Abstract
Background: The ability to promptly sequence whole genomes at a relatively low cost has revolutionized how we study the microbiome. Analyzing whole genome sequencing (WGS) data enables metagenomics at scale. Still, it is a complex process that involves multiple moving parts and can be unintuitive for scientists that do not typically work with this type of data.
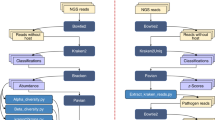
Methods: Thus, to help lower the barrier for less computationally inclined individuals, TAXAPRO, a metagenomics pipeline that accurately assembles organelle genomes from WGS, data was developed. TAXAPRO seamlessly combines WGS analysis tools to create a pipeline that automatically processes raw WGS data and presents information on microorganisms’ diversity and relative abundance.
Results: TAXAPRO was evaluated using gut microbiome data from COVID-19 patients. Analysis performed by TAXAPRO demonstrated a relatively high abundance of Clostridia and Bacteroides genera and a low abundance of Proteobacteria were detected in the gut microbiome of patients hospitalized with COVID-19, consistent with the original findings results derived using a different analysis methodology.
Conclusion: Our results provide evidence that the TAXAPRO workflow dispenses quick and automated microorganism diversity and relative abundance information without the hassle of manually performing the analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Sender, R., Fuchs, S., Milo, R.: Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell 164(3), 337–340 (2016). https://doi.org/10.1016/j.cell.2016.01.013
Cussotto, S., Sandhu, K.V., Dinan, T.G., Cryan, J.F.: The neuroendocrinology of the microbiota-gut-brain axis: a behavioural perspective. Front. Neuroendocrinol. 51, 80–101 (2018). https://doi.org/10.1016/j.yfrne.2018.04.002
Barrett, E., Ross, R.P., O’Toole, P.W., Fitzgerald, G.F., Stanton, C.: γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 113(2), 411–417 (2012). https://doi.org/10.1111/j.1365-2672.2012.05344.x
Yang, N.J., Chiu, I.M.: Bacterial signaling to the nervous system through toxins and metabolites. J. Mol. Biol. 429(5), 587–605 (2017). https://doi.org/10.1016/j.jmb.2016.12.023
David, L.A., Maurice, C.F., Carmody, R.N., et al.: Diet rapidly and reproducibly alters the human gut microbiome. Nature 505(7484), 559–563 (2014). https://doi.org/10.1038/nature12820
Dethlefsen, L., Huse, S., Sogin, M.L., Relman, D.A.: The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6(11), e280 (2008). https://doi.org/10.1371/journal.pbio.0060280
Gaulke, C.A., Sharpton, T.J.: The influence of ethnicity and geography on human gut microbiome composition. Nat. Med. 24(10), 1495–1496 (2018). https://doi.org/10.1038/s41591-018-0210-8
Turnbaugh, P.J., Ley, R.E., Mahowald, M.A., Magrini, V., Mardis, E.R., Gordon, J.I.: An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444(7122), 1027–1031 (2006). https://doi.org/10.1038/nature05414
Cui, L., Zhao, T., Hu, H., Zhang, W., Hua, X.: Association study of gut flora in coronary heart disease through high-throughput sequencing. Biomed. Res. Int. 2017, 3796359 (2017). https://doi.org/10.1155/2017/3796359
Cattaneo, A., Cattane, N., Galluzzi, S., et al.: Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 49, 60–68 (2017). https://doi.org/10.1016/j.neurobiolaging.2016.08.019
Brooks, E.F., Bhatt, A.S.: The gut microbiome: a missing link in understanding the gastrointestinal manifestations of COVID-19? Cold Spring Harb Mol Case Stud. 7(2), a006031 (2021). https://doi.org/10.1101/mcs.a006031
Hussain, I., Cher, G.L.Y., Abid, M.A., Abid, M.B.: Role of gut microbiome in COVID-19: an insight into pathogenesis and therapeutic potential. Front. Immunol. 12, 765965 (2021). https://doi.org/10.3389/fimmu.2021.765965
Yeoh, Y.K., Zuo, T., Lui, G.C.Y., et al.: Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70(4), 698–706 (2021). https://doi.org/10.1136/gutjnl-2020-323020
Zuo, T., Zhang, F., Lui, G.C.Y., et al.: Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 159(3), 944-955.e8 (2020). https://doi.org/10.1053/j.gastro.2020.05.048
Gu, S., Chen, Y., Wu, Z., et al.: Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 71(10), 2669–2678 (2020). https://doi.org/10.1093/cid/ciaa709
Metzker, M.L.: Sequencing technologies — the next generation. Nat. Rev. Genet. 11(1), 31–46 (2010). https://doi.org/10.1038/nrg2626
Hollister, E.B., Riehle, K., Luna, R.A., et al.: Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 3, 36 (2015). https://doi.org/10.1186/s40168-015-0101-x
Pallen, M.J.: Diagnostic metagenomics: potential applications to bacterial, viral and parasitic infections. Parasitology 141(14), 1856–1862 (2014). https://doi.org/10.1017/S0031182014000134
Costalonga, M., Herzberg, M.C.: The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 162(2 Pt A), 22–38 (2014). https://doi.org/10.1016/j.imlet.2014.08.017
Li, C.X., You, Z.X., Lin, Y.X., Liu, H.Y., Su, J.: Skin microbiome differences relate to the grade of acne vulgaris. J. Dermatol. 46(9), 787–790 (2019). https://doi.org/10.1111/1346-8138.14952
Greathouse, K.L., White, J.R., Vargas, A.J., et al.: Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19(1), 123 (2018). https://doi.org/10.1186/s13059-018-1501-6
Shreiner, A.B., Kao, J.Y., Young, V.B.: The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31(1), 69–75 (2015). https://doi.org/10.1097/MOG.0000000000000139
Bull, M.J., Plummer, N.T.: Part 1: the human gut microbiome in health and disease. Integr Med (Encinitas). 13(6), 17–22 (2014)
Sehli, S., Allali, I., Chahboune, R., et al.: Metagenomics approaches to investigate the gut microbiome of COVID-19 patients. Bioinform. Biol. Insights 15, 1177932221999428 (2021). https://doi.org/10.1177/1177932221999428
LaMar, D.: FastQC. Published online (2015). https://qubeshub.org/resources/fastqc
Bolger, A.M., Lohse, M., Usadel, B.: Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15), 2114–2120 (2014). https://doi.org/10.1093/bioinformatics/btu170
Nurk, S., Meleshko, D., Korobeynikov, A., Pevzner, P.A.: MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5), 824–834 (2017). https://doi.org/10.1101/gr.213959.116
Hyatt, D., Chen, G.L., LoCascio, P.F., Land, M.L., Larimer, F.W., Hauser, L.J.: Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11(1), 119 (2010). https://doi.org/10.1186/1471-2105-11-119
Bankevich, A., Nurk, S., Antipov, D., et al.: SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19(5), 455–477 (2012). https://doi.org/10.1089/cmb.2012.0021
Cock, P.J.A., Antao, T., Chang, J.T., et al.: Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25(11), 1422–1423 (2009). https://doi.org/10.1093/bioinformatics/btp163
McKinney, W.: Data structures for statistical computing in Python. In: Proceedings of the 9th Python in Science Conference. Published Online, pp. 56–61 (2010). https://doi.org/10.25080/Majora-92bf1922-00a
Hunter, J.D.: Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9(3), 90–95 (2007). https://doi.org/10.1109/MCSE.2007.55
Waskom, M.L.: Seaborn: statistical data visualization. J. Open Sour. Softw. 6(60), 3021 (2021). https://doi.org/10.21105/joss.03021
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., Madden, T.L.: NCBI BLAST: a better web interface. Nucleic Acids Res. 36(Web Server issue), W5–9 (2008). https://doi.org/10.1093/nar/gkn201
Koutsoumanis, K., Allende, A., Alvarez-Ordóñez, A., et al.: Whole genome sequencing and metagenomics for outbreak investigation, source attribution and risk assessment of food-borne microorganisms. EFSA J. 17(12), e05898 (2019). https://doi.org/10.2903/j.efsa.2019.5898
Dominguez Del Angel, V., Hjerde, E., Sterck, L., et al.: Ten steps to get started in genome assembly and annotation. F1000Res. 7, ELIXIR-148 (2018). https://doi.org/10.12688/f1000research.13598.1
Zhou, Q., Su, X., Ning, K.: Assessment of quality control approaches for metagenomic data analysis. Sci. Rep. 4(1), 6957 (2014). https://doi.org/10.1038/srep06957
Wang, Y., Li, G., Ma, M., et al.: GT-WGS: an efficient and economic tool for large-scale WGS analyses based on the AWS cloud service. BMC Genomics 19(Suppl 1), 959 (2018). https://doi.org/10.1186/s12864-017-4334-x
Berg, G., Rybakova, D., Fischer, D., et al.: Microbiome definition re-visited: old concepts and new challenges. Microbiome. 8(1), 103 (2020). https://doi.org/10.1186/s40168-020-00875-0
Lozupone, C.A., Stombaugh, J.I., Gordon, J.I., Jansson, J.K., Knight, R.: Diversity, stability and resilience of the human gut microbiota. Nature 489(7415), 220–230 (2012). https://doi.org/10.1038/nature11550
Acknowledgments
This research was supported through cloud computational resources provided by the National Institutes of Health Office of Data Science Strategy (NIH ODSS). This research was also supported through computational resources of HPC-MARWAN (www.marwan.ma/hpc) provided by the National Center for Scientific and Technical Research (CNRST), Rabat, Morocco. Hassan Ghazal is a US NIH grant recipient through the H3abionet/H3africa consortium U24HG006941. The authors thank Brigit Shea Sullivan and Alicia Lillich from the NIH Library Editing Service for manuscript editing assistance.
Author information
Authors and Affiliations
Contributions
SS: Conceptualization, Data Curation, Formal Analysis, Investigation, Methodology, Resources, Software, Supervision; ZEO: Visualization, Investigation, Writing – Original Draft Preparation; CE: Data Curation, Formal Analysis, Methodology, Software, Writing – Original Draft Preparation; AIA: Data Curation, Formal Analysis; KAS: Formal Analysis, Investigation; IEJ: Conceptualization; S. Jbara: Formal Analysis, Methodology; A. Afolayan: Investigation, Resources; OIA: Funding Acquisition, Formal Analysis, Investigation, Project Administration, Resources, Supervision, Review & Editing; A. Dillman: Funding Acquisition, Resources, Supervision; H. Ghazal: Conceptualization, Review & Editing. All authors read and approved the final manuscript.
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Sehli, S. et al. (2024). TAXAPRO: A Streamlined Pipeline to Analyze Shotgun Metagenomes. In: Ezziyyani, M., Kacprzyk, J., Balas, V.E. (eds) International Conference on Advanced Intelligent Systems for Sustainable Development (AI2SD’2023). AI2SD 2023. Lecture Notes in Networks and Systems, vol 905. Springer, Cham. https://doi.org/10.1007/978-3-031-52385-4_8
Download citation
DOI: https://doi.org/10.1007/978-3-031-52385-4_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-52384-7
Online ISBN: 978-3-031-52385-4
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)