Abstract
Sexual reproduction provides a valuable tool for strain development and gene identification in fungi, but has been limited in its use because many economically important fungi are only known to reproduce asexually. However, in the past decade a number of publications have reported sexual cycles in supposedly ‘asexual’ fungi, indicating that many presumed asexual fungi might have the potential for sexual reproduction. Methods are therefore described to induce a sexual cycle in various filamentous fungi, including the use of mating-type gene diagnostics to identify compatible mating types in heterothallic ascomycete species, the use of different crossing conditions, and importance of inoculation technique. The conditions for successful mating are often species-specific and it is therefore difficult to give a general protocol. Different methods for the isolation and analysis of ascospore offspring are then described. Finally, ways in which the sexual cycle can be exploited for purposes including gene identification and localization, strain improvement, and gene complementation are covered. It is anticipated that the application of such methodologies will generate sexual progeny with new characteristics or previously unobserved properties.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The vast majority of fungal species are able to undergo sexual reproduction involving the formation of sexual spores via meiosis. This form of reproduction is thought to have many evolutionary advantages, and where present offers a valuable laboratory tool for experimental genetic analysis (Dyer and Paoletti 2005; Aanen and Hoekstra 2007; Lee et al. 2010). However, a surprisingly high minority of fungal species (approximately 20 %) are only known to reproduce by asexual (mitotic) means. This includes many species of industrial importance, notably several Aspergillus and Penicillium species (Dyer and Paoletti 2005; Dyer and O’Gorman 2011). The aim of this chapter is twofold. Firstly, to describe how sexual cycles may be induced in filamentous fungi including ‘fastidious’ species (Kwon-Chung and Sugui 2009), which require very specific conditions, and sexually ‘recalcitrant’ species, where a sexual cycle might not yet have been reported. Secondly, to describe methods by which sexual progeny can be isolated, with further possibilities suggested for progeny analysis. The chapter will focus on filamentous ascomycete species (Pezizomycotina), which represent one of the largest groups in the fungal kingdom, and in particular on members of industrial importance. A final section is included to briefly describe ways in which the sexual cycle can be exploited for purposes including gene identification and localization, strain improvement, and gene complementation.
2 Sexual Reproduction and Breeding Systems in Filamentous Ascomycete Fungi
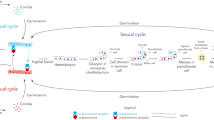
In 1820, microscopic sexual structures in fungi were reported for the first time in a culture of Syzygites megalocarpus (Zygomycota, Mucorales) (Ehrenberg 1820; Idnurm 2011). In 1904, Blakeslee showed that S. megalocarpus is a self-fertile species and also observed the existence of different “sexes” (‘mating types’) in Rhizopus stolonifer (reported as Rhizopus nigricans). Based on his findings he introduced the terms ‘homothallism’ for self-fertile (or self-compatible) and ‘heterothallism’ for self-incompatible (or obligate outcrossing) individuals (Blakeslee 1904). Thus, by definition, individuals of homothallic fungal species can complete the sexual cycle without the need for a mating partner, whereas individuals of heterothallic species require a mating partner of compatible mating type for sexual reproduction to occur. However, it is important to note that homothallic species are not restricted to self-fertility, as individuals normally retain the ability to out-cross under suitable conditions (Dyer et al. 1992; Burnett 2003; Cavindera and Trail 2012). In the case of heterothallic pezizomycete species there are normally only two mating types present. By convention these are now termed MAT1-1 and MAT1-2, although for some species alternative established terminology such as matA and mata (e.g. in Neurospora) or plus ‘+’ and minus ‘−’ (e.g. in Podospora) are used (Dyer et al. 1992; Turgeon and Yoder 2000). Also some heterothallic species have an additional layer of sexual compatibility superimposed on the mating type. Individuals can either be male (M), female (F), or hermaphrodites (MF) with respect to their ability to form sexual mating structures such as ascogonia, protoperithecia, microconidia, and spermatia (Debuchy et al. 2010). For a cross to be successful not only must isolates of opposite mating type be present, but also one mating partner must be able to act as a male and the other as a female. However, this system appears to be restricted to certain taxonomic groupings such as Fusarium and Magnaporthe species (Gordon 1961; Takan et al. 2012). A third reproductive strategy occurring in filamentous fungi was later described, called ‘pseudohomothallism’ (or secondary homothallism) (Dodge 1957). Ascomycetous pseudohomothallic species (e.g. Neurospora tetrasperma, Podospora anserina) develop four spored asci in which most ascospores contain two nuclei, one of each mating type (Raju and Perkins 1994). A typical binucleate ascospore germinates to form a self-fertile mycelium due to the fact that the arising heterokaryic hyphae contains both matA and mata nuclei (Pöggeler 2001).
In heterothallic pezizomycete species sexually compatible haploid strains are normally morphologically indistinguishable and not differentiated into male and female sexes, the mating partners are instead distinguished only by their mating type (Bistis 1998). Extensive studies over the past 25 years have revealed that both sexual identity and later stages of sexual development are controlled in fungi by so called ‘mating-type’ (MAT) genes (Debuchy et al. 2010). In heterothallic pezizomycete fungi there is usually only one MAT locus at which between one and three MAT genes may be present. The DNA sequence of the MAT genes of isolates of opposite mating type, together with other non-coding sequence present at the MAT locus, is highly dissimilar although the regions flanking the MAT locus are highly conserved (Debuchy and Turgeon 2006). Because of the dissimilarity in sequence between the opposite MAT loci, they are referred to as ‘idiomorphs’ instead of alleles to emphasize that the sequences at the same locus are highly dissimilar (Metzenberg and Glass 1990). By convention, MAT1-1 mating-type isolates contain a MAT idiomorph which includes a MAT1-1 gene encoding a protein with a motif called the alpha box, whereas MAT1-2 mating-type isolates contain a MAT idiomorph which includes a MAT1-2 gene encoding a regulatory protein with a DNA-binding domain of the high mobility group (HMG) family. These two idiomorphs are designated MAT1-1 and MAT1-2, respectively. Where more than one MAT gene is present in an idiomorph, each gene within an idiomorph is indicated by the idiomorph symbol followed by a dash and a number, e.g., MAT1-2-1 (Turgeon and Yoder 2000). The organization of MAT genes differs in homothallic species, where both alpha box MAT1-1 and HMG domain MAT1-2 genes are normally present in the same individual; this co-occurrence appears to confer the ability to self-fertilize. The alpha and HMG genes can be tightly linked at a single MAT locus or be present at two distinct MAT loci within the genome (Paoletti et al. 2007; Debuchy et al. 2010).
2.1 Use of MAT Genes as Diagnostic Tools for Induction of a Sexual Cycle
The discovery and characterization of mating-type genes from a diverse range of fungi has provided a major advance in the ability to induce sexual cycles of chosen fungal species in vitro. Fungal MAT genes were first identified using molecular methods from the yeast Saccharomyces cerevisiae (Astell et al. 1981) and since then have been characterized from numerous filamentous ascomycete species (Debuchy and Turgeon 2006; Debuchy et al. 2010).
Although the MAT genes and idiomorphs show considerable sequence divergence overall, it has nevertheless been possible to identify partly conserved alpha box and HMG domain regions of the MAT1-1 and MAT1-2 genes, respectively, encoding homologous 65–80 amino acid regions of the MAT proteins. This has allowed the design of degenerate PCR primers [which can include the alternative base inosine (I) to avoid too high a rate of degeneracy] that can be used to amplify these regions of the MAT genes from species where genome sequence data is lacking. Due to sequence divergence within the Pezizomycotina, degenerate primers often need to be designed for groups of related fungal species because it has proved difficult to design all encompassing pezizomycete MAT degenerate primer sets (Dyer et al. 1995; Arie et al. 1997; Singh et al. 1999; Table 2.1). But once MAT amplicons have been obtained they can then be sequenced to confirm homology to known MAT genes, and if desired the arising sequence can be used to design specific primers (which are likely to be less prone to PCR artefacts than degenerate primers) for use as a MAT diagnostic tool to determine the mating type of isolates. It is noted that the MAT1-2 gene sequence tends to be conserved to a higher extent than MAT1-1 gene sequence, so it might be necessary to obtain MAT1-1 idiomorph sequence via a chromosome walking approach based on inwards sequencing from the SLA and APC genes found in the conserved region bordering the MAT loci (Eagle 2009, C Eagle and PS Dyer unpublished results) or use of TAIL-PCR to amplify whole MAT idiomorph regions (Arie et al. 1997). More recently it has also been possible to use whole genome sequence to identify MAT genes by BLAST analysis and thereby design MAT diagnostic primers set directly. Such MAT diagnostic tests may use different primer pairs for the MAT1-1 or MAT1-2 genes, necessitating two rounds of PCR (see examples in Table 2.1). Alternatively, multiplex PCR-based tests have been designed to allow mating type to be determined using a single PCR (Table 2.1). The latter diagnostic relies on the use of one primer binding to conserved sequence in the flanking regions of isolates of both mating type, together with two further primers that bind to sequence either present in the MAT1-1 or MAT1-2 idiomorph. The location of the latter two primers is designed such that differential size PCR products are generated according to whether isolates are of MAT1-1 or MAT1-2 genotype, thereby allowing the rapid and efficient determination of mating type [for further explanation of rationale see Dyer et al. (2001b) and Paoletti et al. (2005)].
The availability of a PCR-based mating-type detection method is a major aid when dealing with induction of sexual reproduction in heterothallic species. Before the introduction of these MAT diagnostic techniques, isolates had to be crossed in all combinations with each other and when successful mating occurred, mating-type tester strains were selected. With these tester strains, larger sets of isolates were then examined in order to determine their fertility and mating type. Whereas now with the use of MAT-specific primers, the mating type of isolates can be determined prior to crossing on agar media. It is therefore possible to set up, in a more efficient manner, directed crossings between isolates known to be of the opposite mating type. This significantly reduces the amount of effort needed to study for example the presence of a sexual cycle in presumed asexual species or the sexual fertility within a population. Indeed, this approach has led to major breakthroughs over recent years with the induction of a sexual cycle in a series of previously considered ‘asexual’ species when crosses were set up between known MAT1-1 and MAT1-2 isolates. A model example concerns the opportunistic human pathogen Aspergillus fumigatus. This species was described by Fresenius in 1863 and for more than 140 years was only known as an asexual organism (Samson et al. 2009). However, there was accumulating evidence for the presence of a cryptic (i.e., hidden, so far undescribed) sexual cycle. Firstly, studies of the population structure provided evidence for sexual recombination based on the analysis of the association of alleles of five loci (Pringle et al. 2005) and sequence present at three intragenic regions (Paoletti et al. 2005). Secondly, it was possible to identify MAT1-1 and MAT1-2 isolates of A. fumigatus using genomic BLAST searching and degenerate PCR approaches, together with the presence of a series of genes related to sex within the genome (Galagan et al. 2005; Paoletti et al. 2005). A multiplex PCR-based MAT diagnostic test was then developed, and analysis of 290 worldwide clinical and environmental isolates revealed the presence of MAT1-1 and MAT1-2 genotypes in similar proportions (43 % and 57 %, respectively) (Paoletti et al. 2005). The presence of the two mating types in equal frequencies within a population is an indication of sexual reproduction (Milgroom 1996). In a subsequent study, analysis of a population of A. fumigatus strains from five locations in Dublin, Ireland, revealed an almost exact 1:1 ratio of MAT1-1:MAT1-2 isolates (O’Gorman et al. 2009). Furthermore, 88 out of 91 isolates were genetically unique according to a RAPD-PCR DNA fingerprinting study, and a phylogenetic analysis demonstrated that the MAT-1 and MAT1-2 isolates were interleaved when represented on a phylogenetic tree. This provided strong evidence for recent or extant sexual reproduction and led O’Gorman et al. (2009) to set up directed crosses between known MAT1-1 and MAT1-2 isolates on a range of media under a variety of different growth conditions. An exciting result was then obtained when it was found that a sexual cycle, leading to production of cleistothecia containing recombinant ascospores, could be induced when cultures were crossed on oatmeal agar in Parafilm-sealed Petri-dishes which were incubated at 30 °C in darkness for 6–12 months (O’Gorman et al. 2009). More recently, a “supermater” pair of A. fumigatus isolates have been identified which produce abundant cleistothecia containing viable ascospores after only 4 weeks incubation under the same conditions (Sugui et al. 2011). There have subsequently been further examples of the discovery of sexual states in other supposedly ‘asexual’ fungal species using similar directed crosses between known MAT1-1 and MAT1-2 isolates [reviewed by Dyer and O’Gorman (2012)]. These have included notably the description of a sexual state for the industrial workhorse Trichoderma reesei (Seidl et al. 2009), the penicillin producer Penicillium chrysogenum (Böhm et al. 2013), the opportunistic pathogen Aspergillus lentulus (Swilaiman et al. 2013), the aflatoxin producers Aspergillus flavus and Aspergillus parasiticus (Moore 2014), and the starter culture of blue veined cheese Penicillium roqueforti (Ropars et al. 2014; S Swilaiman, J Houbraken, J Frisvad, R Samson and PS Dyer, unpublished results).
It should be cautioned that the presence of the MAT genes alone is insufficient to prove that a sexual stage exists. Given that several hundred other genes are also likely to be required for a functional sexual cycle to occur, it is possible that loss of function of any of these genes could result in reduced fertility or asexuality. For example, a series of over 75 genes have been identified which are required for sexual reproduction in the aspergilli, encompassing processes such as environmental sensing, mating, fruit body formation, and ascospore production. Such ‘sex-related’ genes might be predicted to accumulate deleterious mutations or even be lost in purely asexual species, in which there was no functional constraint on their conservation (Dyer 2007; Dyer and O’Gorman 2012).
3 Methods to Induce Sexual Reproduction in Filamentous Fungi
In this next section methods will be described to induce sexual reproduction in filamentous ascomycete species based on procedures that have been used to obtain successful mating in a variety of fungal species. The first step involves the selection of suitable strains; this is followed by MAT analysis; then selection of suitable agar media, inoculation procedures, and incubation conditions to induce a sexual cycle. Finally after formation of a sexual state, single ascospore isolates should be obtained and these should be examined for evidence of recombination. It is noted that sexual reproduction for many homothallic species can be achieved fairly readily as there is no need for any mating step, so most of the following discussion will apply to heterothallic species which can be more demanding in their sexual requirements.
4 Materials
4.1 Solutions
Sterile distilled water or tap water.
Tween 80: 0.5 g/L, ddH2O to 1 L.
4.2 Agar Media Inducing Sexual Reproduction
Carrot agar (CA): Fresh washed, peeled, diced carrots (400 g) in 400 mL ddH2O. Autoclave at 121 °C for 15 min. After autoclaving, blend the carrots and add additional 500 mL ddH2O. Add ZnSO4·7H2O (0.01 g/L), CuSO4·5H2O (0.005 g/L), agar (20 g/L). Mix well and autoclave at 121 °C for 15 min.
Mixed cereal agar (MCA): Gerber mixed grain cereal (Gerber Products Co., Freemont, Michigan) (50 g/L), agar (20 g/L), in 1 L ddH2O. Mix well and autoclave at 121 °C for 15 min (McAlpin and Wicklow 2005).
Oatmeal agar (OA)Footnote 1:
Version (1): Blend 30 g of oats and add 1 L ddH2O. Boil and let it stand for 1 h. Add ZnSO4·7H2O (0.01 g/L), CuSO4·5H2O (0.005 g/L), agar (20 g/L). Autoclave for 15 min, at 121 °C (Samson et al. 2010).
Version (2): Add 40 g of oats to 1 L tap water. Bring to the boil then lower the heat to just below boiling point (i.e., bubbling gently) for a further 45 min. Then filter through two layers of cheese cloth and restore the volume of the solution to 1 L with tap water and mix thoroughly. Add agar (20 g/L). Autoclave for 15 min at 121 °C with slow cool down and slow release of pressure to prevent media loss (O’Gorman et al. 2009).
Potato dextrose agar (PDA): 200 g sliced potatoes are boiled in 1 L of ddH2O and sieved, add glucose (20 g/L), agar (20 g/L), ZnSO4·7H2O (0.01 g/L), CuSO4·5H2O (0.005 g/L). pH is approximately 5.6. Autoclave for 15 min, at 121 °C (Samson et al. 2010).
Tap water agar (TWA)Footnote 2: Bacto™ agar (15 g/L) in 1 L tap water. Autoclave for 15 min, at 121 °C. Supplement with appropriate natural growth substrate (Dyer et al. 1993).
V8 agar (V8): V8® vegetable juice (Campbell) (175 mL/L), CaCO3 (3 g/L), ZnSO4·7H2O (0.01 g/L), CuSO4·5H2O (0.005 g/L), agar (20 g/L) in 1 L ddH2O. Mix well and autoclave at 121 °C for 15 min (Samson et al. 2010).
5 Methods
5.1 Strain Selection
5.1.1 Identification
The first step in inducing a sexual cycle is the requirement for the correct identification of isolates of the same biological species. In the past, fungal taxonomy was primary based on phenotypic and physiological characters. Nowadays molecular techniques like DNA sequencing are commonly applied for identification purposes. Such data has shown that many well-known ‘species’ are actually species complexes composed of closely related species that might be sexually fertile when crossing within a species, but sexually sterile when attempts are made to cross different species. For example, P. chrysogenum is a complex of five species, namely P. chrysogenum sensu stricto, Penicillium rubens, Penicillium allii-sativi, Penicillium tardochrysogenum, and Penicillium vanluykii (Henk et al. 2011; Houbraken et al. 2012). Using these current taxonomic insights, the main penicillin producer is named P. rubens and in the study of Böhm et al. (2013) all P. chrysogenum strains that were able to reproduce sexually are actually P. rubens. The induction of the sexual cycle would have probably been much more difficult or even impossible if attempts had been made to cross different members of the P. chrysogenum sensu lato complex and might explain why the sexual stage had remained undiscovered up to that point. Similarly, it was discovered that the ‘single’ anamorphic species Pseudocercosporella herpotrichoides is composed of two closely related, but intersterile, species Tapesia (Oculimacula) yallundae and Tapesia acuformis (Dyer et al. 1996). Again, attempts to induce a sexual cycle by erroneously crossing isolates of the different species would have failed in this case.
Therefore, it is essential to verify that isolates to be used in crossing experiments are of the same biological species. The internal transcribed spacer regions (ITS) of the ribosomal gene have been used in many taxonomic studies for species identification and have been selected as fungal barcodes (Schoch et al. 2012). The main advantages of using the ITS locus for identification are the ease of amplification by PCR and the presence of a high number of ITS sequences in the public databases. However, the resolution of this locus is insufficient for species identification of all fungi. For example, closely related species belonging to the industrially important genera Aspergillus, Penicillium, Fusarium, and Trichoderma can share the same ITS barcode. In those cases, it is recommended to sequence other (protein coding) genes. There is no consensus which region to sequence and the choice largely depends on the genus/species being dealt with. Details on molecular and phylogenetic identification methods can be found in Crous et al. (2009).
5.1.2 Origin of Strains
Freshly isolated strains are generally more fertile than strains maintained for longer periods in culture collections, which can be prone to a ‘slow decline’ in fertility following prolonged subculture (Dyer and Paoletti 2005). For example, Paecilomyces variotii strains isolated from heat-treated products proved to be fertile, while older isolates from a culture collection were unable to mate (Houbraken et al. 2008). Similar observations were found in the heterothallic Histoplasma capsulatum. This species lost fertility during laboratory passage and it was suggested that selective pressures may serve to maintain fertility in the environment (Kwong-Chung et al. 1974; Fraser et al. 2007). Furthermore, even when obtaining isolates from the field it is important to be aware that such isolates can exhibit a range of fertility due to various physiological and genetic factors (Dyer et al. 1992). Indeed the same ‘slow decline’ in sexual fertility observed during in vitro culture may be occurring in vivo in natural populations subject to strong selection pressure favoring asexual propagation (Dyer and Paoletti 2005). For example, Sugui et al. (2010) found that most attempted crosses involving the emerging agent of aspergillosis Aspergillus (Neosartorya) udagawae either failed to produce cleistothecia or produced ascospores which did not germinate. Similarly, Swilaiman et al. (2013) found that many clinical isolates of the opportunistic pathogen A. lentulus exhibited low fertility or were sterile in crosses. However, in both of these cases it was possible to detect isolates that successfully crossed to form cleistothecia with ascospores. This illustrates the fact that it is very important to select a number of representative field isolates for crossing studies to ensure that at least some representatives will exhibit sexual fertility if it is present.
5.1.3 Strain Typing
Before the start of the mating experiments, the isolates should ideally be typed using methods such as AFLP (Amplified Fragment Length Polymorphism), SSR (microsatellites or simple sequence repeats), RFLP (Restriction Fragment Length Polymorphism), RAPD (Random Amplified Polymorphic DNA), or MLST (MultiLocus Sequence Typing) DNA fingerprinting. The use of such typing methods allows the selection of independent (non-clonal) strains for crossing purposes, avoiding the error of setting up repeated crosses with different isolates of the same field strain. Typing can also generate insights as to whether there is evidence of recombination among the strains.
5.2 Detection of MAT Genes
In order to increase the likelihood of success and reduce the number of crosses that need to be set up, it is recommended that the mating type of test strains be determined prior to crossing efforts. This enables directed crosses to be set up on agar media (see below) between MAT1-1 and MAT1-2 mating partners that are known to be potentially sexually compatible. For certain species or species groups, mating type-specific primers have been developed that amplify part of either the MAT1-1 or MAT1-2 gene. A selection of primers pairs already published for some important pezizomycete species is given in Table 2.1 together with details of some degenerate primers pairs that should be more broadly applicable to wide groups of species (Dyer et al. 1995; Arie et al. 1997; Singh et al. 1999; Paoletti et al. 2005; Seymour et al. 2005). Especially note that the use of hot-start PCR can greatly increase the chances of success; for example, Singh et al. (1999) were unable to amplify a MAT1-2 region from T. yallundae using standard PCR, but found that the use of hot-start PCR gave very strong amplification of the required product. In cases where no MAT amplicons are obtained, it is recommended that new degenerate primers are designed based on known MAT gene sequence of species that are phylogenetically closely related to the test species.
5.3 Agar Media
A large variety of agar media have been used for the induction of fungal sexual cycles. Generally, media based on natural substrates are more effective than synthetic media and which agar to use largely depends on the species or genus. Some species are fastidious and need specific nutrients, which often mimic their natural growth substrate. For example, the sexual cycle of the cereal pathogen T. yallundae occurs in nature on straw stubble left after harvest (Dyer et al. 2001a) and attempts to induce the sexual cycle in vitro on a range of synthetic media failed. However, it was possible to induce the sexual cycle when MAT1-1 and MAT1-2 isolates were inoculated onto straw segments (especially those with nodes), which were kept moist by being placed on TWA (Dyer et al. 1993). Similarly, it was only possible to induce sex in Thermomyces dupontii (=Talaromyces thermophilus) on natural oat grains rather than synthetic agar media (Pitt 1979; Houbraken et al. 2014), and three dermatophytic Trichophyton species required growth on sterilized baby or rabbit hair (placed on agar) to induce sexual reproduction (Kawasaki et al. 2010). By contrast, for species such as Neurospora crassa it is possible to induce sex on fully synthetic media, one reason why this is used as a model organism (Perkins 1986). The diversity in nutrient requirements for sexual reproduction is illustrated well by members of the genus Aspergillus. Species such as the homothallic Aspergillus nidulans and Aspergillus fischeri reproduce sexually on a fairly wide range of media, including fully synthetic complete media and oatmeal agar (OA) (Paoletti et al. 2007). This contrasts with some heterothallic species which have more exacting demands. Members of the section Flavi (e.g. A. flavus, A. parasiticus) have only been successfully crossed on mixed cereal agar (MCA), while crosses of members of the section Fumigati (A. fumigatus, A. lentulus) have only proved fruitful on OA. In contrast to these two high water activity media, species belonging to section Aspergillus (Eurotium-type ascomata) require a low water activity medium (e.g., malt extract agar with 40 % sucrose) for fruiting body formation (Dyer and O’Gorman 2012). In Fusarium, the standard medium to induce fruiting bodies, called perithecia, is carrot agar. In contrast, attempts with Fusarium keratoplasticum to use this standard medium were unsuccessful while crosses on V8 agar induced the sexual cycle in this species (Covert et al. 2007; Short et al. 2013). In the case of homothallic Giberella zeae sex can be induced by the gentle removal of surface mycelia, followed by treatment with detergent solution (Cavindera and Trail 2012). One intriguing example is that of different Cryptococcus species. Nielsen et al. (2007) found that it was possible to induce the sexual cycle of Cryptococcus neoformans on pigeon guano media, but that this was not possible for the related species Cryptococcus gattii. It was suggested that the ability to undergo sexual reproduction on pigeon guano represented an evolutionary adaptation that allowed ancestral strains of C. neoformans to sweep the globe (Nielsen et al. 2007; Heitman et al. 2014). It is important to note that when making agar media there can be difference among ingredients of different suppliers. For example, different brands of yeast extracts are available and these can have a strong influence on the phenotype of the culture. Furthermore, agar media based on natural ingredients can vary between manufactures and labs, and even within one lab batch to batch differences can occur.
Some ascomycetes may require exogenous vitamins, minerals, or other natural materials for ascomata (ascocarp) production, and these are often not present in synthetic media. This might be one of the explanations why sexual reproduction is more often found on media made from natural substrates such as oatmeal, (mixed) cereals, and cornmeal agar. A fractionation study of V8 juice agar revealed that no single factor was responsible for its utility in inducing sex in C. neoformans, but rather the unique composition of V8 juice provided sustenance for sex, especially the copper content (Kent et al. 2008). Other studies have found it necessary to add compounds to a standard medium to induce sex; for example biotin was added to OA to stimulate mating in P. rubens (Böhm et al. 2013). Meanwhile, certain nutritional auxotrophs of A. nidulans can require supplementation of media to ensure sexual development, e.g., tryptophan, arginine, and riboB2 mutants are self-sterile (Dyer and O’Gorman 2012) and heterologous expression of pyrG can result in reduced fertility (C Scazzocchio pers. comm.; Robellet et al. 2010). An overview of agar media used in mating experiments of selected heterothallic species belonging to either the Eurotiales or Hypocreales as representative groupings is given in Table 2.2.
Thus, the best strategy when attempting to induce sexual reproduction in vitro is to trial a range of agar media, which should include some basal media supplemented with the natural growth substrate of the species in question.
5.4 Incubation Conditions
Besides the nutrient availability, various other environmental factors such as light, temperature, and oxygen determine the success of mating experiments. These factors are genus and in some cases also species-specific. There are numerous reports in the literature of how different environmental conditions influence fungal sexual reproduction, and in the present chapter only some representative examples can be given. For example, Fusarium perithecia are formed in abundance under alternating 12 h/dark and 12 h/light cycles (with both fluorescent and near ultra violet light), whereas perithecia are absent when incubated in darkness (Table 2.2). Similarly the sexual cycle of T. acuformis can be induced under near UV or white light, but not in darkness (Dyer et al. 1996). By contrast, it is necessary to incubate Aspergillus species in darkness to trigger sexual reproduction because light preferentially induces asexual sporulation, reflecting the natural ecology of many Aspergillus species (Mooney and Yager 1990; Han et al. 2003; Dyer and O’Gorman 2012). Incubation temperature also has a strong influence on sexual fertility. For example, Choi et al. (2009) showed that their Fusarium fuijikuroi strains only produced perithecia at 23 °C and none were formed at 18, 26, and 28 °C. However, in Fusarium graminearum, the optimal temperature was 28.5 °C and Fusarium circinatum perithecia were more abundantly produced at 20 °C than at 25 °C (Table 2.2) (Tschanz et al. 1976; Covert et al. 1999). Meanwhile, in Aspergillus and Penicillium, oxygen limitation can induce sexual reproduction (Dyer and O’Gorman 2012), which can be achieved by sealing Petri dishes with Parafilm (Table 2.2).
Thus, the best strategy when attempting to induce sexual reproduction in vitro is to trial a range of growth conditions, which ideally might mimic those encountered in the wild when sexual reproduction occurs. This approach was used to induce sex in plant pathogenic Tapesia species which were known to sexually reproduce in the field in early spring in the UK on exposed straw stubble. It was found that incubation in vitro at low temperatures between 7 and 10 °C under white light could induce sex, but the sexual cycle was inhibited above these temperatures (Dyer et al. 1996).
5.5 Inoculations
For homothallic species, self fertilization can be induced using either point inoculation or spore spread methods (e.g. Paoletti et al. 2007; Todd et al. 2007; Cavindera and Trail 2012). In the case of heterothallic species, several methods have been described in the literature for crossing on agar media. We have summarized three different methods below. In the first, the “barrage zone” method, strains of opposite mating type are inoculated in close proximity on an agar medium. During incubation, the strains grow towards each other. Fruiting body formation then mainly occurs in the barrage zone (Fig. 2.1a, b), but sometimes also towards the centre or on the opposite periphery of the colony (Fig. 2.1c). Interestingly, this method can also be used to promote outcrossing in homothallic species, especially when using complementary auxotrophic strains [see Todd et al. (2007) and Cavindera and Trail (2012) for further details]. In the second, the “mixed culture” method, spore suspensions of the same concentration are prepared for isolates of opposite mating type. These suspensions are mixed together and then used to inoculate the agar medium. This results in intermingled growth of both partners from an early stage allowing close sexual interaction. Finally, in Fusarium, Neurospora, and other pyrenomycete fungi, a third “fertilization” crossing technique is also sometimes applied. In this method, a strain of one mating type is cultured on an agar medium to allow development of protoperithecia. After growth, the culture is then fertilized (so-called ‘spermatization’) with a spore-suspension of the opposite mating partner. This fertilization method can also be used in heterothallic Botrytis species that produce sclerotia, which can be spermatized by being soaked with conidia of the opposite mating type (Faretra et al. 1988). Crosses with strains of the same mating type should be used as controls.
Mating experiments between P. variotii strains. (a–c) Detail image of a potato dextrose agar plate where two P. variotii strains of opposite mating types were inoculated on each side of the Petri-dish. In a and b, ascomata are formed in the middle, in c, ascomata are also present on the opposite side of the colony. (d, e) Higher-magnification view of the ascomata. (f) Micrograph of the ascomata showing asci with ascospores. (g) Similar as f, but also showing the presence of heat-sensitive conidia (arrows); isolation of the ascospores, which are heat-resistant, can be achieved by applying the “heat treatment method”. Scale bars = 10 μm
5.5.1 Barrage Zone Method
Cultures of opposite mating types are crossed in Petri dishes containing an agar medium inducing recombination. The following protocol is based on O’Gorman et al. (2009).
-
1.
Prepare single spore isolates of each isolate and incubate under conditions inducing sporulation.
-
2.
Harvest the spores of each isolate in sterile water containing 0.05 % Tween 80.
-
3.
Inoculate 1.0–2.5 μL of each spore suspension (e.g., containing 500 spores) onto the agar surface about 4 cm apart and perpendicular to aliquots of spore-suspensions of the opposite mating type. This configuration created four interaction/barrage zones as colonies grew.
-
4.
Seal, if required, with Petri dishes with Parafilm and incubate at conditions inducing recombination.
-
5.
Regularly check the cultures on the production of fruiting bodies.
5.5.2 Mixed Culture Method
Agar slants are used in the protocol mentioned below and this method is useful when the agar medium needs be incubated for a long time. However, this method can be adopted for agar plates as well. In that case the agar media can be sealed with Parafilm in order to induce fruiting body formation and prevent drying out of the plates. The following protocol is based on Horn et al. (2009c).
-
1.
Grow the fungal strains under conditions inducing sporulation.
-
2.
Harvest the spores in sterile water containing 0.05 % Tween 80.
-
3.
Dilute or concentrate (e.g., by centrifugation) the spore suspensions until a concentration of 5 × 105 spores per mL is obtained.
-
4.
Mix the spore suspensions of strains of opposite mating types. Spore suspensions of single isolates can be used as negative controls.
-
5.
Inoculate the slants with 10 μL spore suspensions on a medium inducing a sexual cycle (production of sclerotia).
-
6.
Incubate the slants with loose caps until sclerotia are produced.
-
7.
Enclose the caps of the slants and enclose the slants in sealed plastic bags to prevent desiccation.
-
8.
Allow prolonged incubation and regularly check the cultures for the production of fruiting bodies.
5.5.3 Fertilization Method
The fertilization method below is derived from standard protocols used for Fusarium species (Klittich and Leslie 1988).
-
1.
Grow the fungal strains of one mating type under conditions inducing sporulation.
-
2.
Harvest the spores in sterile water containing 0.05 % Tween 80.
-
3.
Spread the cultures of one mating type on a suitable agar media.
-
4.
Incubate the agar plates until the agar is covered with fungal growth. Plates can be checked for evidence of ascogonia and protoperithecial formation.
-
5.
Fertilize the cultures by dispensing 1 mL of spore-suspension in 0.05 % Tween 80 carrying at least 5 × 105 conidia from the opposite strains.
-
6.
Work the spore suspension into the mycelia with a glass rod until the suspension is absorbed. Self-fertilizations can be made by substituting sterilized 0.05 % Tween 80 solution for the spore suspension. Any excess conidial suspension can be removed using a sterile Pasteur pipette.
-
7.
Seal, if required, with Petri dishes with Parafilm and incubate at conditions inducing recombination.
-
8.
Regularly check the cultures for the production of fruiting bodies.
5.6 Single Ascospore Cultures
In heterothallic ascomycete species, a successful mating experiment will lead to production of ascomata (ascocarps) (Fig. 2.1d, e) containing asci and ascospores (Fig. 2.1f, g). Single ascospores cultures then need to be obtained for further analysis. The method of isolation of the ascospores depends on the way the ascospores are produced (e.g., in a closed ascoma or “open” perithecium) and other features of the species in question (e.g., degree of heat resistance). Four commonly used methods are described below.
5.6.1 Direct Isolation
The easiest method to obtain single ascospores cultures is by transferring ascospores directly from the fruiting body. This method can be applied in, e.g. Fusarium, where ascospores are produced in a perithecium and ooze out to form a prominent spore mass. This method can also be adapted for closed fruiting bodies as described by Todd et al. (2007) for A. nidulans.
-
1.
Examine the agar plates using a stereomicroscope and isolate ascospores direct from a spore mass if present with a sharp needle. If necessary suspend the ascospores in 0.01 % Tween 20 or 0.05 % Tween 80 solution. In the case of closed fruiting bodies, these should be individually picked up and rolled across the surface of 4 % TWA plates to remove adhering conidia and hyphae [see Todd et al. (2007) for a detailed description]. The latter ascomata can then be transferred to a microfuge tube and crushed to release the ascospores. These should then be suspended in 0.01 % Tween 20 or 0.05 % Tween 80 solution.
-
2.
Transfer the ascospores onto a clear agar medium (e.g. water agar), spreading or using a decimal dilution series if a spore suspension is used.
-
3.
Incubate overnight (or longer as need be) at a temperature allowing germination of the ascospores.
-
4.
Locate single ascospores using a dissecting microscope and transfer these or hyphal tips from germinating ascospores to separate agar plates. Transfers can be efficiently made using a fine, flattened platinum wire mounted on the end of glass tubing. Such wires can be flame-sterilized and cool down rapidly.
5.6.2 Heat Treatment
The second method is based on a heat treatment of the ascospores. In some genera, ascospores are markedly more heat-resistant than the (heat-sensitive) asexual spores. This feature can be used to generate single ascospore isolates free of contaminating conidia and hyphae. For some species this heat treatment step is also required to trigger ascospore germination; without this ascospores do not germinate at standard growth temperatures (Perkins 1986; O’Gorman et al. 2009). This method has been applied for example in Neurospora, Paecilomyces, and Aspergillus sect. Fumigati species.
-
1.
Transfer the fruiting bodies from the agar plate into microfuge tubes containing 0.05 % Tween 80 or 0.01 % Tween 20 (ideally buffered at pH 6 with, e.g., 0.01 M sodium phosphate).
-
2.
Agitate the suspension with glass beads in order to break open the ascomata and asci, or simply crush the ascomata on the side of the tube before thoroughly mixing in the buffered Tween.
-
3.
Check by microscopy whether the majority of ascospores are released into the suspension. If not, repeat step 2.
-
4.
Heat-treat the suspension in a water bath or on a thermal cycler (time and temperature is species-dependent). Note that most conidia are eliminated by a treatment of 10 min at 60 °C.
-
5.
Make a decimal dilution in 0.05 % Tween 80 or 0.01 % Tween 20.
-
6.
Spread plate 0.1 mL of each dilution on agar plates (duplicate).
-
7.
Incubate at a suitable temperature allowing ascospore germination.
-
8.
After incubation, pick individual young germinating colonies from the highest dilutions (preferable) or locate single ascospores using a dissecting microscope as described above.
5.6.3 Ejected Ascospores
Many species forcibly eject ascospores from the ascomata. The fact that ascospores are shot out of the ascomata provides a convenient way to collect pure ascospores, free of contaminating hyphae and conidia (Dyer et al. 1993). However, some caution must be exercised as some species (notably lichen-forming fungi) eject ascospores in packets, so it can prove difficult to isolate individual ascospores. There are two related methods available according to how the ejected ascospores are trapped, as follows.
Method 1
-
1.
Pour a thin layer of clear agar (e.g. water agar) into the top (i.e. uppermost) lid of a Petri dish.
-
2.
Place the lid over a Petri dish containing either individual ascoma or sets of ascomata (transferred on agar to keep the cultures moist).
-
3.
Incubate the cultures for a suitable period (e.g., overnight or up to 48 h) during which time ascospores will be ejected onto the overlying agar.
-
4.
Inspect the lids containing the ascospores using a dissecting microscope.
-
5.
Subculture either ascospores or hyphal tips from germinating ascospores as described above.
Method 2
-
1.
Aliquot 3 mL of 0.05 % Tween 80 or 0.01 % Tween 20 into the bottom of a sterile 5 mL Petri dish.
-
2.
Attach either an individual ascoma or sets of ascomata to the lid of the Petri dish using Vaseline (petroleum jelly), ensuring that some of the underlying growth media is included to prevent desiccation of the ascoma.
-
3.
Incubate the cultures for a suitable period (e.g., overnight or up to 48 h) during which time ascospores will be ejected into the underlying liquid.
-
4.
Spread plate 0.5 mL aliquots of the Tween solution onto 9 cm Petri dishes containing a suitable clear media, and leave the dishes left to dry.
-
5.
Incubate further and inspect the plates using a dissecting microscope.
-
6.
Subculture either ascospores or hyphal tips from germinating ascospores as described above.
5.6.4 Isolation from Sclerotial Fruiting Bodies
If the fruiting bodies are firm and sclerotial, the following method can be used.
-
1.
Harvest the fruiting bodies by adding 10 mL 0.05 % Tween 80 to the culture (slant or plate).
-
2.
Scrape of the agar surface with a transfer loop.
-
3.
Filter the suspension through a 100 μm filter.
-
4.
Transfer the retained sclerotia/stromata to a vial.
-
5.
Vortex the suspension followed by decanting to remove residual conidia. Repeat this procedure at least five times.
-
6.
Filter the suspension onto Whatman #4 filter paper.
-
7.
Clean the stromata used for obtaining single-ascospore cultures further by vortexing 1–2 min in 10 mL sterile water containing 1 g glass beads (200–350 μm diameter).
-
8.
Remove from the beads.
-
9.
Vortex and decant twice with water.
-
10.
Dissect sclerotia/stromata with a micro-scalpel under the stereomicroscope.
-
11.
Transfer the ascospores onto an agar medium (e.g. water agar) as described above.
-
12.
Incubate overnight at a temperature allowing germination.
-
13.
Locate single ascospores using a (dissecting) microscope and transfer to separate agar plates.
5.7 Analysis of Progeny
A selection of the collected single-ascospore isolates should be assessed, to confirm that recombination has occurred in the case of putative outcrossing. Ascospore analysis can also be used to confirm the breeding system of the species in question. Various DNA fingerprinting techniques, such as RAPD and AFLP analysis, together with use of MLST markers and segregation of the MAT locus itself have been used to evaluate variation in the parents and progeny of a cross (e.g. Murtagh et al. 2000; Seymour et al. 2005; O’Gorman et al. 2009; Horn et al. 2009c; Swilaiman et al. 2013). It would be expected that self-fertilization via a homothallic breeding system would lead to uniformity in the ascospore progeny, whereas heterothallism and outcrossing would lead to genetic variation among the offspring (Murtagh et al. 2000). Indeed, a 1:1 segregation of mating types among the ascospore progeny is a clear indication of the presence of a heterothallic breeding system. However, it is cautioned that some ascospore progeny will be identical due to the fact that each set of eight ascospores in an ascus is composed of four sets of sister ascospores, derived from a mitotic division post-meiosis and tetrad formation.
6 Utilization of the Sexual Cycle as a Tool for Gene Identification and Manipulation
Once a reliable method has been developed to induce the sexual cycle in a given fungal species, it can then be used as a valuable laboratory tool for a range of applications such as classical genetic analysis, strain improvement, and as a complement to modern genetic manipulation experiments. A comprehensive description of the applications and uses of the sexual cycle is beyond the scope of the present chapter. Instead brief mention will now be made of ways in which the sexual cycle can be used in the context of fungal gene identification and transformation relevant to current accompanying chapters.
6.1 Genetics of Traits of Interest and Gene Identification
For many studies of gene transformation, candidate genes will have been identified at the onset of studies. However, in some cases the gene(s) responsible for a particular trait (and associated phenotype) of interest might be unknown. When studying such a trait it is very useful to know at the onset of gene manipulation studies whether that trait has a monogenic (i.e., determined by a single gene) or polygenic (i.e., determined by several genes) basis. This can influence the design of subsequent experimental and gene transformation studies. The sexual cycle provides an ideal tool to determine the genetic basis of a trait of interest because crosses can be set up between parents which differ in that trait. The ascospore progeny can then be collected and assessed for the trait of interest, with different patterns predicted in the frequency of offspring according to the genetic basis of the trait. In the case of a monogenic trait determined by a single dominant gene, it would be expected that haploid ascospore progeny will show a 1:1 segregation pattern for that trait. This can ideally be confirmed by backcrossing to the relevant parent. Conversely, if a trait has a polygenic basis with several genes segregating simultaneously then a more complex pattern of inheritance will be evident, with the progeny often failing to show distinct classes but rather a continuous distribution of phenotypes between the two parents (Caten 1979; Dyer et al. 2000).
Once the genetic basis of a trait is known, various techniques exploiting the sexual cycle are then available to locate and try and identify the specific gene or genes of interest, or other genetic causal factor(s). Examples include firstly the use of classical genetic mapping techniques. Genetic maps now available for almost 30 fungal species, although these vary in their coverage (Foulongne-Oriol 2012). By using two- and three-point crossing data it should be possible to locate the position of the gene between two known markers on a genetic map (Perkins 1986) and then chromosome walking and bioinformatic approaches can be used to try and identify specific genes. This topic has recently been reviewed by Foulongne-Oriol (2012) and examples of the use of mapping of Mendelian traits are provided in the review of Hall (2013). Secondly, a method termed ‘bulk segregant analysis’ (BSA) can be used to identify DNA marker(s) linked to a region of the genome responsible for a particular phenotype based on analysis of sexual progeny (Michelmore et al. 1991). This involves making pooled bulks of DNA from the progeny based on the presence or absence of the phenotype of interest. These pooled samples can either then be screened for the presence of DNA markers (e.g., PCR fingerprints) or subjected to next generation sequencing (NGS). In theory the only differences between the pools should arise from the genetic marker of interest together with regions of the genome linked to that marker. Examples of the application of BSA to filamentous fungi using molecular markers include the work of Chun and Lee (1999), Jurgenson et al. (2002), Jin et al. (2007), Lewis et al. (2007), and Dettman et al. (2010). Meanwhile, BSA has been applied in conjunction with NGS by Lambreghts et al. (2009), Pomraning et al. (2011), and Nowrousian et al. (2012). A third method for gene localization based on the analysis of sexual progeny is the technique of quantitative trait loci (QTL) analysis. The QTL method is especially suitable for providing insights into the genetic basis of more complex polygenic traits and can provide an estimation of the number of genes contributing to a particular trait and the identification of regions of the genomes linked to a particular trait (Miles and Wayne 2008). Hall (2013) has described how QTL mapping can be applied to genetic analysis of Neurospora species, including a review of the various mapping methods, and readers are referred there for further details. Recent examples of QTL analysis in filamentous fungi include those of Christians et al. (2011) in A. nidulans and Turner et al. (2011) in Neurospora. This method has the pre-requirement of parents which differ genetically with respect to the trait of interest, together with the presence of a dense genetic map. Thus, QTL mapping is only applicable to certain studies. It is also cautioned that although both BSA and QTL approaches can involve considerable work, they most often end with the identification of a genome region of interest with various candidate genes, rather than the actual identification of specific genes. Thus, further work is normally required after such studies.
6.2 Gene Manipulation by Sexual Reproduction: Strain Improvement and Gene Complementation
Other chapters in this book describe how the genetic composition of fungi can be manipulated by various methods of gene transformation. Although often overlooked, the sexual cycle can be also used as an efficient method for gene transformation.
The sexual cycle can be used to combine together genes of interest by crossing parents with the individual gene(s) and then selecting for ascospore offspring showing the desirable mixture of genes. For example, it might be desirable in certain gene function studies to produce mutant strains with multiple gene deletions. This can be achieved by lengthy rounds of transformation and marker recycling (Yoon et a1. 2011). However, the sexual cycle can provide an efficient alternative method because strains with complementary gene deletions can simply be crossed together. The sexual progeny are then screened for the presence or absence of the genes (e.g. by PCR assay) and isolates containing the desired combinations of multiple gene deletion selected for further study. In a parallel fashion, Böhm et al. (2013) illustrated how such an approach could be used for industrial strain improvement, with sexual progeny of P. chrysogenum screened for isolates that exhibited high penicillin titre but lack of a contaminating secondary metabolite chrysogenin. More broadly, sexual crosses can be set up to generate novel genetic diversity, allowing the offspring to be screened for isolates exhibiting, for example, either enhanced or decreased production of a particular metabolite, a phenomenon known as ‘transgressive segregation’ (Rieseberg et al. 1999).
Finally, the sexual cycle can be used for gene complementation purposes. When gene deletion has been shown to lead to a particular loss (or gain) of function, it is often required to then return the original gene back to the mutant strain to show that the original phenotype can be restored. This can be problematic if a limited number of selectable markers are available. However, by crossing the gene deletion strain with a strain containing the wild-type allele, it is possible to reintroduce the wild-type gene (Paoletti et al. 2007). If a consistent correlation between the presence of the wild-type gene (this can be screened for by PCR) and the presence of the restored phenotype can be demonstrated, or progeny which recapitulate the original genotype can be shown to exhibit the wild-type phenotype, then the role of the gene has been proven, i.e., proof of gene function through gene restoration by sexual crossing.
7 Conclusions and Outlook
Many industrially and clinically important fungal species were once thought to reproduce only asexually. Using a selection of the methods listed above, it has been shown that certain of these species may also reproduce sexually. For some other species a sexual state has still never been observed, although analyses of molecular markers indicate recombination. The term heterothallic is used for outcrossing species where a sexual state has been observed, and we suggest the term “proto-heterothallic” for such asexual species where genetic evidence, such as the presence of complementary MAT loci, indicates the presence of a sexual cycle. Similarly, for species with a homothallic mating type organization lacking a sexual state we propose the term “proto-homothallic”.
In this manuscript, we describe tools for the induction of a sexual state in heterothallic fungi. Based on the review, we show that no single protocol can secure induction of a sexual state in heterothallic fungi, not even in species belonging to the same genus, e.g., Aspergillus. There are likely to be more surprises waiting as we observe a ‘sexual revolution’ in fungi (Dyer and O’Gorman 2011). Recently, sclerotia production was induced in the proto-heterothallic species Aspergillus niger. The formation of sclerotia, thought to be sterile fruiting bodies in certain Aspergillus species, can be induced by inoculating A. niger onto fruits such as raisins, blueberries, cranberries, mulberries, apricot, prune, and mango on a CYA agar (Frisvad et al. 2014). By following (one of the) methods mentioned above and applying these specific growth conditions, a sexual state might be discovered in the near future for this biotechnologically important species. Finally, experimental crossings in vitro do not strictly reflect what happens in natura. For example, heat-resistant ascospores of the heterothallic P. variotii frequently spoil pasteurised fruit drinks and other food products, indicating a common occurrence of these ascospores and therefore the sexual state in nature. However, incubation times up to 6 weeks and specific agar media (PDA) are needed to induce sexual recombination in the lab (Houbraken et al. 2008).
Notes
- 1.
Commercially made oatmeal agar (OA) is available from certain manufacturers, but in our experience this is not able to induce sex in demanding species. Instead, it is best to prepare OA in house. Different brands of oats can be used and these have can have an effect on the mating. Commonly used brands are Pinhead oatmeal (Odlums, Ireland) and Quaker Oats. For Sordaria and Chaetomium species and Penicillium rubens (P. chrysogenum) this medium needs to be supplemented with biotin (6.4 μg/L) to induce sex (Böhm et al. 2013).
- 2.
In our experience Bacto™ agar is less prone to condensation problems than some cheaper, less pure, commercial agars.
References
Aanen DK, Hoekstra RF (2007) Why sex is good: on fungi and beyond. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA (eds) Sex in fungi: molecular determination and evolutionary principles. ASM, Washington, pp 527–534
Arabatzis M, Velegraki A (2013) Sexual reproduction in the opportunistic human pathogen Aspergillus terreus. Mycologia 105:71–79
Arie T, Christiansen SK, Yoder OC, Turgeon BG (1997) Efficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG box. Fungal Genet Biol 32:118–130
Astell CR, Ahlstrom-Jonasson L, Smith M, Tatchell K, Nasmyth KA, Hall BD (1981) The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell 27:15–23
Barrs VR, van Doorn TM, Houbraken J, Kidd SE, Martin P, Pinheiro DM, Richardson M, Varga J, Samson RA (2013) Aspergillus felis sp. nov., an emerging agent of invasive aspergillosis in humans, cats and dogs. PLoS One 8:e64871
Bistis GN (1998) Physiological heterothallism and sexuality in euascomycetes: a partial history. Fungal Genet Biol 23:213–222
Blakeslee AF (1904) Sexual reproduction in the Mucorineae. Proc Natl Acad Sci U S A 40:205–319
Böhm J, Hoff B, O’Gorman CM, Wolfers S, Klix V, Binger D, Zadra I, Kürnsteiner H, Pöggeler S, Dyer PS, Kück U (2013) Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc Natl Acad Sci U S A 110:1476–1481
Burnett J (2003) Fungal populations and species. Oxford University Press, New York
Caten CE (1979) Quantitative genetic variation in fungi. In: Thompson JN, Thoday JM (eds) Quantitative genetic variation. Academic, New York, pp 35–59
Cavindera B, Trail F (2012) Role of Fig1, a component of the low-affinity calcium uptake system, in growth and sexual development of filamentous fungi. Eukaryot Cell 11:978–988
Choi H-W, Kim J-M, Hong S-K, Kim WG, Chun S-C, S-H Y (2009) Mating types and optimum culture conditions for sexual state formation of Fusarium fujikuroi isolates. Mycobiology 37:247–250
Christians JK, Cheema MS, Vergara IA, Watt CA, Pinto LJ, Chen N, Moore MM (2011) Quantitative trait locus (QTL) mapping reveals a role for unstudied genes in Aspergillus virulence. PLoS One 6:e19325
Chun SJ, Lee Y-H (1999) Genetic analysis of a mutation on appressorium formation in Magnaporthe grisea. FEMS Microbiol Lett 173:133–137
Covert SF, Aoki T, O’Donnell K, Starkey D, Holliday A, Geiser DM, Cheung F, Town C, Strom A, Juba J, Scandiani M, Yang XB (2007) Sexual reproduction in the soybean sudden death syndrome pathogen Fusarium tucumaniae. Fungal Genet Biol 44:799–807
Covert SF, Briley A, Wallace MM, McKinney VT (1999) Partial MAT-2 gene structure and the influence of temperature on mating success in Gibberella circinata. Fungal Genet Biol 28:43–54
Crous PW, Verkleij GJM, Groenewald JZ, Samson RA (eds) (2009) Fungal biodiversity. CBS laboratory manual series 1. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands
Darbyshir HL, van de Vondervoort PJI, Dyer PS (2013) Discovery of sexual reproduction in the black aspergill. Fungal Genet REp 60(Suppl):687
Debuchy R, Berteaux-Lecellier V, Silar P (2010) Mating systems and sexual morphogenesis in ascomycetes. In: Borkovich KA, Ebbole DJ (eds) Cellular and molecular biology of filamentous fungi. ASM, Washington, pp 501–535
Debuchy R, Turgeon BG (2006) Mating-type structure, evolution, and function in euascomycetes. In: Kües U, Fischer R (eds) The mycota I: growth, differentiation and sexuality. Springer, Berlin, Germany, pp 293–323
Dettman JR, Anderson JB, Kohn LM (2010) Genome-wide investigation of reproductive isolation in experimental lineages and natural species of Neurospora: identifying candidate regions by microarray-based genotyping and mapping. Evolution 64:694–709
Dodge BO (1957) Rib formation in ascospores of Neurospora and questions of terminology. Bull Torrey Bot Club 84:182–188
Dyer PS (2007) Sexual reproduction and significance of MAT in the aspergilli. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA (eds) Sex in fungi: molecular determination and evolutionary principles. ASM, Washington, pp 123–142
Dyer PS, Bateman GL, Wood HW (2001a) Development of apothecia of the eyespot pathogen Tapesia on cereal crop stubble residue in England. Plant Pathol 50:356–362
Dyer PS, Furneaux PA, Douhan G, Murray TD (2001b) A multiplex PCR test for determination of mating type applied to the plant pathogens Tapesia yallundae and Tapesia acuformis. Fungal Genet Biol 33:173–180
Dyer PS, Hansen J, Delaney A, Lucas JA (2000) Genetic control of resistance to the DMI fungicide prochloraz in the cereal eyespot pathogen Tapesia yallundae. Appl Environ Microbiol 66:4599–4604
Dyer PS, Ingram DS, Johnstone K (1992) The control of sexual morphogenesis in the Ascomycotina. Biol Rev 67:421–458
Dyer PS, Nicholson P, Lucas JA, Peberdy JF (1995) Genetic control of sexual compatibility in Tapesia yallundae, 79. Abstracts, eighteenth fungal genetics conference, Asilomar. University of California, USA
Dyer PS, Nicholson P, Lucas JA, Peberdy JF (1996) Tapesia acuformis as a causal agent of eyespot disease of cereals and evidence for a heterothallic mating system using molecular markers. Mycol Res 100:1219–1226
Dyer PS, Nicholson P, Rezanoor HN, Lucas JA, Peberdy JF (1993) Two-allele heterothallism in Tapesia yallundae, the teleomorph of the cereal eyespot pathogen Pseudocercosporella herpotrichoides. Physiol Mol Plant Pathol 43:403–414
Dyer PS, O’Gorman CM (2011) A fungal sexual revolution: Aspergillus and Penicillium show the way. Curr Opin Microbiol 14:649–654
Dyer PS, O’Gorman CM (2012) Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev 36:165–192
Dyer PS, Paoletti M (2005) Reproduction in Aspergillus fumigatus: sexuality in a supposedly asexual species? Med Mycol 43(Suppl 1):S7–S14
Eagle CE (2009) Mating-type genes and sexual potential in the Ascomycete genera Aspergillus and Penicillium. PhD thesis, University of Nottingham, UK
Ehrenberg CG (1820) Syzygites, eine neue Schimmelgattung, nebst Beobachtungen über sichtbare Bewegung in Schimmeln. Verhandl. Gesamte Naturf., Freunde, Berlin, pp 98–109
Faretra F, Antonacci E, Pollastro S (1988) Sexual behaviour and mating system of Botryotinia fuckeliana, teleomorph of Botrytis cinerea. J Gen Microbiol 134:2543–2550
Foulongne-Oriol M (2012) Genetic linkage mapping in fungi: current state, applications, and future trends. Appl Microbiol Biotechnol 95:891–904
Fraser JA, Stajich JE, Tarcha EJ, Cole GT, Inglis DO, Sil A, Heitman J (2007) Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot Cell 6:622–629
Fresenius G (1863) Beiträge zur Mykologie. H.L. Brönner, Frankfurt, Germany
Frisvad JC, Petersen LM, Lyhne K, Larsen TO (2014) Formation of sclerotia and production of indoloterpenes by Aspergillus niger and other species in section Nigri. PLoS One 9:e94857
Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman J, Batzoglou S, Lee SL, Batürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren B (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115
Gordon WL (1961) Sex and mating type in relation to the production of perithecia by certain species of Fusarium. Proc Can Phytopathol Soc 28:11
Hall C (2013) Quantitative genetics in Neurospora. In: Kasbekar DP, McClusky K (eds) Neurospora genomics and molecular biology. Caister Academic, Norfolk, UK, pp 65–84
Han KH, Lee DB, Kim JH, Kim MS, Han KY, Kim WS, Park YS, Kim HB, Han DM (2003) Environmental factors affecting development of Aspergillus nidulans. J Microbiol 41:34–40
Heitman J, Carter DA, Dyer PS, Soll DR (2014) Sexual reproduction of human fungal pathogens. In: Casadevall A, Mitchell AP, Berman J, Kwon-Chung KJ, Perfect JR, Heitman J (eds) Fungal pathogens. Cold Spring Harbour Laboratory Press, New York
Henk DA, Eagle CE, Brown K, van den Berg MA, Dyer PS, Peterson SW, Fisher MC (2011) Speciation despite globally overlapping distributions in Penicillium chrysogenum: the population genetics of Alexander Fleming’s lucky fungus. Mol Ecol 20:4288–4301
Hoff B, Pöggeler S, Kück U (2008) Eighty years after its discovery, Fleming’s Penicillium strain discloses the secret of its sex. Eukaryot Cell 7:465–470
Horn BW, Moore GG, Carbone I (2009a) Sexual reproduction in Aspergillus flavus. Mycologia 101:423–429
Horn BW, Moore GG, Carbone I (2011) Sexual reproduction in aflatoxin-producing Aspergillus nomius. Mycologia 103:174–183
Horn BW, Olarte RA, Peterson SW, Carbone I (2013) Sexual reproduction in Aspergillus tubingensis from section Nigri. Mycologia 105:1153–1163
Horn BW, Ramirez-Prado J, Carbone I (2009b) The sexual state of Aspergillus parasiticus. Mycologia 101:275–280
Horn BW, Ramirez-Prado JH, Carbone I (2009c) Sexual reproduction and recombination in the aflatoxin-producing fungus Aspergillus parasiticus. Fungal Genet Biol 46:169–175
Houbraken J, Frisvad JC, Seifert KA, Overy DP, Tuthill DM, Valdez JG, Samson RA (2012) New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia 29:78–100
Houbraken J, Varga J, Rico-Munoz E, Johnson S, Samson RA (2008) Sexual reproduction as the cause of heat resistance in the food spoilage fungus Byssochlamys spectabilis (anamorph Paecilomyces variotii). Appl Environ Microbiol 74:1613–1619
Houbraken J, de Vries RP, Samson RA (2014) Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol 86:199–249
Idnurm A (2011) Sex determination in the first-described sexual fungus. Eukaryot Cell 10:1485–1491
Jin Y, Allen S, Baber L, Bhattarai EK, Lamb TM, Versaw WK (2007) Rapid genetic mapping in Neurospora crassa. Fungal Genet Biol 44:455–465
Jurgenson JE, Zeller KA, Leslie JF (2002) Expanded genetic map of Gibberella moniliformis (Fusarium verticillioides). Appl Environ Microbiol 68:1972–1979
Kawasaki M, Anzawa K, Wakasa A, Takeda K, Mochizuki T, Ishizaki H, Hemashettar B (2010) Matings among three teleomorphs of Trichophyton mentagrophytes. Jpn J Med Mycol 51:143–452
Kent CR, Ortiz-Bermúdez P, Gilies SS, Hull CM (2008) Formulation of a defined V8 medium for induction of sexual development of Cryptococcus neoformans. Appl Environ Biol 74:6248–6253
Kerényi Z, Moretti A, Waalwijk C, Olah B, Hornok L (2004) Mating type sequences in asexually reproducing Fusarium species. Appl Environ Microbiol 70:4419–4423
Klittich CJR, Leslie JF (1988) Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuroi). Genetics 118:417–423
Kwon-Chung KJ, Sugui JA (2009) Sexual reproduction in Aspergillus species of medical or economic importance: why so fastidious? Trends Microbiol 17:481–487
Kwon-Chung KJ, Weeks RJ, Larsh HW (1974) Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. Am J Epidemiol 99:44–49
Lambreghts R, Shi M, Belden WJ, deCaprio D, Park D, Henn MR, Galagan JE, Baştürkmen M, Birren BW, Sachs MS, Dunlap JC, Loros JL (2009) A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics 181:467–781
Lee SC, Ni M, Li W, Shertz C, Heitman J (2010) The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev 74:298–340
Lewis ZA, Shiver AL, Stiffler N, Miller MR, Johnson EA, Selker EU (2007) High-density detection of restriction-site-associated DNA markers for rapid mapping of mutated loci in Neurospora. Genetics 177:1163–1171
López-Villavicencio M, Aguileta G, Giraud T, de Vienne DM, Lacoste S, Couloux A, Dupont J (2010) Sex in Penicillium: combined phylogenetic and experimental approaches. Fungal Genet Biol 47:693–706
McAlpin CE, Wicklow DT (2005) Culture media and sources of nitrogen promoting the formation of stromata and ascocarps in Petromyces alliaceus (Aspergillus section Flavi). Can J Microbiol 51:765–771
Metzenberg RL, Glass NL (1990) Mating type and mating strategies in Neurospora. Bioessays 12:53–59
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A 88:9828–9832
Miles CM, Wayne M (2008) Quantitative trait locus (QTL) analysis. Nat Educ 1:208
Milgroom MG (1996) Recombination and the multilocus structure of fungal population. Annu Rev Phytopathol 34:457–477
Mooney JL, Yager LN (1990) Light is required for conidiation in Aspergillus nidulans. Genes Dev 4:1473–1482
Moore GG (2014) Sex and recombination in aflatoxigenic Aspergilli: global implications. Front Microbiol 5:32
Murtagh GJ, Dyer PS, Crittenden PD (2000) Sex and the single lichen. Nature 404:564
Nielsen KA, De Obaldia AL, Heitman J (2007) Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot Cell 6:949959
Nowrousian M, Teichert I, Masloff S, Kück U (2012) Whole-genome sequencing of Sordaria macrospora mutants identifies developmental genes. G3. Genes Genomes Genetics 2:261–270
Nováková A, Hubka V, Dudová Z, Matsuzawa T, Kubátová A, Yaguchi T, Kolařík M (2014) New species in Aspergillus section Fumigati from reclamation sites in Wyoming (U.S.A.) and revision of A. viridinutans complex. Fung Div 64(1):253–274. doi:10.1007/s13225-013-0262-5
O’Gorman CM, Fuller HT, Dyer PS (2009) Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474
Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis JP, Latgé JP, Denning DW, Dyer PS (2005) Evidence for sexuality in the opportunistic human pathogen Aspergillus fumigatus. Curr Biol 15:1242–1248
Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, Archer DB, Dyer PS (2007) Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol 17:1384–1389
Perkins DD (1986) Hints and precautions for the care, feeding and breeding of Neurospora. Fungal Genet Newsl 33:36–41
Pitt JI (1979) The genus Penicillium and its teleomorph states Eupenicillium and Talaromyces. Academic, London, UK
Pöggeler S (2001) Mating-type genes for classical strain improvements of ascomycetes. Appl Microbiol Biotechnol 56:589–601
Pomraning KR, Smith KM, Freitag M (2011) Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1. Eukaryot Cell 10:724–733
Pringle A, Baker DM, Platt JL, Wares JP, Latgé JP, Taylor JW (2005) Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886–1899
Raju NB, Perkins DD (1994) Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora and Podospora. Dev Genet 15:104–118
Ramirez-Prado JH, Moore GG, Horn BW, Carbone I (2008) Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet Biol 45:1292–1299
Raper KB, Fennell DI (1965) The genus Aspergillus. Williams & Wilkins Co, Baltimore
Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83:363–372
Robellet X, Oestreicher N, Guitton A, Vélot C (2010) Gene silencing of transgenes inserted in the Aspergillus nidulans alcM and/or alcS loci. Curr Genet 56:341–348
Ropars J, López-Villavicencio M, Dupont J, Snirc A, Gillot G, Coton M, Jany J-L, Coton E, Giraud T (2014) Induction of sexual reproduction and genetic diversity in the cheese fungus Penicillium roqueforti. Evol Appl 7:433–441
Rydholm C, Dyer PS, Lutzoni F (2007) DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot Cell 6:868–874
Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B (2010) Food and indoor fungi. CBS laboratory manual Series 2. CBS KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands
Samson RA, Varga J, Dyer PS (2009) Morphology and reproductive mode of Aspergillus fumigatus. In: Latgé JP, Steinbach WJ (eds) Aspergillus fumigatus and Aspergillosis. ASM, Washington, pp 7–13
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246
Seidl V, Seibel C, Kubicek CP, Schmoll M (2009) Sexual development in the industrial workhorse Trichoderma reesei. Proc Natl Acad Sci U S A 106:13909–13914
Seymour FA, Crittenden PD, Dickinson MJ, Paoletti M, Montiel D, Cho L, Dyer PS (2005) Breeding systems in the lichen-forming fungal genus Cladonia. Fungal Genet Biol 42:554–563
Short DP, O’Donnell K, Thrane U, Nielsen KF, Zhang N, Juba JH, Geiser DM (2013) Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet. Biol 53:59–70
Singh G, Dyer PS, Ashby AM (1999) Intra-specific and inter-specific conservation of mating-type genes from the discomycete plant-pathogenic fungi Pyrenopeziza brassicae and Tapesia yallundae. Curr Genet 36:290–300
Steenkamp ET, Wingfield BD, Coutinho TA, Zeller KA, Wingfield MJ, Marasas WF, Leslie JF (2000) PCR-based identification of MAT-1 and MAT-2 in the Gibberella fujikuroi species complex. Appl Environ Microbiol 66:4378–4382
Sugui JA, Losada L, Wang W, Varga J, Ngamskulrungroj P, Abu-Asab M, Chang YC, O'Gorman CM, Wickes BL, Nierman WC, Dyer PS, Kwon-Chung KJ (2011) Identification and characterization of an Aspergillus fumigatus “supermater” pair. MBio 2:e00234–11
Sugui JA, Vinh DC, Nardone G, Shea YR, Chang YC, Zelazny AM, Marr KA, Holland SM, Kwon-Chung KJ (2010) Neosartorya udagawae (Aspergillus udagawae), an emerging agent of aspergillosis: how different is it from Aspergillus fumigatus? J Clin Microbiol 48:220–228
Swilaiman SS, O’Gorman CM, Balajee SA, Dyer PS (2013) Discovery of a sexual cycle in Aspergillus lentulus, a close relative of A. fumigatus. Eukaryot Cell 12:962–969
Takada M, Udagawa S (1988) A new species of heterothallic Talaromyces. Mycotaxon 31:417–425
Takan JP, Chipili J, Muthumeenakshi S, Talbot NJ, Manyasa EO, Bandyopadhyay R, Sere Y, Nutsugah SK, Talhinhas P, Hossain M, Brown AE, Sreenivasaprasad S (2012) Magnaporthe oryzae populations adapted to finger millet and rice exhibit distinctive patterns of genetic diversity, sexuality and host interaction. Mol Biotechnol 50:145–158
Todd RB, Davis MA, Hynes MJ (2007) Genetic manipulation of Aspergillus nidulans: meiotic progeny for genetic analysis and strain construction. Nat Protoc 2:811–821
Turner E, Jacobson DJ, Taylor JW (2011) Genetic architecture of a reinforced, postmating reproductive isolation barrier between Neurospora spceies indicates evolution via natural selection. PLoS Genet 7:e1002204
Tschanz AT, Horst RK, Nelson PE (1976) The effect of environment on sexual reproduction of Gibberella zeae. Mycologia 68:327–340
Turgeon BG, Yoder OC (2000) Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet Biol 31:1–5
Yoon J, Maruyama J, Kitamoto K (2011) Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl Microbiol Biotechnol 89:747–759
Acknowledgments
J.H. thanks Tineke van Doorn and Richard Summerbell for the discussions on the term proto-heterothallic.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Houbraken, J., Dyer, P.S. (2015). Induction of the Sexual Cycle in Filamentous Ascomycetes. In: van den Berg, M., Maruthachalam, K. (eds) Genetic Transformation Systems in Fungi, Volume 2. Fungal Biology. Springer, Cham. https://doi.org/10.1007/978-3-319-10503-1_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-10503-1_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-10502-4
Online ISBN: 978-3-319-10503-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)