diagnostic techniques to study the diversity of peri-implantitis microbiota has brought new insights on this subject. The overall results of these studies suggest that the structure of the microbial community of peri-implantitis might not be as similar to the subgingival microbiota of periodontitis, as previously thought. In addition, putative pathogens other than those associated with periodontal diseases may play a role in the onset and progression of peri-implant infection. However, it is important to note that these findings come from a limited number of studies evaluating relatively reduced numbers of samples. Therefore, further investigations on the diversity of peri-implant microbiota would be helpful in order to establish a better comparison between periodontal and peri-implant biofilms and could greatly contribute to define more effective preventive and therapeutic strategies for peri-implant diseases.
References
Abusleme L, Hong BY, Dupuy AK, Strausbaugh LD, Diaz PI (2014) Influence of DNA extraction on oral microbial profiles obtained via 16S rRNA gene sequencing. J Oral Microbiol 23:1–7
Al-Radha AS, Pal A, Pettemerides AP, Jenkinson HF (2012) Molecular analysis of microbiota associated with peri-implant diseases. J Dent 40:989–998
Atieh MA, Alsabeeha NH, Faggion CM Jr, Duncan WJ (2013) The frequency of peri-implant diseases: a systematic review and meta-analysis. J Periodontol 84:1586–1598
Becker W, Becker BE, Newman MG, Nyman S (1990) Clinical and microbiologic findings that may contribute to dental implant failure. Int J Oral Maxillofac Implants 5:31–38
Case RJ, Boucher Y, Dahllöf I, Holmström C, Doolittle WF, Kjelleberg S (2007) Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol 73:278–288
Charalampakis G, Abrahamsson I, Carcuac O, Dahlén G, Berglundh T (2014) Microbiota in experimental periodontitis and peri-implantitis in dogs. Clin Oral Implants Res 25:1094–1098
Cilia V, Lafay B, Christen R (1996) Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol Biol Evol 13:451–461
da Silva ES, Feres M, Figueiredo LC, Shibli JA, Ramiro FS, Faveri M (2014) Microbiological diversity of peri-implantitis biofilm by Sanger sequencing. Clin Oral Implants Res 25:1192–1199
Dabdoub SM1, Tsigarida AA, Kumar PS (2013) Patient-specific analysis of periodontal and peri-implant microbiomes. J Dent Res 92(12 Suppl):168S–175S
de Lillo A, Booth V, Kyriacou L, Weightman AJ, Wade WG (2004) Culture-independent identification of periodontitis-associated Porphyromonas and Tannerella populations by targeted molecular analysis. J Clin Microbiol 42:5523–5527
Diaz PI, Dupuy AK, Abusleme L, Reese B, Obergfell C, Choquette L, Dongari-Bagtzoglou A, Peterson DE, Terzi E, Strausbaugh LD (2012) Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol Oral Microbiol 27:182–201
Eke PI, Braswell LD, Fritz ME (1998) Microbiota associated with experimental peri-implantitis and periodontitis in adult macaca mulatta monkeys. J Periodontol 69:190–194
Faveri M, Mayer MP, Feres M, de Figueiredo LC, Dewhirst FE, Paster BJ (2008) Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol 23:112–118
Faveri M, Gonçalves LF, Feres M, Figueiredo LC, Gouveia LA, Shibli JA, Mayer MP (2011) Prevalence and microbiological diversity of archaea in peri-implantitis subjects by 16S ribosomal RNA clonal analysis. J Periodontal Res 46:338–344
França LT, Carrilho E, Kist TB (2002) A review of DNA sequencing techniques. Q Rev Biophys 35:169–200
Green PM, Giannelli F (1994) Direct sequencing of PCR-amplified DNA. Mol Biotechnol 1:117–124
Harrington CT, Lin EI, Olson MT, Eshleman JR (2013) Fundamentals of pyrosequencing. Arch Pathol Lab Med 137:1296–1303
Harris LG, Meredith DO, Eschbach L, Richards RG (2007) Staphylococcus aureus adhesion to standard micro-rough and electropolished implant materials. J Mater Sci Mater Med 18:1151–1156
Heitz-Mayfield LJ (2008) Diagnosis and management of peri-implant diseases. Aust Dent J 53(Suppl 1):43–48
Heitz-Mayfield LJ, Lang NP (2010) Comparative biology of chronic and aggressive periodontitis vs. peri-implantitis. Periodontology 2000 53:167–181
Heydenrijk K, Meijer HJA, van der Reijden WA, Raghoebar GM, Vissink A, Stegenga B (2002) Microbiota around root-form endosseous implants: a review of the literature. Int J Oral Maxillofac Implants 17:829–838
Hiyari S, Bennett KM (2011) Dental diagnostics: molecular analysis of oral biofilms. J Dent Hyg 85:256–263
Hudetz D, Ursic Hudetz S, Harris LG, Luginbühl R, Friederich NF, Landmann R (2008) Weak effect of metal type and ica genes on staphylococcal infection of titanium and stainless steel implants. Clin Microbiol Infect 14:1135–1145
Hultin M, Gustafsson A, Hallström H, Johansson LA, Ekfeldt A, Klinge B (2002) Microbiological findings and host response in patients with peri-implantitis. Clin Oral Implants Res 13:349–358
Koyanagi T, Sakamoto M, Takeuchi Y, Ohkuma M, Izumi Y (2010) Analysis of microbiota associated with peri-implantitis using 16S rRNA gene clone library. J Oral Microbiol 24:1–7
Koyanagi T, Sakamoto M, Takeuchi Y, Maruyama N, Ohkuma M, Izumi Y (2013) Comprehensive microbiological findings in peri-implantitis and periodontitis. J Clin Periodontol 40:218–226
Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL (2006) Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol 44:3665–3673
Kumar PS, Mason MR, Brooker MR, O’Brien K (2012) Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J Clin Periodontol 39:425–433
Lee KH, Maiden MF, Tanner AC, Weber HP (1999) Microbiota of successful osseointegrated dental implants. J Periodontol 70:131–138
Leonhardt A, Renvert S, Dahlén G (1999) Microbial findings at failing implants. Clin Oral Implants Res 10:339–345
Leonhardt A, Bergström C, Lekholm U (2003a) Microbiologic diagnostics at titanium implants. Clin Implant Dent Relat Res 5:226–232
Leonhardt A, Dahlén G, Renvert S (2003b) Five-year clinical, microbiological, and radiological outcome following treatment of peri-implantitis in man. J Periodontol 74:1415–1422
Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA (2004) Methanogenic archaea and human periodontal disease. Proc Natl Acad Sci U S A 101:6176–6681
Matarazzo F, Ribeiro AC, Feres M, Faveri M, Mayer MP (2011) Diversity and quantitative analysis of Archaea in aggressive periodontitis and periodontally healthy subjects. J Clin Periodontol 38:621–627
Mombelli A (1993) Microbiology of the dental implant. Adv Dent Res 7:202–206
Mombelli A (1999) Prevention and therapy of peri-implant infections. In: Lang NP, Karring T, Lindhe J (eds) Proceedings of the 3rd European workshop on periodontology. Quintessence Book, Berlin, pp 281–303
Mombelli A, Décaillet F (2011) The characteristics of biofilms in peri-implant disease. J Clin Periodontol 38(Suppl 11):203–213
Mombelli A, Lang NP (1998) The diagnosis and treatment of periimplantitis. Periodontology 2000 17:63–76
Mombelli A, Mericske-Stern R (1990) Microbiological features of stable osseointegrated implants used as abutments for overdentures. Clin Oral Implants Res 1:1–7
Mombelli A, van Oosten MA, Schurch E Jr, Land NP (1987) The microbiota associated with successful or failing osseointegrated titanium implants. Oral Microbiol Immunol 2:145–151
Mombelli A, Marxer M, Gaberthüel T, Grunder U, Lang NP (1995) The microbiota of osseointegrated implants in patients with a history of periodontal disease. J Clin Periodontol 22:124–130
Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE (2001) Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770–3783
Persson GR, Renvert S (2013) Cluster of bacteria associated with peri-implantitis. Clin Implant Dent Relat Res (in press)
Persson GR, Samuelsson E, Lindahl C, Renvert S (2010) Mechanical non-surgical treatment of peri-implantitis: a single-blinded randomized longitudinal clinical study. II. Microbiological results. J Clin Periodontol 37:563–573
Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J (2009) Metagenomic pyrosequencing and microbial identification. Clin Chem 55:856–866
Pontoriero R, Tonelli MP, Carnevale G, Mombelli A, Nyman SR, Lang NP (1994) Experimentally induced peri-implant mucositis. A clinical study in humans. Clin Oral Implants Res 54:254–259
Prober JM, Trainor GL, Dam RJ et al (1987) A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science 238(4825):336–341
Quirynen M, Vogels R, Peeters W, van Steenberghe D, Naert I, Haffajee A (2006) Dynamics of initial subgingival colonization of ‘pristine’ peri-implant pockets. Clin Oral Implants Res 17:25–37
Rams TE, Balkin BE, Roberts TW, Molzan AK (2013) Microbiological aspects of human mandibular subperiosteal dental implants. J Oral Implantol 39:714–722
Rastogi G, Coaker GL, Leveau JH (2013) New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol Lett 348(1):1–10
Renvert S, Roos-Jansåker AM, Lindahl C, Renvert H, Rutger Persson G (2007) Infection at titanium implants with or without a clinical diagnosis of inflammation. Clin Oral Implants Res 18:509–516
Rutar A, Lang NP, Buser D, Bürgin W, Mombelli A (2001) Retrospective assessment of clinical and microbiological factors affecting periimplant tissue conditions. Clin Oral Implants Res 12:189–195
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467
Sanz M, Newman MG, Nachnani S, Holt R, Stewart R, Flemmig T (1990) Characterization of the subgingival microbial flora around endosteal sapphire dental implants in partially edentulous patients. Int J Oral Maxillofac Implants 5:247–253
Shchipkova AY, Nagaraja HN, Kumar PS (2010) Subgingival microbial profiles of smokers with periodontitis. J Dent Res 89:1247–1253
Shibli JA, Martins MC, Lotufo RFM, Marcantonio E Jr (2003) Microbiologic and radiographic analysis of ligature-induced peri-implantitis with different dental implant surfaces. Int J Oral Maxillofac Implants 18:383–390
Shibli JA, Melo L, Ferrari DS, Figueiredo LC, Faveri M, Feres M (2008) Composition of supra-and subgingival biofilm of subjects with healthy and diseased implants. Clin Oral Implants Res 19:975–982
Silverstein LH, Kurtzman D, Garnick JJ, Schuster GS, Steflik DE, Moskowitz ME (1994) The microbiota of the peri-implant region in health and disease. Implant Dent 3:170–174
Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr (1998) Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144
Tabanella G, Nowzari H, Slots J (2009) Clinical and microbiological determinants of ailing dental implants. Clin Implant Dent Relat Res 11:24–36
Tillmanns HWS, Hermann JS, Tiffee JC, Burgess AV, Meffert RM (1998) Evaluation of three different dental implants in ligature-induced peri-implantitis in the beagle dog. Part II. Histology and microbiology. Int J Oral Maxillofac Implants 13:59–68
Vianna ME, Conrads G, Gomes BP, Horz HP (2006) Identification and quantification of archaea involved in primary endodontic infections. J Clin Microbiol 44:1274–1282
Wade WG (2011) Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol 38(Suppl 11):7–16
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Zitzmann NU, Berglundh T (2008) Definition and prevalence of peri-implant diseases. J Clin Periodontol 35(8 Suppl):286–291
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Faveri, M., Figueiredo, L.C., Shibli, J.A., Pérez-Chaparro, P.J., Feres, M. (2015). Microbiological Diversity of Peri-Implantitis Biofilms. In: Donelli, G. (eds) Biofilm-based Healthcare-associated Infections. Advances in Experimental Medicine and Biology, vol 830. Springer, Cham. https://doi.org/10.1007/978-3-319-11038-7_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-11038-7_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-11037-0
Online ISBN: 978-3-319-11038-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)