Abstract
Segmentation of 2D echocardiography (2DE) images is an important prerequisite for quantifying cardiac function. Although deep learning can automate analysis, variability in image quality and limitations in model generalisability can result in inaccurate segmentations. We present an automated quality control (QC) methodology to identify invalid segmentations, and propose post-processing techniques to automatically correct erroneous segmentations. A workflow was developed to utilise a deep learning model, trained using the CAMUS dataset, for segmenting all frames within apical two-chamber and four-chamber 2DE images from an independent dataset containing 91 participants (28 females; 51 healthy controls and 40 patients with mixed cardiac pathologies). Single- and multi-frame QC and post-processing techniques were applied, and subsequently validated against manual QC in a sample of 50 randomly selected participants. Cardiac indices derived from the automated segmentations using 2DE were compared to reference values obtained through expert manual analysis on the same subjects. Single-frame QC improved the proportion of usable frames from 76% to 96%. Multi-frame QC indicated failures in 53% of the images, and while the resulting specificity was 96%, correction only achieved a sensitivity of 42% with respect to manual assessment. The exclusion of the rejected images resulted in improvements in the reliability between predicted and manual measurements. These results demonstrated that applying automated QC to deep learning segmentation methods can enhance the reliability of 2DE segmentations.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
2D echocardiography (2DE) is a highly accessible imaging modality that allows non-invasive and real-time examination of the geometry, motion, and deformation of the heart. Accurate segmentation of cardiac structures including the left ventricular (LV) cavity (LV\(_{cav}\)), myocardium (LV\(_{myo}\)), and left atrium (LA), is crucial for deriving clinical indices used to assess cardiac function. Recent studies have shown the feasibility of using deep learning models to automatically segment the left heart from 2DE, substantially accelerating image analysis [16].
Despite numerous efforts to create robust deep learning models for cardiac segmentation, the ability to perform effectively across different datasets remains a major challenge [5]. This issue is exemplified by ongoing concerns regarding the accuracy and presence of erroneous segmentations when employing such models in cross-dataset segmentation tasks. Incorporating post-processing can improve flawed segmentations, such that a larger pool of valuable images can be used for clinical analysis [10].
While minor segmentation issues can generally be resolved with simple post-processing operations, correcting some faults can be challenging due to ambiguity in the images. Combining automated quality control (QC) methodology with segmentation processes can help to address errors and improve the reliability and accuracy of the generated segmentations [14]. QC eliminates the need for time-consuming and subjective manual review processes and can highlight prevalent failure modes in deep learning algorithms. This can be used to apply targeted improvements to automated segmentations, which may result in more accurate and reliable predictions of standard clinical indices.
Various QC methodologies and metrics have been proposed, including abnormality detection [14], real-time Dice Similarity Coefficients (DSC) [12], reverse classification accuracy [11], Bayesian uncertainty-based methods [3, 13], and convexity scores [20]. Despite these advancements, no open-source QC methodologies are currently available.
To our knowledge, this study presents the first publicly accessible automated and quality-controlled workflow for left heart segmentation from 2DE images using a state-of-the-art deep learning model. Single-frame QC criteria were used to identify appropriate post-processing steps, and multi-frame QC criteria were applied to assess the reliability of standard cardiac indices derived from the segmentations. The dependability of the workflow was assessed by comparing the generated cardiac indices with those derived from expert manual analysis.
2 Methods
The proposed QC workflow consists of automated left heart segmentation, single-frame QC assessment, followed by post-processing procedures and multi-frame QC analysis. Subsequently, this workflow is utilised for the calculation of routine clinical cardiac indices. A schematic representation of this workflow is depicted in Fig. 1.
2.1 Datasets
CAMUS. The publicly available CAMUS dataset [7] comprised apical two-chamber (A2CH) and four-chamber (A4CH) 2DE views acquired from 500 patients at end-diastole (ED) and end-systole (ES). All images were acquired using a GE Vivid E95 ultrasound scanner (GE Vingmed Ultrasound, Horten, Norway), with a GE M5S probe (GE Healthcare, US). Expert manual labels were available for LV\(_{cav}\), LV\(_{myo}\), and LA on the ED and ES frames.
CARDIOHANCE. The private CARDIOHANCE dataset consists of echocardiograms for 91 participants conducted at the University of Auckland. Ethical approval for this study was granted by the Health and Disability Ethics Committee of New Zealand (17/CEN/226), and all research was performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from each participant. This dataset comprised 51 healthy controls (20 females) and 40 patients (8 females) with mixed cardiac pathologies. The primary diagnoses were: cardiac amyloidosis (11), LV hypertrophy (10), aortic regurgitation (7), dilated cardiomyopathy (5), hypertrophic cardiomyopathy (4), heart transplant (2), and coronary artery disease (1). 2DE image sequences were available with both the LV\(_{cav}\) and LA clearly visible. The images were obtained with a Siemens ACUSON SC2000 ultrasound scanner, equipped with a 4Z1c matrix array transducer (Siemens Medical Solutions, Issaquah, WA, USA).
2.2 Deep Learning Model for Segmentation
A self-configuring segmentation framework (2D nnU-Net (v2) [4]), was trained on the CAMUS dataset to segment the LV\(_{cav}\), LV\(_{myo}\), and LA. This dataset was split into a 90/10 training/testing ratio, and the model was trained for 100 epochs using five-fold cross-validation. The nnU-Net model was used to perform cross-dataset segmentations of the left heart on all images in the CARDIOHANCE dataset for which no expert labels were available.
2.3 Single-frame Quality Control and Post-processing
Following cross-dataset segmentation, various issues in the segmentations became apparent, including missing or duplicated structures and the presence of holes or discontinuities within or between structures. To address this, a single-frame QC assessment was implemented to identify and flag erroneous segmentations. Subsequently, the flagged segmentations were post-processed.
Initially, mean centroids of LV\(_{cav}\) and LA were computed across the entire image sequence, excluding any frames that were flagged during the QC assessment. Structures lacking the mean centroid were removed, thereby eliminating redundant structures. The LV\(_{myo}\) was identified by including only structures bordering LV\(_{cav}\). Additionally, any holes within or between structures were filled. The issue of missing structures could not be resolved by simple post-processing.
2.4 Multi-frame Quality Control
Cycle Selection. In clinical practice, image sequences often consist of multiple cardiac cycles. In this study, we defined a cardiac cycle as the duration between two consecutive ED points. The ED and ES points were taken as the locations of the maximum and minimum LV\(_{cav}\) areas in the area-time curve, respectively. The cardiac cycle was selected based on having the fewest flagged frames and the highest contrast-to-noise ratio [9] between the myocardium and blood pool.
Structural Information. After selecting the best cycle from each sequence, frames with a disconnected LV\(_{myo}\) or LA extending beyond the imaging field of view, indicative of partial cutoff, were identified and flagged. If two or more frames were flagged within the selected cardiac cycle, the cycle was excluded.
Area-Time Curve Analysis. Area-time curves were generated for the LV\(_{cav}\) and LA across the entire cardiac cycle to differentiate between well-segmented and poorly-segmented cycles. A population prior was established as a reference, based on the selected cycles from all cases in the CARDIOHANCE dataset. Using the ES timings from all cycles, an average ES point was computed to temporally align the area-time curves for each subject. Subsequently, all curves were normalised, thereby accounting for variations in heart size, and a mean curve was computed for both LV\(_{cav}\) and LA.
The similarity between the original area-time curves of each image and the reference curve was assessed using Dynamic Time Warping (DTW) [1]. DTW measures the cumulative Euclidean distance between curves after DTW alignment. Images were excluded if the DTW distance for either the LV\(_{cav}\) or LA exceeded thresholds of 1 and 2, respectively. These thresholds were empirically chosen, with the threshold for the LA being larger due to greater variation observed in the area-time curves of the LA compared to those of LV\(_{cav}\).
Validation. To validate the multi-frame QC, an expert observer manually reviewed segmentations from one cardiac cycle for 50 randomly selected participants in order to identify any erroneous segmentations or temporal incoherencies. Sensitivity and specificity were computed based on the results of the automated QC and manual review.
2.5 Calculation of Clinical Indices
LV end-diastolic volume (EDV) and end-systolic volume (EDV) were computed from ED and ES segmentations using the biplane method of disks summation [2], allowing for LV ejection fraction (EF) determination. The contours extracted from the segmentations of the LV\(_{cav}\) at ED and ES were used to compute the endocardial LV global longitudinal strain (GLS). Furthermore, the maximum area of the LA was calculated.
Expert Manual Analysis. Alongside the calculation of clinical indices from the predicted segmentations, a sonographer carried out manual analysis of the 2DE images using TOMTEC-ARENA 2.31 2D CPA (TOMTEC Imaging Systems GmbH, Unterschleißheim, Germany). This analysis included LV EDV, ESV, EF, GLS assessment, and measurement of the maximum LA area.
2.6 Statistics
The agreement between the clinical indices derived from the predicted segmentations and expert manual analysis on the images in the CARDIOHANCE dataset was quantified using the average measure, two-way mixed effects intraclass correlation coefficient (ICC) [6]. Paired-sample t-tests were performed to identify statistically significant differences (p-values < 0.05). All tests were conducted using Python [18] v3.10, with the Pingouin [17] and SciPy [19] packages.
3 Results
3.1 Evaluation of Deep Learning Segmentation Performance
The segmentation performance on the CAMUS test set, measured by DSC, Hausdorff Distance (HD) and Mean Absolute Distance (MAD), is presented in Table 1. The LV\(_{cav}\) achieved the highest DSC compared to LV\(_{myo}\) and LA, while the HD and MAD were lowest for the LV\(_{cav}\). The LA exhibited the highest variability across all metrics.
3.2 Single-frame Quality Control and Post-processing
Table 2 presents the percentages of flagged frames per failure mode identified through single-frame QC. Before post-processing, 20750 frames passed, representing 76% of the 27406 frames in the CARDIOHANCE dataset. The number of passed frames increased to 26233 after post-processing, accounting for 96% of the total number of frames.
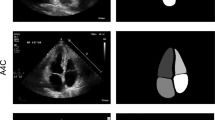
The main reason for flagging frames before post-processing was the presence of holes within the LV\(_{cav}\) (9.4%) or between LV\(_{cav}\) and LV\(_{myo}\) (10.0%). After post-processing, frames were mainly flagged due to the presence of multiple distinct LV\(_{myo}\) structures (3.9%). Figure 2 illustrates examples of post-processing procedures.
Examples demonstrating the effect of post-processing (PP) on segmentations. Top row: 2D echocardiography (2DE) images; middle row: segmentations before PP; bottom row: segmentations after PP. The examples showcase different scenarios, including (A) filling holes in the left atrium, and (B) filling holes in the left ventricular cavity (LV\(_{cav}\)), (C) removing redundant structures and filling holes between LV\(_{cav}\) and myocardium (LV\(_{myo}\)), and (D) unimproved unconnected LV\(_{myo}\).
3.3 Multi-frame Quality Control
Table 3 presents a comparison between clinical indices derived from predicted segmentations and expert manual analysis using TOMTEC software, both before and after exclusion based on multi-frame QC assessment. The comparison revealed significant biases in all indices before and after exclusion, with the exception of LV EF after exclusion. After QC exclusion, all ICC values increased, indicating enhanced reliability between the segmentations and TOMTEC measurements.
In total, 53% of all images were excluded through the multi-frame QC assessment. Manual validation further demonstrated a sensitivity of 42% and a specificity of 96% for excluding images based on both structural faults and temporal inconsistencies.
4 Discussion
The 2D nnU-Net model that was trained in this study outperforms all methods listed on the CAMUS challenge websiteFootnote 1 for LV\(_{cav}\), LV\(_{myo}\) and LA segmentations. Despite its good performance on the CAMUS test set, several issues arose during cross-dataset segmentation on the CARDIOHANCE dataset. Single-frame QC analysis, presented in Table 2, revealed a substantial improvement in the availability of valid frames after post-processing. Only a small fraction of the frames still exhibited an issue with unconnected cardiac labels after post-processing, as depicted in Fig. 2D. To address this issue, it may be beneficial to incorporate a statistical shape model in the post-processing step.
After multi-frame QC, over half of all images were excluded. For the acceptable images, there was a low sensitivity of 42% with respect to manual assessment, indicating that a considerable number of flawed segmentations remained undetected, which could be problematic if used in a clinical setting. On the other hand, a high specificity of 96% was observed, indicating that only a small proportion of accurately segmented images was flagged erroneously. To enhance sensitivity in detecting faults and abnormalities, additional QC criteria can be introduced, potentially including structural properties like convexity and simplicity of anatomical structures [8, 20].
Table 3 demonstrates a significant underestimation of the LV EDV and ESV, as well as the maximum LA areas. This discrepancy could be caused by apical undersegmentation in the LV\(_{cav}\). This can be attributed to the inherent limitations of the expert manual labels in the CAMUS dataset, which have also been associated with volume underestimation [20]. Moreover, the LV GLS values were found to be overestimated compared to the values obtained with TOMTEC in both A2CH and A4CH views, possibly due to the presence of apical undersegmentation in the LV\(_{cav}\). This undersegmentation can result in higher GLS measurements [15].
The exclusion of flagged images resulted in an improved agreement of LV GLS and LA maximum area in A4CH views with respect to expert manual analysis. Initially categorised as poor (ICC < 0.5) and moderate (0.5 \(\le \) ICC < 0.75) reliability, the LV GLS and LA maximum area measurements were reassessed as exhibiting moderate and good (0.75 \(\le \) ICC < 0.9) reliability, respectively, after excluding the indices of flagged images. However, poor reliability persisted in the measurements of LV GLS in A2CH views (ICC = 0.445).
In this study, the manual determination of multi-frame QC criteria raises concerns regarding optimality and dataset-specific applicability. It may therefore be beneficial to explore more generalised criteria, reducing dependence on qualitative manual threshold determination. Furthermore, the utilisation of a single average population prior in area-time curve analysis overlooks potential variations in curve shapes between healthy controls and patients with cardiac pathologies. Future studies could investigate the use of population priors tailored to specific pathologies, thereby enhancing the effectiveness of the analysis.
5 Conclusion
In this study, we have presented an automated QC workflow for left heart analysis from 2DE, with the objective of enhancing the accuracy and reliability of automatically segmented images when applied to cross-dataset segmentation. The implementation of QC, which identifies, flags and applies corrections to faulty segmentations, is shown to be crucial for improving the reliability of the derived cardiac indices when compared to expert manual analysis. Application of the workflow resulted in an improvement in the proportion of usable frames from 76% to 96%. By releasing the code associated with the proposed automated QC workflow, we aim to facilitate collaborative efforts and further developments by the community.
References
Berndt, D.J., Clifford, J.: Using dynamic time warping to find patterns in time series. In: KDD Workshop. vol. 10, pp. 359–370. Seattle, WA, USA (1994)
Folland, E.D., Parisi, A.F., Moynihan, P.F., Jones, D.R., Feldman, C.L., Tow, D.E.: Assessment of left ventricular ejection fraction and volumes by real-time, two-dimensional echocardiography. A comparison of cineangiographiy and radionuclide techniques. Circulation 60(4), 760–766 (1979). https://doi.org/10.1161/01.cir.60.4.760
Hann, E., Gonzales, R.A., Popescu, I.A., Zhang, Q., Ferreira, V.M., Piechnik, S.K.: Ensemble of deep convolutional neural networks with Monte Carlo dropout sampling for automated image segmentation quality control and robust deep learning using small datasets. In: Papież, B.W., Yaqub, M., Jiao, J., Namburete, A.I.L., Noble, J.A. (eds.) MIUA 2021. LNCS, vol. 12722, pp. 280–293. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-80432-9_22
Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021). https://doi.org/10.1038/s41592-020-01008-z
Keshavan, A., Datta, E., M. McDonough, I., Madan, C.R., Jordan, K., Henry, R.G.: Mindcontrol: a web application for brain segmentation quality control. NeuroImage 170, 365–372 (2018). https://doi.org/10.1016/j.neuroimage.2017.03.055
Koo, T.K., Li, M.Y.: A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 15(2), 155–163 (2016). https://doi.org/10.1016/j.jcm.2016.02.012
Leclerc, S., et al.: Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. IEEE Trans. Med. Imaging 38(9), 2198–2210 (2019). https://doi.org/10.1109/TMI.2019.2900516
Leclerc, S., et al.: Deep learning segmentation in 2D echocardiography using the CAMUS dataset: automatic assessment of the anatomical shape validity. In: International Conference on Medical Imaging with Deep Learning - Extended Abstract Track, London, United Kingdom (2019)
Meyers, B., Brindise, M., Kutty, S., Vlachos, P.: A method for direct estimation of left ventricular global longitudinal strain rate from echocardiograms. Sci. Rep. 12(1), 4008 (2022). https://doi.org/10.1038/s41598-022-06878-1
Painchaud, N., Skandarani, Y., Judge, T., Bernard, O., Lalande, A., Jodoin, P.M.: Cardiac segmentation with strong anatomical guarantees. IEEE Trans. Med. Imaging 39(11), 3703–3713 (2020). https://doi.org/10.1109/TMI.2020.3003240
Robinson, R., et al.: Automated quality control in image segmentation: application to the UK Biobank cardiovascular magnetic resonance imaging study. J. Cardiovasc Magn. Reson. 21(1), 18 (2019). https://doi.org/10.1186/s12968-019-0523-x
Robinson, R., et al.: Real-time prediction of segmentation quality. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 578–585. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_66
Roy, A.G., Conjeti, S., Navab, N., Wachinger, C.: Inherent brain segmentation quality control from fully ConvNet Monte Carlo sampling. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11070, pp. 664–672. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00928-1_75
Ruijsink, B., et al.: Fully automated, quality-controlled cardiac analysis from CMR. JACC Cardiovasc. Imaging 13(3), 684–695 (2020). https://doi.org/10.1016/j.jcmg.2019.05.030
Smiseth, O.A., Donal, E., Penicka, M., Sletten, O.J.: How to measure left ventricular myocardial work by pressure-strain loops. Eur. Heart J. Cardiovasc. Imaging 22(3), 259–261 (2020). https://doi.org/10.1093/ehjci/jeaa301
Thrall, J.H., Li, X., Li, Q., Cruz, C., Do, S., Dreyer, K., Brink, J.: Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J. Am. Coll. Radiol. 15(3, Part B), 504–508 (2018). https://doi.org/10.1016/j.jacr.2017.12.026
Vallat, R.: Pingouin: statistics in Python. J. Open Source Softw. 3(31), 1026 (2018). https://doi.org/10.21105/joss.01026
Van Rossum, G., Drake, F.L.: Python 3 Reference Manual. CreateSpace, Scotts Valley, CA (2009)
Virtanen, P., et al.: SciPy 1.0 Contributors: SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). https://doi.org/10.1038/s41592-019-0686-2
Zhang, X., et al.: Generalizability and quality control of deep learning-based 2D echocardiography segmentation models in a large clinical dataset. Int. J. Cardiovasc. Imaging (2022). https://doi.org/10.1007/s10554-022-02554-7
Acknowledgements
We gratefully acknowledge the participants of the CARDIOHANCE study for volunteering their time, and the staff at the Centre for Advanced MRI at the University of Auckland for their expertise and assistance with the imaging components of this study.
Funding
This study was funded by the Health Research Council of New Zealand (programme grant 17/608).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
Our code is publicly available on GitHub: https://github.com/bgeven/AQC-left-heart-segmentation
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Geven, B.W.M. et al. (2024). Automated Quality-Controlled Left Heart Segmentation from 2D Echocardiography. In: Camara, O., et al. Statistical Atlases and Computational Models of the Heart. Regular and CMRxRecon Challenge Papers. STACOM 2023. Lecture Notes in Computer Science, vol 14507. Springer, Cham. https://doi.org/10.1007/978-3-031-52448-6_10
Download citation
DOI: https://doi.org/10.1007/978-3-031-52448-6_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-52447-9
Online ISBN: 978-3-031-52448-6
eBook Packages: Computer ScienceComputer Science (R0)