Abstract
Cardiovascular disease is the main cause of mortality world-wide, its early prediction and early diagnosis are fundamental for patients with this mortal illness. Cardiovascular disease is a real threat for the Health Systems worldwide, mainly because it has become the diagnosis that claim a significant number of lives around the world. Currently, there is a growing need from health entities to integrate the use of technology. Cardiovascular disease identification systems allow the identification of diseases associated with the heart, allowing the early identification of Cardiovascular Diseases (CVD) for an improvement in the quality of life of patients.
According to the above, the predictive models of CVD have become a common research field, where the implementation of feature selection techniques and models based on artificial intelligence provide the possibility of identifying, in advance, the trend of patients who may suffer from a disease associated with the heart.
Therefore, this paper proposes the use of feature selection techniques (Information Gain) with the variation of artificial intelligence techniques, such as neural networks (Som, Ghsom), decision rules (ID3, J48) and Bayesian networks (Bayes net, Naive Bayes) with the purpose of identifying the hybrid model for the identification of cardiovascular diseases.
It was used the data set “Heart Cleveland Disease Data Set” with the same test environment for all the cases, in order to establish which of the mentioned techniques achieves the higher value of the accuracy metric when it comes to identify patients with heart disease. For the development of the tests, 10-fold Cross-Validation was used as a data classification method and 91.3% of the accuracy was obtained under the hybridization of the selection technique “information gain” with the training technique J48.
This study is supported by the Colombian Government through Minciencias grant No. 860 International Studies for Doctorate and the Bicentennial Doctoral Excellence Scholarship Program.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The 42% of deaths from cardiovascular diseases are related to ischemic heart disease, and 34% to cerebrovascular diseases. It is estimated that in 2015 17.7 million people died from this cause, which represents 31% of the deaths registered worldwide.
According to reports from the World Health Organization (WHO) and the National Health Organization (ONS), cardiovascular diseases along with cancer, diabetes and Chronic lung are identified increasing diseases, cataloging them as the main causes of death worldwide, being the cardiovascular one the most relevant cause.
Nowadays, there is a growing need from health entities to integrate the use of technology to their processes. The Cardiovascular disease identifier (CDI) facilitate the identification of early heart diseases that can be analyzed with technology, improving the life of patients.
Moreover, there are data set for cardiovascular diseases. It is important to identify the clinical features that will take part in the evaluation process, in order to make a real model related to daily lives; and to select the techniques that are going to be implemented like preprocessing, characteristic features, classification and post classification.
The purpose of this paper is to make a comparison of supervised techniques of artificial intelligence that were applied to a model in order to predict cardio-vascular diseases. For this study, it was selected the Heart Disease Cleveland Data set. This document is composed by six sections, the first one is related to introduction; Sect. 2 exposes previous work according to the study; Sect. 3 details some definitions and important works; Sect. 4 describes the methodology developed; Sect. 5 explains the conclusions and Sect. 6 details the conclusions and future work.
2 Related Work
The literature review evidenced that some models analyze specific approaches from the technological view, like Machine Learning techniques, [1], comparative studies of cardiovascular risk techniques [2, 3], cardiovascular monitoring system [4]. Other versions, although not exhaustive, are limited to evaluate classification techniques in terms of precision for the prediction of cardiovascular disease via data set [5,6,7]. However, no evidence was found related to perform a comparative study implementing a model that hybridizes any selection technique with a training technique. In [8], it is implemented computer-assisted techniques to provide a fast and accurate tool to identify a patient’s ECG signals. Diagnostic techniques are compared, using data preprocessing, feature engineering, classification and application.
In [9] an end-to-end model is performed by integrating feature extraction and classification into learning algorithms, which not only simplifies the data analysis process, but also shows a good-level evaluated accuracy.
In [10], an extraction of multivariate clinical characteristics of ACS patients recorded in a database registry was implemented. The author and used machine learning algorithms to develop several models with measurable performance to predict heart attacks in patients.
3 Machine Learning Techniques for Classification
3.1 Artificial Neural Networks
According to … Artificial neural networks attempt to capture the essence of biological processes and apply them to new computational models. The artificial neuron is a simplified model of a biological neuron. The interconnection between neurons decides the flow of information in the network and together with the weights and output functions of each neuron define the global behavior of the artificial neural network. It means that neural networks are made up of a large number of linear and non-linear computational elements (neurons) that are complexly interrelated and organized in layers.
Self-Organizing Map Neural Networks.
It is an efficient unsupervised neural algorithm that allows the projection of data from a multidimensional space, in a two-dimensional grid called “map”, qualitatively preserving the topology of the original set. In 1982, the author T. Kohonen presented a network model called self-organizing maps or SOM. The purpose was to demonstrate that an external stimulus (input) is capable to force the formation of maps, assuming a given structure and a functional description. According to (…) the most important characteristic of the SOM is that it learns to classify the data throughout an unsupervised learning algorithm. A SOM learns to classify the training data without any type of external control.
Growing Hierarchical Self Organizing Maps Neuronal Networks.
According to [11], Growing Hierarchical Self Organizing Maps Neuronal Networks or GHSOM is a hierarchical and dynamic structure, developed to overcome the weaknesses and problems that SOM presents. The GHSOM structure consists of multiple layers made up of several independent SOMs whose number and size are determined during the training phase. The adaptation growth process is controlled by two parameters that determine the depth of the hierarchy and the breadth of each map. Therefore, these two parameters are the only ones that have to be initially set in GHSOM. The adaptation growth process is controlled by two parameters that determine the depth of this type of maps are born as an improved version of the SOM architecture. According to [12] there are two purposes for the GHSOM architecture:
-
SOM has a fixed network architecture, which means that the number of units of use as well as the distribution of the units have to be determined before the training.
-
Input data that are naturally hierarchical should be represented in a hierarchical structure for the clarity of its representation. GHSOM uses a hierarchical, multi-layered structure, where each layer is made up of a number of independent SOMs. Only a SOM is used in the first layer of the hierarchy.
For every unit on the map, a SOM could be added to the next layer of the hierarchy. This principle is repeated with the third level of the map and the other layers of the GHSOM, as shown in Fig. 1.
3.2 Bayesian Networks
Bayesian logic was created by the mathematician Thomas Bayes in 1763. It is based on statistics and conditional probabilities to predict the future. It is an unsupervised technique of data classification. Bayesian statistical principles have been useful for the early identification of Cardiovascular Diseases. The result of the application of Bayesian systems is presented in the form of conditional probability relations, instead of rules or signatures. In conclusion, Bayesian Networks are powerful tools as decision models and reasoning under uncertainty.
Naive Bayes.
According to [11] it is a descriptive and predictive classification technique based on the analysis probability theory by T. Bayes [13]. This theory assumes an asymptotically infinite sample size and statistical independence among independent variables, referring with it to attributes, not the class. With these conditions, the probability distributions of each class can be calculated to establish the relationship between the attributes (independent variables) and the class (dependent variable).
3.3 Decision Rules
Supervised learning methods based on decision trees are one of the most popular methods in the Artificial Intelligence field, to deal with the classification problem (…). A classification tree is made up of nodes, branches, and leaves. Each node represents a decision about the values of a particular attribute. The first node in the tree is known as the root node. Finally there are the terminal nodes or leaves in which a decision is made about the class to assign. Thus, when classifying a new case, the values of the attributes will have to be compared with the decisions made in the nodes, following the branch that matches said values in each test or decision. A terminal or leaf node is reached that predicts the class for the case in question. A decision tree can also be viewed as a set of if-then rules.
ID3.
Each internal node of the tree contains a decision on one of the attributes, whose value will depend on the path to follow to classify an example, and each leaf contains a class label. Thus, the classification of an example is carried out by going through the tree from the root to one of the leaves that will determine its class. Initially, the algorithm takes the entire data set (.) Despite its simplicity and low computational cost, ID3 has significant drawbacks, some of which are corrected by its successor C4.5. The most obvious are the inability to work with continuous attributes and handle missing values.
J48.
The j48 algorithm is a version of the classic ID3 decision tree algorithm proposed by Quinlan (.). Decision trees are considered to be part of the supervised classification methods, which means that they have a dependent variable or class and the objective of the classifier is to determine the value of said class for new cases.
The process to start a tree begins with the root node, the one with all the examples or training cases associated to it. The first step is to select the variable or attribute from which the original training sample is going to be divided. This process is recursive, that is, once the variable with which the greatest homogeneity is obtained with respect to the class of child nodes has been determined, the analysis is performed again for each of the child nodes.
3.4 Data Set
Many researchers have commonly focused on the use of the Heart Disease Cleve-land Data-set for the simulation and analysis of CVD, due to the advantages that it offers in terms of variety and data purification, compared to others of the same family and other organizations. An evidence of the use of the Heart Disease Data set is the increasing number of cites and references in papers, conference proceedings, among others in the past five years, like is shown in Table 1.
As a conclusion, the initial stage of data selection for this study, it is used the Heart Disease Cleveland Data set, as an input to test the proposed model. This database consists of a total of 76 features, but all published experiments refer to the use of a subset of only 14 features and 303 records.
Therefore, we have used the already processed UCI Cleveland data set avail- able on the Kaggle website for our analysis. The full description of the 14 attributes used in the work is mentioned in Table 2.
3.5 Selection of Features
In recent years, a large number of data sets have been published on the Internet and are available in repositories online that facilitate the access to the information by the scientific community. Machine learning methods demand to analyze a large number of features. To solve this problem, feature selection algorithms have become a necessary element in the learning process. The author (…) defines it as the process of detecting the relevant characteristics and discarding the irrelevant ones, its aim is to reduce the size of the input data facilitating the processing and analysis of the information. The ability to use the feature selection is important to make an effective analysis, because the data contains information that is not required to generate a model. The feature selection and Information gain techniques are presented in detail below, since these are the basis of the model proposed in this study.
Information Gain (Info.Gain).
It is a filter-based feature selection technique. It is also known as information gain and is used to identify the level of relevance or ranking of the characteristics of a data collection. Equation No. defines the level of relevance. The attribute with the highest information gain is chosen as the split attribute for node N. This attribute minimizes the information needed to classify the pairs in the resulting partition and reflects the least randomness or impurity in these partitions.
Table 3 details the results of implementing the aforementioned technique to the data set, handling order by relevance from highest to lowest.
4 Methodology
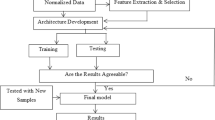
The model proposed in this study trains an artificial intelligence technique (SOM, GHSOM, BAYESIAN NETWORKS, NAIVE BAYES, C4.5, ID3) that will perform automatically the data stream classification process. Such a technique is capable of identifying a patient’s tendency to suffer from cardiovascular disease, regardless of whether new instance types are generated. For the validation of the model, several simulation scenarios were implemented in which three phases were comprised: training, classification and calculation of metrics. The Heart Disease Cleveland Data set was selected. Then, it was applied the load balancing by data instances through the implementation of the Synthetic Minority Over Sampling Technique-SMOTE. Subsequently, the INFO.GAIN technique was applied in order to identify the most relevant characteristics of the data set and categorize them by relevance, and then to run the training with the aforementioned artificial intelligence techniques (Fig. 2).
In the classification phase, the cross-validation technique was applied to 10 folds using the previously mentioned data set The Heart Disease Cleveland Data set Train) where the sampling technique (SMOTE) and the INFO.GAIN feature selection technique were applied. Finally, the data is classified, based on the map generated in the training process and the new data subset. In the final phase, different performance metrics like sensitivity, specificity, precision and accuracy were calculated. It allowed to determine the efficiency of the proposed model.
Fundamentals of the Cardiovascular Disease Identifier Test.
There is a wide variety of diseases that increase with time. Many of them are new, becoming an unknown agent for identification systems. For this reason, there is no 100% of effectiveness in the cardiovascular disease identifier (CDI). Plus, there are bad practices in the use of the Information and Communication Technology, ICT. In other words, there are several factors that can influence negatively in the CDI decision making of cardiovascular diseases, for example identifying a patient with a cardiac pathology as a healthy person.
To evaluate the performance of CDI, four (4) metrics associated to the nature of the event have been identified, as shown in the Fig. 3:
A CDI is more efficient when, during the cardiovascular disease it presents higher hit rates, that is, the percentage of true negatives and true positives tends to 100% and consequently, it presents low failure rates which means, that the percentage of false positives and false negatives tends to 0%. According to the above, it is concluded that a perfect CDI is the one that can detects all disease trends, without generating a false alarm. The author (…) exposes that:
-
True Positive (TP): the model correctly predicts the positive class.
$$ Sensibility = TP/(TP + FN) $$(1) -
True Negative (TN): the model correctly predicts the negative class.
$$ Specificity = TN/(TN + FP) $$(2) -
False Positive (FP): the model incorrectly predicts the positive class when it is actually negative.
$$ Accuracy = \left( {TP + TN} \right)/\left( {TP + FP + FN + TN} \right) $$(3) -
False Negative (FN): the model incorrectly predicts the negative class when it is actually positive.
$$ Precision = TP/(TP + FP) $$(4)
Performance Metrics.
A confusion matrix, also known as an error matrix, is a summary table used to evaluate the performance of a classification model. The number of correct and incorrect predictions are summarized with the count values and broken down by each class. In this study, statistical performance metrics are used to measure the behavior of the CDI related to the classification process, according to the exposed by (…).
-
Sensibility: The ability of an CDI to identify “true positive” results.
-
Specificity: the ability of an CDI to measure the proportion of”true negatives” that have been correctly identified.
-
Accuracy: it is the degree of closeness of the measurements of a quantity (X) to the value of the real magnitude (Y); That is, the proportion of true results (both true positives and true negatives). 100% accuracy means that the measured values are exactly the same as the given values.
-
Precision: it the ratio of true positives against all positive results.
5 Results
The development of this research involved the use of six (6) sets of experimental tests. The test sets used the Info.gain feature selection technique. The different experiments were carried out on a DELL LATITUDE 3470 computer with an Intel Core i7 6500U processor at 2.5 Ghz, 8 GB of DDR 3 Ram at 2400 MHz and NVIDIA GeForce 920 M video card with 2 GB DDR3. Each experiment was carried out 10 times, allowing to obtain the values of the metrics that allowed to evaluate the quality of the processes, with its particular metric of quality and standard deviation. More details are shown in Table 4.
6 Conclusions and Future Work
Studies oriented to the detection of cardiovascular diseases are a great benefit to guarantee a patient inclination of suffering from cardiac diseases, with high accuracy provided after applying classifications techniques.
The INFO.GAIN feature selection method, using the J48 training and classification technique, give 93.5% of precision, 91.3% of accuracy, 92.7% of sensitivity and 91.2% of specificity, evidencing to be the most efficient model for the identification of cardiovascular diseases. For future work, the authors propose the creation of an own data set that can be trained and compared to the Heart Disease Cleveland Data set, identifying the accurate, efficiency, precision and sensibility of the data.
References
Kutyrev, K., Yakovlev, A., Metsker, O.: Mortality prediction based on echocardiographic data and machine learning: CHF, CHD, aneurism, ACS Cases. Procedia Comput. Sci. 156, 97 (2019)
Chu, D., Al Rifai, M., Virani, S.S., Brawner, C.A., Nasir, K., Al-Mallah, M.: The relationship between cardiorespiratory fitness, cardiovascular risk factors and atherosclerosis. Atherosclerosis 304, 44–52 (2020)
Xue, Y., et al.: Efficacy assessment of ticagrelor versus clopidogrel in Chinese patients with acute coronary syndrome undergoing percutaneous coronary intervention by data mining and machine-learning decision tree approaches. J. Clin. Pharm. Ther. 45(5), 1076–1086 (2020)
Rezaianzadeh, A., Dastoorpoor, M., et al.: Predictors of length of stay in the coronary care unit in patient with acute coronary syndrome based on data mining methods. Clin. Epidemiol. Glob. Health 8(2), 383–388 (2020)
Kitchenham, B., et al.: Systematic literature reviews in software engineering–a systematic literature review. Inform. Softw. Technol. 51(1), 7–15 (2009)
Kandasamy, S., Anand, S.: Cardiovascular disease among women from vulnerable populations: a review. Can. J. Cardiol. 34(4), 450–457 (2018)
Retnakaran, R.: Novel biomarkers for predicting cardiovascular disease in patients with diabetes. Can. J. Cardiol. 34(5), 624–631 (2018)
Strodthoff, N., Strodthoff, C.: Detecting and interpreting myocardial infarction using fully convolutional neural networks. Physiol. Meas. 40(1), 015001 (2019)
Idris, N.M., Chiam, Y., et al.: Feature selection and risk prediction for patients with coronary artery disease using data mining. Med. Biol. Eng. Compu. 58(12), 3123–3140 (2020)
Kramer, A., Trinder, M., et al.: Estimating the prevalence of familial hypercholes-terolemia in acute coronary syndrome: a systematic review and meta-analysis. Can. J. Cardiol. 35(10), 1322–1331 (2019)
Leung, K.: Ming: Naive Bayesian classifier. Financ. Risk Eng. 2007, 123–156 (2007)
Barletta, V., et al.: A Kohonen SOM architecture for intrusion detection on in-vehicle communication networks. Appl. Sci. 10(15), 5062 (2020)
Larry Bretthorst, G.: An introduction to parameter estimation using Bayesian probability theory. In: Fougère, P.F. (ed.) Maximum Entropy and Bayesian Methods, pp. 53–79. Springer Netherlands, Dordrecht (1990). https://doi.org/10.1007/978-94-009-0683-9_5
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Comas-Gonzalez, Z., Mardini-Bovea, J., Salcedo, D., De-la-Hoz-Franco, E. (2023). Comparison of Supervised Techniques of Artificial Intelligence in the Prediction of Cardiovascular Diseases. In: Degen, H., Ntoa, S., Moallem, A. (eds) HCI International 2023 – Late Breaking Papers. HCII 2023. Lecture Notes in Computer Science, vol 14059. Springer, Cham. https://doi.org/10.1007/978-3-031-48057-7_4
Download citation
DOI: https://doi.org/10.1007/978-3-031-48057-7_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-48056-0
Online ISBN: 978-3-031-48057-7
eBook Packages: Computer ScienceComputer Science (R0)