Abstract
Laser ablation electrospray ionization (LAESI) is an ambient ionization technique for mass spectrometry that is capable of performing direct spatial imaging on biological specimens with minimal-to-no sample preparation, ensuring experimental conditions that can maintain viability. Mass spectrometry imaging (MSI) by LAESI has accomplished utility in mapping the spatial distribution of small molecules including metabolites and lipids in a wide variety of biological samples under ambient conditions, ranging from sectioned animal and live plant tissues to living microbial colonies and small cohorts of cells. In this chapter, we provide a brief introduction to LAESI and offer practical guidance on performing MSI using this technique. The focus here is to discuss the main steps of custom-building a LAESI setup, to perform multidimensional imaging of tissues and cells, and to demonstrate the utility of LAESI MSI under native or native-like conditions. As recent commercialization has extended this new analytical resource to a broader user base, we anticipate two- and three-dimensional MSI by LAESI to benefit basic and applied research.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Mass spectrometry imaging
- Electrospray ionization
- LAESI
- Mass spectrometry
- Metabolites
- Lipids
- Peptides
- Proteins
- Atmospheric pressure
- Ambient
- Two-dimensional
- Three-dimensional
- Depth profiling
- Single-cell analysis
- Tissue imaging
- Bacteria
- Mid-IR laser
- Laser ablation
- O–H vibrations
- Resonant water excitation
- In vivo imaging
1 Introduction
Laser ablation electrospray ionization (LAESI) is an ambient ionization technique for mass spectrometry (MS) that is well suited for the investigation of biological tissues and cells (Nemes and Vertes 2007). As the underlying principles and operation of the technology have been the topic of various recent reviews (Nemes and Vertes 2012; Wu et al. 2013; Monge et al. 2013), visualized experiments (Nemes and Vertes 2010a; Shrestha and Vertes 2010), and book chapters (Vertes et al. 2013; Nemes and Vertes 2010b, 2015; Li et al. 2015b), the discussion here is limited to a brief introduction of the method followed by protocols to allow for performing LAESI MS imaging (MSI). using a custom-built system as well as the LAESI DP-1000, the commercialized platform.
LAESI is designed to utilize the substantial amount of water that is natively present in or exogenously added to the sample of interest such as a biological specimen. Sampling is accomplished by fast mid-infrared (mid-IR) ablation using 2940 nm as the excitation wavelength . At this wavelength, light energy is efficiently coupled into water-rich samples via resonant excitation of the O–H vibrations, predominantly in water, typically the most abundant component of biological samples. Based on fast imaging experiments (Apitz and Vogel 2005), the sudden deposition of energy gives rise to ablation at atmospheric pressure , which proceeds via three overlapping stages. In the first few hundred nanoseconds, fast surface evaporation and phase explosion produce an ablation plume that rapidly expands away from the surface of the sample. As expansion of this plume is slowed by collisions with ambient gas molecules, the plume eventually halts and collapses back onto the sample, causing the buildup of significant pressure locally in the sample surface. The last stage of the process is driven by the relaxation of this pressure through the ejection of neutral material, projecting particulate matter over hundreds of microseconds and several millimeters to centimeters above the sample surface.
Immediately after ablation, these ejected neutral projectiles are captured in charged droplets generated by an electrospray source to convert molecules of the sample to gas-phase ions that can be analyzed by a mass spectrometer. Based on the formation of multiply charged droplets and little-to-no fragmentation, the process by which ions are generated from the charged droplets in LAESI is thought to be similar to those in electrospray ionization (ESI ) (Nemes et al. 2012; Nemes and Vertes 2007). These mechanisms lend utility to convert a broad range of biomolecules to intact molecular ions, ranging from small metabolite s to large proteins (Shrestha et al. 2013). As LAESI-generated ions have been found to have indistinguishable internal energy to those produced by traditional ESI (Nemes et al. 2012), laser ablation practically extends classical, soft ESI to microprobe operation with in situ and in vivo operation.
Furthermore, sampling by mid-IR laser ablation and ionization by ESI allows for flexibility in meeting select analytical and biological research needs. There is flexibility in the physical dimensions of interrogation, the resolution of a spatial imaging experiment, and the types of biomolecules of interest that can be pursued. By adjusting the laser light fluence and number of laser pulses, the extent of ablation is readily tunable usually with cylindrical voxel dimensions between ~500 μm diameter × ~500 μm depth (multiple laser shots), or ~100 nL, and ~20 μm diameter × ~20 μm depth (single laser pulse), or ~5 pL, using classical light-focusing optical elements (‘conventional’ LAESI) and sharpened optical fibers (fiber-optic LAESI), respectively. These metrics have fostered applications in the domains of spatial profiling (Nemes and Vertes 2015), lateral imaging in two dimensions (Nemes et al. 2010; Li et al. 2015a; Nemes et al. 2008) and with single-cell resolution (Shrestha et al. 2010b; Shrestha et al. 2011), as well as subcellular interrogations (Stolee and Vertes 2013; Stolee et al. 2012). Furthermore, repeated ablation at a particular location (pixel) on the sample enables depth profiling of the chemical composition in tissues (Nemes et al. 2008). The combination of depth profiling and two-dimensional (2D) MSI laid the foundation for three-dimensional (3D) MSI (Nemes et al. 2009) in situ without the need for tissue sectioning and subsequent 2D imaging of the sections, a feature that is unique to LAESI (Nemes and Vertes 2012).
MSI using mid-IR LAESI offers complementary performance to contemporary MS technologies. The use of water as the natural light-absorbing medium minimizes signals in the low-mass region of the LAESI mass spectrum (<m/z 500), extending classical ultraviolet matrix-assisted laser ablation desorption/ionization (MALDI) to small molecules, particularly primary and secondary metabolite s, lipids , and drugs. Besides LAESI, various other ambient ionization sources have demonstrated a continually expending array of success for MSI . Representative examples include but are not limited to desorption electrospray ionization (DESI), electrospray-assisted laser desorption ionization (ELDI), and liquid extraction surface analysis (LESA) mass spectrometry. For these and other ambient ionization MSI techniques, readers are referred to recent reviews in the field (Wu et al. 2013; Nemes and Vertes 2012). In what follows, we discuss the use of LAESI for spatial interrogations of biomolecules in samples with no or limited sample preparation.
2 Applications
The integration of three different technologies—these are laser ablation, electrospray ionization, and mass spectrometry—harbors broad operational space in LAESI, fostering niche applications in MSI. These include analysis with in situ and in vivo operation, microprobe-type chemical analysis for lateral and depth profiling , single-cell measurements, and 3D MSI (see references earlier). Representative applications are summarized in Table 1. Here, we review methods and provide useful comments, or tips, toward a successful LAESI MSI experiment .
2.1 LAESI MSI Using Classical Optics
In the conventional configuration, LAESI employs classical light-diffraction elements to steer and focus the mid-IR laser beam and an electrospray source to ionize ablated biomolecules. The schematics of the LAESI M SI platform are shown in Fig. 1. The relative positioning of the ablation plume and electrospray source determines the efficiency of their intersection, which in turn controls the efficiency of ion production. The positioning of this intersection volume relative to the inlet of the mass spectrometer defines the ion collection efficiency for mass analysis, evaluated as the signal-to-noise ratio in the LAESI mass spectrum. Hence, LAESI experiments typically begin with the careful initialization of the configuration, encompassing various interconnected variables for ionization. Critical variables are the angle of laser ablation (θ LA; cf. Fig. 1) and absolute distances between the sample and electrospray (d S−ES), laser ablation plume and the electrospray plume (d LA−ES), and the laser ablation plume and the MS inlet (d LA−MS). Spray potential, electrospray flow rate, and angle of ion collection settings are secondary variables that are adjusted afterward. To accomplish a required set of performance metrics, iterative refinement is recommended for these parameters.
LAESI ion source for a successful MSI experiment. The schematic highlights the primary experimental variables, overlaid on the picture of the LAESI DP-1000 ion source (Protea Biosciences). Key: MS, mass spectrometer inlet (extension tube); d LA−MS, distance between laser ablation plume and mass spectrometer inlet; d LA−ES, distance between laser ablation plume and electrospray emitter tip; d ES−MS, distance between electrospray tip and mass spectrometer inlet; d S−ES, distance between sample and electrospray axis; X–Y–Z, independent axes of translation; f, working distance of mid-IR light-focusing optical element; θ LA, angle of incidence for mid-IR light from the sample surface; and R X/Y/Z , imaging resolution in the X/Y/Z direction
Molecular imaging by LAESI relies on the coordination of spatially resolved microsampling and mass spectrometric analysis of the generated ions. Typically, the sample is rastered in the focal point of the mid-IR laser beam across a predefined interrogation area in the X–Y directions (D X/Y ), desired spatial resolution (R X/Y/Z ), and accumulation times (dwell time, τ X/Y ) in the X and Y lateral directions during 2D imaging. An additional set of variables in the Z direction extends imaging to three dimensions. Hence, the total amount of time required for imaging is given by \( {t}_{2D}=\left({D}_X{D}_Y{\tau}_X{\tau}_Y\right)/\left({R}_X{R}_Y\right) \) in two dimensions and by \( {t}_{3D}=\left({D}_X{D}_Y{D}_Z{\tau}_X{\tau}_Y{\tau}_Z\right)/\left({R}_X{R}_Y{R}_Z\right) \) in three dimensions. Synchronization between these variables spatially and temporally has been automated in the LAESI DP-1000® (Protea Biosciences, Morgantown, WV, USA), the commercialized LAESI platform that houses all components necessary for mid-IR laser ablation, electrospray ionization, sample translation, equipment for cooling/heating, and specialized software analysis tools. These instrumental components are fine-controlled by a dedicated software (LAESI Desktop Software; Protea Biosciences) to allow for thorough optimization of the experimental parameter space. LAESI DP-1000 is capable of interfacing to various types of mass spectrometers using an adapter assembly (see MS extension tube in Fig. 1), and the resulting data sets are analyzed using ProteaPlot (Protea Biosciences), a dedicated imaging software.LAESI MS demonstrates increasing utility in mapping the distribution of metabolite s and lipids from different tissues (plant to animal) and bacterial and fungal species (Lin et al. 2014; Walsh et al. 2012; Razunguzwa et al. 2014; Boyce et al. 2014) such as Bacillus cereus, Staphylococcus aureus, Pseudomonas aeruginosa, Aspergillus fumigatus, and Arbuscular mycorrhizal fungi spores, many of which are pathogenic for humans.
2.2 High-Resolution MSI with Optical Fibers
To accomplish different imaging resolutions for particular applications, as shown in Fig. 2, the size of the ablation crater is tuned depending on the physicochemical properties of the tissue (e.g., water content and tensile strength) and the laser (e.g., fluence and pulse width). Conventional optics for mid-IR light-focusing such as CaF2 or ZnSe lenses have successfully led to imaging resolutions between ~150–500 μm, capable of deciphering major anatomical regions in tissues. In cases, where finer features such as cell-to-cell differences need to be distinguished, higher resolution is needed; a resolution of ~25–50 μm is required to resolve neighboring cells in many epidermal tissues of plants. Furthermore, because cells are the fundamental units of organization in these samples, it is logical to use cells as the actual pixels for MSI. To realize imaging with 10–50 μm resolution, germanium oxide (GeO2) optical fibers have been used as the mid-IR light-focusing element in a near-field configuration. The optical fiber was etched down to a few micrometers and positioned proximally to the cell of interest to ablate areas smaller than single cells (Shrestha and Vertes 2009). Consecutive ablation of single cells in a preselected area using this fiber allowed for cell-by-cell imaging using LAESI (Shrestha et al. 2011; Li et al. 2015). Figure 2 provides the workflow and decision process for two applications demanding different resolutions.
Experimental workflow for MSI using LAESI (Left). Key workflow steps (Middle). In the traditional setting, conventional mid-IR light-focusing optical elements are used to obtain resolutions between ~150–500 μm (pixel size). Typical samples in this setting range from plant and animal tissues to bacterial colonies (Right). To extend the resolution to single cells, the mid-IR light-focusing optic is replaced with a fiber optic, allowing for local ablation with ~10–50 μm resolution. By utilizing the coordinates of each cell to define pixels of the raster pattern, the entire tissue is imaged cell by cell
2.3 Ion Mobility Separation Coupled to LAESI MSI
Recent studies demonstrated that the addition of a separation step prior to mass analysis of LAESI-generated ions enhances the signal-to-noise ratio and facilitates molecular identifications (Li et al. 2015a; Shrestha and Vertes 2014). Ion mobility mass spectrometry (IMS) has sufficiently high spectral resolution and speed to be able to analyze the complex mixture of ions that is generated within seconds of a LAESI experiment. The combination of LAESI and IMS extends measurements to a new domain (LAESI IMS MSI), where it becomes possible to distinguish isobaric ions and structural isomers that are not separated by mass-to-charge ratio (m/z) in the traditional approach. In combination with IMS , LAESI MSI raises an opportunity to identify and map the spatial distribution of isobaric molecules that can have very different biological functions (Nielen and van Beek 2014).
3 Materials and Protocols
In this section, we describe the essential steps to custom-build a LAESI interface and to utilize it for 2D and 3D MSI of tissues of animals and plants. Representative examples are shown for coronal brain sections of the rat (Rattus norvegicus) and the zebra plant (Aphelandra squarrosa) in Fig. 3. We present progressive advances in refining the resolving power to the level of individual cells. Using the LAESI DP-1000, we demonstrate that MSI is significantly simplified using various automated features and software that are provided by this platform.
Examples of MSI in 2D and 3D using a custom-built LAESI system (Top). 2D imaging of a coronal rat brain section (R. norvegicus) revealed heterogeneous distribution of glycerophosphoethanolamine 36:2 (m/z 728.559) across the tissue with accumulation in the corpus callosum (Bottom). 3D imaging of the zebra plant leaf (A. squarrosa) uncovered different molecular distributions laterally and cross-sectionally in the tissue. For example, chlorophyll a (green-blue, m/z 893.546) was more abundant in the mesophyll layer, whereas kaempferol/luteolin (yellow-red, m/z 287.049) accumulated in the veins of the plant with higher concentration in the second-to-third layer of the tissue (Images were adapted with permission from (Nemes et al. 2009). Copyright 2009 American Chemical Society.)
3.1 2D and 3D LAESI MSI
3.1.1 Materials
-
1.
Set up an electrospray source as follows. Supply the electrospray solution at 200–400 nL/min through a tapered-tip stainless steel emitter (e.g., MT320-50-5-5; New Objective, Woburn, MA, USA) using a syringe pump (e.g., Physio 22; Harvard Apparatus, Holliston, MA, USA). Connect the emitter to a stable high-voltage power supply capable of providing regulated ~3000 V (e.g., PS350; Stanford Research Systems, Sunnyvale, CA, USA).
-
2.
Position the tip of the electrospray emitter ~10 mm on-axis from the inlet of a mass spectrometer (d MS−ES) using a three-axis translation stage.
-
3.
Mount the sample on a motorized three-axis translation stage (e.g., LTA-HS; Newport, Irvine, CA, USA) ~13 mm below the electrospray emitter (d S−ES). Ensure that the sample surface is appreciably uniform in topography (flat), water content, and tissue strength across the surface that is desired to be imaged.
-
TIP: Note the locations of fibrous veins in plants, bones in animal tissues, and dried areas as these areas will likely generate little-to-no signal. This information can be helpful when interpreting results.
-
-
4.
Focus a laser beam of 2940 nm wavelength, <100 ns pulse duration, and 2–20 Hz frequency on the sample surface by steering it through a plano-convex lens using appropriate mirrors. In our experiments, we used a Nd:YAG laser with an optical parametric oscillator to generate the light beam (e.g., Opolette 100; Opotek Inc., Carlsbad, CA, USA) and gold-coated mirrors (Thorlabs, Newton, NJ, USA) to focus the laser light through a CaF2 lens with f = 50 mm (Thorlabs) onto the sample.Footnote 1 WARNING: Direct exposure of Class IV laser beams can cause permanent eye damage. Always wear appropriate protective eyewear that block the beam at the operating wavelength and perform these experiments in appropriate and designated laser areas. Consult your local safety officer and laser health and safety rules and regulations for details.
-
TIP: Set the distance between the lens and sample to the focal length of the lens before focusing the laser beam to establish a good starting point for fine-tuning ablation in later steps.
-
-
5.
Generate the electrospray by applying ~3000 V to the emitter and supplying 50% methanol (or acetonitrile) with 0.1% acetic/formic acid for the positive ion mode and 50% methanol (or acetonitrile) with 0.1% ammonium acetate (or ammonium hydroxide) for the negative ion mode at the above stated flow rate using a syringe pump.
-
WARNING: High voltage poses electric shock hazard. Properly shield all conductive surfaces, and ensure appropriate d ES−MS (at least ~10 mm) to avoid electrical breakdown to the mass spectrometer.
-
WARNING: Follow standard safety protocols to work with solvents and reagents. As a general guide, avoid direct contact or inhalation of solvents to prevent irritation to the skin, eyes, and the respiratory tract.
-
-
6.
Interface the LAESI ion source to a mass spectrometer that has an atmospheric pressure (AP) interface . We have successfully adapted multiple custom-built LAESI systems and different LAESI DP-1000s to various types of mass spectrometers ranging from time-of-flight and ion trap instruments to instruments incorporating ion mobility separation (refer to publications earlier).
-
7.
(Optional) In preparation for MSI of animal tissues, utilize a cryostat microtome (e.g., CM1800; Leica Microsystems Inc., Buffalo Grove, IL, USA) to prepare ~20–150 μm-thick sections of tissue and immediately freeze these section onto glass slides to avoid the loss of native water.
-
WARNING: Always handle cold surfaces using a pair of appropriate protective gloves.
-
3.1.2 Setup Initialization
-
1.
Coarse-adjust the optical beam path to yield ablation marks of desired size on the sample. The dimension of the ablation area will determine the maximal imaging resolution.
-
TIP: Start with a thermosensitive (burn) paper as the sample (e.g., S-18642; Uline, Pleasant Prairie, WI, USA) to monitor the optical path and establish conditions yielding ablation marks of ~250 μm circular diameter.1 Inspect the ablation marks for symmetry (circular vs. ellipsoidal burn marks). Afterward, the thermosensitive paper should be replaced with the sample to refine the setup (e.g., for an animal tissue section).
-
-
2.
Obtain a LAESI mass spectrum for the sample, and select a biomolecular ion of interest which will be used to optimize the setup.
-
TIP: It is useful to select two different ions (m/z peaks), one with high initial abundance to coarse-adjust the setup, and the other with lower abundance for fine tuning. Inspect the ablation marks to avoid obtaining increasing S/N at the expense of lower resolution, unless acceptable otherwise.
-
-
3.
Fine-tune all parameters of the LAESI setup (including the source geometry, voltages, and optics) to maximize ion production for the biomolecular ions of interest. During this step, it is important to utilize a pristine tissue area at each X–Y–Z coordinate between consecutive conditions.
-
TIP: Continuously raster the sample in the X–Y dimensions manually or by using the motorized translation stage (line scan function).
-
-
4.
For 3D MSI , determine the resolution of depth profiling for the tissue by inspecting the depth of the ablation crater under a microscope (upright or stereo). The depth of the ablation crater determines the depth resolution (R Z ) during 3D MSI.
-
TIP: Fine-adjust the laser fluence while inspecting the ablation crater in the lateral (X–Y) and the cross-sectional (Z) directions using the microscope.
-
3.1.3 Perform 2D/3D LAESI MSI
-
1.
Mount a fresh sample on the translation stage.
-
TIP: For plant tissues, use tape to immobilize the sample on a microscope glass slide, and mount the slide into a slide holder held by the translation stage. For animal tissues, thaw-mount the tissue by allowing it to thaw for a few seconds followed by prompt refreezing on a substrate cooled by a Peltier stage.2
-
-
2.
Determine the area of interest on the sample, and calculate the amount of time required for imaging, t 2D for 2D and t 3D for 3D imaging. Based on this time, calculate the amount of electrospray solvent that will be necessary to perform the imaging experiment, and also determine the size of MS data that will be generated.
-
3.
To ensure that the experiment will be performed without obstruction, inspect that the translation stage has a free travel range in the X, Y, and Z directions, the syringe supplying the electrospray solvent contains a sufficient volume of the solvent and the corresponding piston has adequate translation range, and the PC controlling the mass spectrometer has sufficient disk space. Ensure that the MS instrument is able to collect data for the entire time.
-
4.
(Important) Depending on the sample, ablated particles may deposit on the electrospray emitter , at which point direct ESI may elevate the background signal for an extended amount of time. This can be readily avoided by refining the laser pulse energy, increasing d S−ES, and/or occasionally cleaning the surface of the electrospray emitter.
-
5.
To perform MSI in two dimensions, execute the following steps in sync: turn on the mid-IR laser, start rastering the sample surface in the X–Y plane, and simultaneously acquire MS data while delivering multiple laser shots at each location, i.e., X–Y coordinate (pixel).
-
6.
To perform MSI in three dimensions, utilize single laser pulses and register the corresponding mass spectra individually for each laser pulse.
-
TIP: Utilize at least 2–3-times faster data acquisition rates than the repetition rate of the mid-IR laser source. In our experiments, we found 0.2 Hz repetition rate and 1–2 Hz mass spectrometric data acquisition rate particularly useful during the development of 3D MSI. Note that the LAESI DP-1000 provides a commercial source for 3D LAESI MSI.
-
-
7.
To finish the experiment, turn off the mid-IR laser, the data collection, the electrospray source, and the syringe pump.
-
TIP: Rinse the electrospray emitter with water–methanol mixture.
-
-
8.
Visualize the molecular distribution of detected ions by correlating their selected-ion peak areas to the coordinates of analysis, X–Y in 2D MSI and X–Y–Z in 3D MSI, where Z corresponds to individual laser pulses delivered in the Z direction.
-
EXAMPLE: Representative imaging results are provided in Fig. 3 using the coronal section of the rat brain for a 2D and the leaf of the zebra plant (A. squarrosa) for a 3D experiment. Comparison of the optical image with the 2D ion image reveals that glycerophosphoethanolamine was accumulated in the corpus callosum of the brain. The example for 3D imaging demonstrates that chlorophyll a and kaempferol/luteolin have a vastly different lateral and cross-sectional distribution in the leaf.
-
3.2 Imaging Bacterial Colonies Using the LAESI DP-1000
The following protocol outlines molecular imaging of living microbial colonies on agar using the LAESI DP-1000 system.
3.2.1 Imaging Bacterial Colonies Using the LAESI DP-1000
-
1.
Install the LAESI DP-1000 system on a compatible mass spectrometer (Thermo Fisher Scientific and Waters mass spectrometers), and activate the data acquisition and analysis software packages.
-
2.
Pour agar close to the brim of the Petri dish. After solidification of the agar, plate the bacteria and incubate them until desired amount of growth has occurred.
3.2.2 Optimization of the Ion Source
-
1.
Place the bacterial culture plate on the sample tray of the DP-1000. Using interactive mode, adjust the stage in the Z dimension to d S−ES = ~15 mm.
-
2.
Set d MS−ES to 7 mm. At this distance, the d LA−ES measurement will be 2 mm.
-
TIP: Good starting parameters for the electrospray are: 4000 V spray potential; 1 μL/min flow rate; 50% methanol with 0.1% acetic acid in positive ion mode.
-
-
3.
Use the interactive mode to fine-tune the focusing distance (f, Fig. 1).
-
TIP: It is recommended to empirically determine the focal distance by translating the focusing lens until the sample is in visual focus (in line with the camera) and by monitoring the mass spectrometric signal intensity upon delivering the laser pulses.
-
-
4.
Select the appropriate laser fluence .
-
TIP: Recommended starting parameters for the laser include 80% energy (~800 μJ) and 10 pulses at 10 Hz repetition rate for 2D imaging of bacterial colonies.
-
3.2.3 Data Acquisition
-
1.
To start an imaging experiment, set up a project in the control software (LAESI Desktop Software) of the LAESI DP-1000. Take a photograph of the sample using the onboard wide-angle camera and define a desired imaging area by using a grid.
-
2.
Set the spatial resolution of the grid using the LAESI Desktop Software. A setting of ~200 μm provides a robust initial estimate.
-
3.
Save experimental parameters includi ng analysis locations, electrospray conditions, stage temperature, and laser settings, in the project for archive use. After saving the project, start the analysis. MS data can be acquired manually or automatically with the LAESI DP-1000. When the analysis is complete, the LAESI DP-1000 will automatically turn off the LAESI subsystems by default. The data acquisition can be stopped automatically (e.g., after a pre-defined time period in the software) or terminated manually.
3.2.4 Data Analysis
-
1.
Use ProteaPlot to visualize molecular images for the detected ions of interest.
-
TIP: Experiment with background subtraction, photographic and molecular image overlay, spectral comparison, and data export capabilities in ProteaPlot to highlight features of interest and en hance data analysis.
-
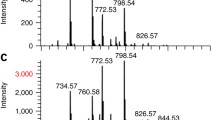
EXAMPLE: A representative ex ample is provided for the human pathogen Serratia marcescens in Fig. 4. For this experiment, a bacterial colony of S. marcescens was grown on LBK agar and imaged in two dimensions using the LAESI DP-1000. The positive ion mode mass spectrum (m/z 150–2000) shows a complex molecular makeup with hundreds of metabolite s. The molecular images were constructed for six selected molecular species in the colony using the ProteaPlot software .
Fig. 4 MSI of bacterial colonies using the LAESI DP-1000 (Top). Photographs of Serratia marcescens bacterial colony growing on LBK agar area before and after LAESI. Ablation craters are visible after the analysis (Bottom). A representative mass spectrum acquired during LAESI MSI of the colony. Insets (a) to (e) correspond to the molecular image of various mass-selected ions that were detected in this spectrum (see m/z values in spectrum). Ion intensity increases from blue to red color. Using accurate mass and tandem MS, the signal labeled (a) at m/z 324.5 was identified as prodigiosin, a secondary metabolite pigment that provides the characteristic red color of this bacterial species
-
3.3 Imaging Cell by Cell
Last, this section describes a cell-by-cell imaging protocol for a monolayer of single cells using a custom-built LAESI system that is equipped with an optical fiber (cf. Fig. 2). A representative image of the optical fiber setup is shown in Fig. 5.
LAESI MSI with single-cell resolution (Top). The tip of an etched optical fiber transmitting mid-IR light is brought proximally to individual cells of interest to perform local ablation. A two-camera system measures distances and angles of incidence (not shown). Critical parameters requiring careful optimization are labeled. Key: d S−F , distance between sample (cell) and fiber tip (Bottom). To obtain molecular images on a cohort of cells, the LAESI analysis is performed on every cell of the population, one cell at a time, using the sharpened optical fiber. As an example, the ion image of cyanidin (m/z 287) is overlaid on the optical image of an onion epidermal tissue, demonstrating that this ion correlates with purple pigmentation (Images were adapted with permission from (Shrestha and Vertes 2009; Shrestha et al. 2011). Copyrights 2009 and 2011 American Chemical Society)
3.3.1 Materials
-
1.
Set up an electrospray source, mid-IR laser, three-axis translation stage, and mass spectrometer in function as described in Sect. 3.1.
-
2.
Use a germanium oxide (GeO2) optical fiber transmitting in the mid-IR spectral range (e.g., 450 μm core diameter HP fiber; Infrared Fiber Systems Inc., Silver Spring, MD, USA) and sapphire blade (e.g., KITCO Fiber Optics, Virginia Beach, VA, USA) for cleaving this optical fiber.
-
3.
Use a fiber micromanipulator (e.g., MN-151; Narishige, Tokyo, Japan) and other miscellaneous optical components (Thorlabs) as needed.
-
4.
For visualization of the tissue in perpendicular directions, employ a long-working-distance microscope system consisting of two independent cameras (e.g., K1 CentriMax; Edmund Optics Inc., Barrington, NJ, USA).
-
5.
Prepare a mock sample such as the abaxial or adaxial epidermis layers peeled off from a leaf (e.g., Peace lily), mounted on a clean microscope slide, and attached to the three-axis translation stage.
3.3.2 Preparing the Optical Fiber
-
1.
Remove the plastic coat of the optical fiber by dipping it into heated 1-methyl-2-pyrrolidinone (reagent grade) until the coat turns soft, allowing to peel off the coat with a lint-free tissue. Minor debris needs to be washed off by rinsing with methanol. Score the ends of the fiber using a sapphire blade, and gently snap the ends off to attain clean, blunt tip ends.
-
TIP: For the next step, it is important to ensure that the optical fiber has smooth ends. Repeat this step if needed. Inspect the quality of the fiber ends under a microscope.
-
-
2.
Chemically sharpen (etch) one end of the fiber using 1–2% nitric acid (reagent grade) as follows: To produce a fiber with ~20 μm tip diameter, immerse the end of the fiber vertically in the nitric acid solution ~0.3–0.5 mm below the meniscus until the fiber tip automatically detaches from the acid surface. Remove the sharpened tip and rinse it with deionized water prior to use. The other end of the fiber should be left unmodified.
3.3.3 Setup for LAESI Single-Cell MSI
-
1.
Mount the fiber on a fiber chuck (e.g., BFT-5; Siskiyou Corporation, Grants Pass, OR, USA) using a bare fiber chuck (e.g., BFC300; Siskiyou). A schematic of the setup is shown in Fig. 5 (top panel).
-
2.
Couple the mid-IR laser beam into the fiber by focusing it onto the blunt end of the fiber using a plano-convex CaF2 lens.
-
3.
Position the sharpened end of the fiber a few millim eters from the sample surface at an incidence angle θ LA of ~30°.
-
TIP: Use a dual-camera system to aid positioning of the fiber and to prevent accidental damages to the fiber tip. Should the tip break off, dismount the optical fiber, and re-etch its end to the desired tip dimension.
-
-
4.
Deliver the mid-IR laser pulses at high repetition rate (e.g., 10 Hz) to individual cells using the sharpened optical fiber to cause localized ablation, usually with dimensions <50 μm in diameter. Gently lower the fiber tip to a distance from the surface of the tissue (d S−F ) of ~20 μm to ablate the cell of interest. It is common that rupture of the cell wall occurs after a few tens of laser pulses.
-
TIP: Optimize the position of the fiber tip with respect to the mass spectrometer to maximize the signal-to-noise ratio for biomolecules of interest.
-
-
5.
Maneuver the sharpened end of the fiber tip in front of the mass spectrometer orifice by using a micromanipulator at an angle θ LA of ~45° and adjust the distance/position between the optical fiber tip and the cell surface (d S−F ) to obtain the best signal. Be careful not to break the tip.
-
6.
Turn on the electrospray source, the mass s pectrometer, and the mid-IR laser to begin data collection.
-
TIP: Step-by-step instructions on single-cell analysis using LAESI are discussed in greater detail elsewhere (Shrestha et al. 2010a).
-
3.3.4 Cell-by-Cell LAESI MSI
-
1.
Perform successive ablations on individual selected cells across a pre-defined area of the tissue by positioning the sharpened optical fiber tip proximally to the cells of interest and coupling the mid-IR light into the optical fiber.
-
TIP: This process can be performed manually. The sample can be readily positioned using a manual translation stag e while observing single cells with the long-working-distance microscope. This process can also be automated based on the coordinates of the single cells.
-
-
2.
Optimize the repetition rate of the laser and its energy output to obtain the highest signal-to-noise (S/N) ratio from each sample pixel at a desired dwell time (e.g., 4 s).
-
TIP: (1) For single-cell analysis, m ake sure to tune the laser parameters to obtain the best mass spectrum with minimal damage to the neighboring cells. (2) For automated cell-by-cell imaging, identify coordinates around the centroids of single cells using an image processing software (e.g., MetaMorph; Olympus Corporation, Center Valley, PA, USA) and supply the coordinate data to a stage-control program to sequentially position the cells under the optical fiber tip for analysis. A protocol for automated cell-by-cell imaging is described in detail elsewhere (Li et al. 2015b).
-
-
3.
To finish the experiment, turn off all the subsystems and stop the data acquisition.
-
4.
Visualize the molecular distribution of the detected ions across the cell population, cell by cell, by correlating peak intensities/areas to the coordinates of each analyzed cell. Normalizing ion intensities to a common ion or the total ion signal aids the interpretation of the results. Molecular images are constructed by overlaying false colors representing peak intensities on the microscopic image of cells.
-
EXAMPLE: Figure 5 demonstrates the cell-by-cell imaging of A. cepa epidermal cells using a sharpened optical fiber for LAESI. Upon overlaying the distribution of the cyanidin ion (m/z 287) with the optical image of the epidermis, a correlation is apparent between this compound and cells that have purple pigmentation.
-
Notes
- 1.
Er:YAG lasers are widely used to produce this type of laser beam, e.g., in IR-MALDI MS applications.
References
Apitz I, Vogel A (2005) Material ejection in nanosecond Er: YAG laser ablation of water, liver, and skin. Appl Phys A 81(2):329–338
Boyce G, Walsh C, Seeley E, Morton J, Kilby G (2014) Fatty acid and lipid profiling of Arbuscular mycorrhizal fungi with LAESI-MS. In: The annual meeting of the American Society for Mass Spectrometry, Baltimore
Kiss A, Smith DF, Reschke BR, Powell MJ, Heeren RMA (2014) Top-down mass spectrometry imaging of intact proteins by laser ablation ESIFT-ICR MS. Proteomics 14(10):1283–1289. doi:10.1002/pmic.201300306
Li H, Smith BK, Márk L, Nemes P, Nazarian J, Vertes A (2015a) Ambient molecular imaging by laser ablation electrospray ionization mass spectrometry with ion mobility separation. Int J Mass Spectrom 375:681
Li H, Smith BK, Shrestha B, Márk L, Vertes A (2015b) Automated cell-by-cell tissue imaging and single-cell analysis for targeted morphologies by laser ablation electrospray ionization mass spectrometry. In: He L (ed) Methods in molecular biology, Mass spectrometry imaging of small molecules. Humana Press, New York, pp 117–127
Lin D, H G, King J, Cichewicz R (2014) Investigation of an unprecedented natural non-enzymatic reaction with laser ablation electrospray ionization (LAESI) mass spectrometry technology. In: The Annual Meeting of the American Society of Mass Spectrometry, Baltimore
Monge ME, Harris GA, Dwivedi P, Fernandez FM (2013) Mass spectrometry: recent advances in direct open air surface sampling/ionization. Chem Rev 113(4):2269–2308. doi:10.1021/cr300309q
Nemes P, Vertes A (2007) Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem 79(21):8098–8106. doi:10.1021/ac071181r
Nemes P, Vertes A (2010a) Atmospheric-pressure molecular imaging of biological tissues and biofilms by LAESI mass spectrometry. J Vis Exp 43, e2097
Nemes P, Vertes A (2010b) Laser ablation electrospray ionization for atmospheric pressure molecular imaging mass spectrometry. In: Rubakhin SS, Sweedler JV (eds) Mass spectrometry imaging, Principles and protocols. Humana, New York, pp 159–171. doi:10.1007/978-1-60761-746-4_9
Nemes P, Vertes A (2012) Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. TrAC Trends Anal Chem 34:22–34
Nemes P, Vertes A (2015) Laser ablation electrospray ionization mass spectrometry: mechanisms, configurations, and imaging applications. In: Domin M, Cody R (eds) Ambient ionization mass spectrometry. Royal Society of Chemistry, Cambridge, p 348
Nemes P, Barton AA, Li Y, Vertes A (2008) Ambient molecular imaging and depth profiling of live tissue by infrared laser ablation electrospray ionization mass spectrometry. Anal Chem 80(12):4575–4582
Nemes P, Barton AA, Vertes A (2009) Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal Chem 81(16):6668–6675
Nemes P, Woods AS, Vertes A (2010) Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal Chem 82(3):982–988
Nemes P, Huang H, Vertes A (2012) Internal energy deposition and ion fragmentation in atmospheric-pressure mid-infrared laser ablation electrospray ionization. Phys Chem Chem Phys 14(7):2501–2507
Nielen MWF, van Beek TA (2014) Macroscopic and microscopic spatially-resolved analysis of food contaminants and constituents using laser-ablation electrospray ionization mass spectrometry imaging. Anal Bioanal Chem 406(27):6805–6815. doi:10.1007/s00216-014-7948-8
Razunguzwa TT, Henderson HD, Reschke BR, Walsh CM, Powell MJ (2014) Laser-ablation electrospray ionization mass spectrometry (LAESIs-MS): ambient ionization technology for 2D and 3D molecular imaging. In: Domin M, Cody R (eds) Ambient ionization mass spectrometry. Royal Society of Chemistry, Cambridge, pp 462–481
Shrestha B, Vertes A (2009) In situ metabolic profiling of single cells by laser ablation electrospray ionization mass spectrometry. Anal Chem 81(20):8265–8271. doi:10.1021/ac901525g
Shrestha B, Vertes A (2010) Direct analysis of single cells by mass spectrometry at atmospheric pressure. J Vis Exp 43, e2144. doi:10.3791/2144
Shrestha B, Vertes A (2014) High-throughput cell and tissue analysis with enhanced molecular coverage by laser ablation electrospray ionization mass spectrometry using ion mobility separation. Anal Chem 86(9):4308–4315
Shrestha B, Nemes P, Nazarian J, Hathout Y, Hoffman EP, Vertes A (2010a) Direct analysis of lipids and small metabolites in mouse brain tissue by AP IR-MALDI and reactive LAESI mass spectrometry. Analyst 135(4):751–758. doi:10.1039/b922854c
Shrestha B, Nemes P, Vertes A (2010b) Ablation and analysis of small cell populations and single cells by consecutive laser pulses. Appl Phys A 101(1):121–126
Shrestha B, Patt JM, Vertes A (2011) In situ cell-by-cell imaging and analysis of small cell populations by mass spectrometry. Anal Chem 83(8):2947–2955
Shrestha B, Javonillo R, Burns JR, Pirger Z, Vertes A (2013) Comparative local analysis of metabolites, lipids and proteins in intact fish tissues by LAESI mass spectrometry. Analyst 138(12):3444–3449. doi:10.1039/c3an00631j
Stolee JA, Vertes A (2013) Toward single-cell analysis by plume collimation in laser ablation electrospray ionization mass spectrometry. Anal Chem 85(7):3592–3598. doi:10.1021/ac303347n
Stolee JA, Shrestha B, Mengistu G, Vertes A (2012) Observation of subcellular metabolite gradients in single cells by laser ablation electrospray ionization mass spectrometry. Angew Chem Int Ed 51(41):10386–10389. doi:10.1002/anie.201205436
Vertes A, Nemes P, Shrestha B, Barton AA, Chen Z, Li Y (2008) Molecular imaging by Mid-IR laser ablation mass spectrometry. Appl Phys A 93(4):885–891
Vertes A, Shrestha B, Nemes P (2013) Direct metabolomics from tissues and cells: new approaches for small molecule and lipid characterization. In: Wevers R, Lutz N, Sweedler JV (eds) Methodologies for metabolomics, Experimental strategies and techniques. Cambridge University Press, Cambridge, pp 140–158
Walsh C, Henderson H, Boyce G, Sexstone A, Gibson L, Powell M (2012) Mass spectrometric profiling of bacterial and mammalian cells with LAESI-MS. In: The Annual meeting of the American Society for Mass Spectrometry, Vancouver
Wu CP, Dill AL, Eberlin LS, Cooks RG, Ifa DR (2013) Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev 32(3):218–243. doi:10.1002/mas.21360
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Shrestha, B., Walsh, C.M., Boyce, G.R., Nemes, P. (2016). Microprobe MS Imaging of Live Tissues, Cells, and Bacterial Colonies Using LAESI. In: Cramer, R. (eds) Advances in MALDI and Laser-Induced Soft Ionization Mass Spectrometry. Springer, Cham. https://doi.org/10.1007/978-3-319-04819-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-04819-2_8
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-04818-5
Online ISBN: 978-3-319-04819-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)