Abstract
Breast cancer continues to be one of the most lethal cancer types, mainly affecting women. However, thanks to the utilization of deep learning approaches for breast cancer detection, there has been a considerable boost in the performance in the field. The loss function is a core element of any deep learning architecture with a significant influence on its performance. The loss function is particularly important for tasks such as breast mass segmentation. For this task, challenging properties of input images, such as pixel class imbalance, may result in instability of training or poor detection results due to the bias of the loss function toward correctly segmenting the majority class. Inspired by the success of sample-level loss functions, we propose a hybrid loss function incorporating both pixel-level and region-level losses, where the breast tissue density is used as a sample-level weighting signal. We refer to the proposed loss as Density-based Adaptive Sample-Level Prioritizing (Density-ASP) loss. Our motivation stems from the observation that mass segmentation becomes more challenging as breast density increases. This observation makes density a viable option for controlling the effect of region-level losses. To demonstrate the effectiveness of the proposed Density-ASP, we have conducted mass segmentation experiments using two publicly available datasets: INbreast and CBIS-DDSM. Our experimental results demonstrate that Density-ASP improves segmentation performance over the commonly used hybrid losses across multiple metrics.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
1 Introduction
Despite significant progress in breast cancer screening over the last decades, breast cancer remains one of the most fatal cancer types among women [1]. Mammography is the most common screening tool for breast cancer detection, which has been shown to reduce mortality rate [4]. Automated breast cancer detection using mammography could help to reduce the cost of a second reader [2, 3] while at the same time increasing the chance of early detection.
Powered by the well-proven effectiveness of deep learning, recent research work on abnormality detection has achieved promising results. However, these methods are still restricted by limitations, such as pixel class imbalance [5], which can adversely affect results in various tasks such as mass segmentation. These limitations have many times their root in the design of the loss function. In most cases, using a hybrid loss function – the weighted sum of different loss functions – has shown to be more beneficial compared to non-hybrid loss (using only one type of loss function). Although using the de-facto hybrid loss [5,6,7]–a weighted sum of Dice [8] and Binary Cross Entropy (BCE) [9] losses–has been shown to provide stability and robustness, the success of recent work on adaptive loss weighting strategies indicates that hybrid loss could be significantly improved by a sample-level design. Following the design of the ASP loss [10], which uses the mass ratio in the loss weighting strategy, we propose to employ the breast tissue density associated with each sample in the loss weighting strategy.
The rationale behind selecting breast tissue density as the sample-level signaling feature for hybrid loss stems from the observation that breast density (which represents the composition of fat, fibrous, and glandular tissue) is correlated with mass segmentation’s difficulty. In the case of automatic detection from a single view, higher tissue density might be mistaken for an abnormality and increase the false positive rate. Therefore, the breast tissue density of each sample could provide valuable information during training. How to best leverage the information conveyed by each density category for training purposes is an important research question which we attempt to explore in this paper by introducing a region-level loss term in the hybrid loss. The benefit of the region-level loss term lies in the observation that comparison between the regions (rather than pixels) could result in the reduction of false positive and false negative rates by considering the dependencies between (via including surrounding pixels in the calculation of the loss) the pixels.
The Density-ASP loss function proposed in this paper consists of pixel-level and region-level losses. In this paper, loss functions such as Dice and BCE that consider pixels independently in the calculation of the loss are referred to as pixel-level losses. On the other hand, the loss functions that take the dependencies between the pixels into consideration are referred to as region-level losses. For the pixel-level loss term, we have combined Dice [5] and BCE [9]. The combination of these losses has been shown to help to address the issue of pixel class imbalance and increase training stability [6]. For the region-level loss term, we have combined Structural Similarity Index (SSIM) [11] and Region Mutual Information (RMI) [12]. It should be noted that the term hybrid applies to Density-ASP as well as each of the pixel-level and region-level terms (as they consist of two losses in their own categories). Instead of employing fixed weights [6, 7] for each loss term, following the ASP loss methodology [10], we propose to use the ACR breast density category as an indicative signal for prioritizing the region-level loss term over the pixel-level loss term and vice versa. Therefore, the region-level loss is an adaptive loss term that will be prioritized for samples with higher density. Using AU-Net [6], which is a modern and effective variation of U-Net [14], as the baseline architecture, the Density-ASP loss has been evaluated on two benchmark datasets for mass segmentation: INbreast [15], and CBIS-DDSM [16]. The results of our experiments illustrate that Density-ASP loss provides considerable performance improvements compared to commonly used hybrid losses.
The contributions of this paper are four-fold:
-
Incorporating both pixel-level and region-level losses in the Density-ASP loss function.
-
Employing breast tissue density as a prioritizing signal for adaptive sample-level prioritizing loss function for mass segmentation on whole mammograms.
-
Evaluating Density-ASP on two benchmark datasets, INbreast and CBIS-DDSM.
-
Quantitatively analyzing and comparing the findings of our experimental results for Density-ASP loss with the traditional hybrid loss for the baseline approach and state-of-the-art mass segmentation methods.
In the following sections, we first review related work in the field. The proposed method is then explained in detail. Our experimental results, analysis, and comparison with state-of-the-art methods are presented next. Finally, we provide our conclusions and discuss directions for future research.
2 Related Work
Recently, deep learning-based approaches have shown great promise in abnormality detection in medical images, with many studies achieving more accurate mass segmentation results compared to traditional approaches. In this section, we aim to briefly review the related work in deep learning-based approaches for breast mass segmentation, categorizing them into two groups: breast mass segmentation in whole mammograms and loss functions for binary segmentation of medical images.
2.1 Mass Segmentation on Whole Mammograms
The majority of breast mass segmentation approaches fall into one of the following categories based on the type of input they receive: region of interest (RoI) and whole mammogram. RoI-based mass segmentation approaches [17] have different properties, challenges, and strategies compared to methods using whole mammograms [6, 7]; thus, in this section, the primary focus is on reviewing related work in the latter category.
Inspired by [18], one of the pioneer deep learning-based approaches for segmentation, Ronneberger et al. proposed U-Net [14], which is a fully convolutional symmetric encoder-decoder architecture that is instrumental for segmentation tasks with limited training data. This property of U-Net makes it specifically favorable for medical image segmentation where data scarcity is a relatively common limitation. U-Net combines low-level location information from the encoder with high-level semantic information from the decoder.
Thanks to the effectiveness of U-Net, a new wave of variations for different medical tasks has emerged [19,20,21,22,23,24,25], continuing to push the performance boundaries of medical image segmentation. In this context, the method proposed in [26] introduced a similar encoder-decoder architecture (leveraging dense blocks) where multi-scale information is utilized in the network. To enhance the performance of the network without additional parameters, atrous convolution [27] with various sample rates was used in the last encoder block. [28] is another U-Net-based approach based on the idea of utilizing a densely-connected network in the encoder and a CNN with attention gates in the decoder. Another line of research within the scope of multi-scale studies is [29], where the generator was designed as an improved version of U-Net. Before sending the segmentation results to the discriminator, multiscale results were created for three critics with different scales in the discriminator. Ravitha et al. [30], developed an approach employing the error of the outputs of intermediate layers relative to the ground truth labels as a supervision signal to boost model performance.
In [6], the authors introduced an attention-guided dense-up-sampling asymmetric encoder-decoder network (AU-Net) with an intermediate up-sampling block which includes a channel-wise attention mechanism designed to leverage the beneficial information presented in both low and high-level features. To mitigate the problem of relatively low performance of U-Net approach on small-size masses, [7] proposed to use a selective receptive field module with two parts, one for generating several receptive fields with different sizes and one for selecting the appropriate size of the receptive field. AU-Net has been chosen as the baseline model in this study.
2.2 Loss for Medical Image Segmentation
The choice of a suitable loss function, conveying the desired objectives of the task performed by a network, has a tremendous impact on the training process and overall performance of the network. Among the previously introduced losses for segmentation, while some consider the pixels independent entities, others seek to take regional information into consideration to capture the dependencies between the pixels. The first group is generally regarded as pixel-level losses, and the latter as region-level losses in the literature. Considering that both categories are relevant to this research, we provide a concise summary of related studies in both groups, starting with pixel-level approaches and emphasizing the ones proposed for the medical domain.
Binary segmentation could be considered as the classification of pixels into positive (foreground) and negative (background) classes. A common loss function for this task is the BCE loss [9] (Eq. 1) which penalizes the discrepancy between predicted and ground truth classes for all pixels. Weighted Binary Cross Entropy [31] and Balanced Cross Entropy [32] are two BCE variants that differentiate between the effect of false positives and false negatives through weighting coefficients. Focal loss [33] further improved BCE by changing the magnitude of the loss according to the hardness of the examples based on the confidence of the model. Dice loss [8] is suitable for addressing the pixel class imbalance problem [34], formulated as the ratio of correctly classified pixels to the total number of positive pixels in the prediction and ground truth masks (Eq. 2). Tversky loss [35] provides a way to control the contribution of the false positive and the false negative terms in the Dice loss by weighting these terms.
All the aforementioned losses belong to the pixel-level category (i.e., they consider the pixels independently). While providing effective training signals for the network, they neglect to consider the relationship among pixels, which could provide a considerable boost, notably for cases with irregularity in shapes. Initially proposed for image quality assessment, SSIM [11] has been incorporated in the segmentation loss for medical image segmentation [13] and has inspired several region-level losses. SSL [36] and RMI [12] are two examples of region-level losses developed for segmentation. It should be noted that both of these losses consider a fixed-size window around each pixel as the region (a region is defined for each pixel) rather than a fixed location (a region is a fixed location in a grid) in the ground truth and the prediction as utilized in [37, 38].
SSIM [11] uses luminance, contrast, and structure in measuring the differences between two regions. Inspired by the influence of the structural term in the SSIM, which has the potential to be customized for segmentation purposes, the authors of Structural Similarity Loss (SSL) [36] proposed to weight the cross-entropy of every two pixels based on the structural error (error between two image regions which indicates the degree of linear correlation) while ignoring pixels with low error and emphasizing on pixels with high error by thresholding the error rate. With the goal of maximizing the structural similarity between images, RMI [12] first converts the region around a center (pixel) to a multi-dimensional point (for a 3*3 region, it will be a 9D point) and then maximizes the MI between multidimensional distributions.
Several compound losses [33, 35, 39] have been proposed to reap the benefits of different losses by combining two or more of them. Combo loss [39] has been proposed to control the contribution of false positive and false negative by a weighting strategy in the BCE loss term where the total loss is a weighted sum of BCE and Dice loss. In adaptive sample-level prioritizing loss, we have proposed a novel approach to weight the loss terms (Dice and BCE) dynamically. This is performed in an adaptive manner by controlling the influence of each loss according to each sample using the ratio of the mass to image size as a weighting signal. It should be noted that ASP has three versions: quantile-based, cluster-based, and learning-based. In the quantile-based ASP, the images are groups based on the quantile to which the ratio of the mass belongs. In the cluster-based version, the category of an image is identified according to the K-means clustering of the ratios. Finally, the learning-based ASP is a parametrized version of the ASP loss.
We follow the same dynamic weighting strategy as ASP loss [10]. However, instead of utilizing the size of the mass, we opt to utilize the breast tissue density, which is mostly related to the difficulty of the segmentation. In addition, instead of solely using and weighting pixel-level losses, we introduce a combination of hybrid pixel-level and region-level losses.
3 Methodology
In this study, both hybrid pixel-level and region-level loss functions are utilized. Therefore, an overview of hybrid pixel-level \(L_{HP}\) and region-level \(L_{HR}\) losses are provided in the following sections.
3.1 Hybrid Pixel-Level Loss
The standard hybrid pixel-level loss, commonly used for mass segmentation in mammograms, is defined as a weighted sum of BCE and Dice loss, as shown below:
Here \(y\) and \(\hat{y}\) represent the ground truth and the predicted segmentation masks, respectively. \(\alpha \) and \(\beta \) (could be relative, for instance, formulated as \(\beta = 1- \alpha \) ) are the weighting parameters in the hybrid loss denoted as \(L_{HP}\) in Eq. 3. While the cross entropy loss (Eq. 1) includes correctly classified positive and negative pixels, the Dice loss (Eq. 2) incorporates only correctly classified positive pixels, which makes it more suitable in the presence of considerable pixel class imbalance. The combination (Eq. 3) of the two losses has been shown to provide a better learning signal. In particular, it has been reported that adding BCE to the Dice loss helps to mitigate the unstable training associated with using only the Dice loss [33, 34]. On the other hand, adding the Dice loss to BCE helps to improve the performance of the model on datasets with pixel class imbalance compared to using BCE alone.
3.2 Hybrid Region-Level Loss
Region-level losses aim to incorporate the context to which a pixel belongs in the loss calculation by representing each pixel with its own value and the neighboring pixels’ values. In this paper, two of the region-level losses, SSIM and RMI, have been selected and are represented in the following:
Here, \(Y_m\) and \(\hat{Y_m}\) are the multi-dimensional points constructed using a centering pixel and the neighboring pixels in a surrounding square. \(S\) and \(\hat{S}\) are the support sets corresponding to the ground truth and prediction masks, respectively. \(f (y)\) and \(f(\hat{y})\) represent the probability density functions for the ground truth and prediction masks, respectively. The \(f (y, \hat{y})\) captures the joint PDF. The implementation details of the RMI loss are available in [12]. The second region-level loss used in this paper is SSIM-based loss in Eq. 5.
Here, \(Y_p\) and \(\hat{Y_p}\) represent patches in the ground truth and the prediction masks. \(\mu \) and \(\sigma \) are the mean and variance for the corresponding patches, respectively. \(\sigma _{y\hat{y}}\) is the covariance of the two patches. More details (including the selection of \(C1\) and \(C_2\)) are available in [11] .Finally, the hybrid region-level loss is presented in Eq. 6.
In the hybrid region-level loss \(L_{HR}\) (Eq. 6), \(L_{RMI}\) and \(L_{SSIM}\) are the RMI and SSIM losses, respectively. The hyperparameters \(\eta \) and \(\gamma \) represent the weighting coefficients.
3.3 Density-Adaptive Sample-Level Prioritizing Loss
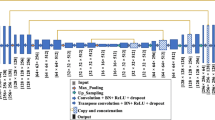
While the aforementioned hybrid pixel-level loss is quite effective, we propose extending it by using an adaptive weighting strategy based on the idea of ASP [10]. The resulting hybrid loss is a combination of region-level and pixel-level losses instead of using only pixel-level losses. We propose using breast tissue density as the sample-level signal for the extended hybrid loss’s prioritizing strategy. In the following, the proposed framework for the Density-ASP loss is explained. Given a training set of \(N\) images and the corresponding segmentation masks, the baseline method learns a mapping between an input image to its segmented counterpart using the training data. In this study, AU-Net was selected as the baseline method; the architecture for AU-Net is presented in Fig. 1a. For the encoder and decoder, ResUnit and the basic decoder proposed in AU-Net have been used. The details of the AU block, basic decoder, and ResUnit encoder are presented in [6]. The Density-ASP loss requires the breast tissue density for each sample. The standard ACR density, which is available in both datasets, was used in this study. ACR breast density reflects the composition of the fat, fibrous, and glandular tissue in four categories.
There are noticeable differences in the appearance of the breast within different density categories in mammography images. Generally, the complexity of the texture increases as density increases. This provides meaningful distinguishing information for the loss function to prioritize the pixel-level or region-level terms in the loss function based on the density of each sample. The more complex the texture is (i.e., higher density category), the more important the contribution of the region-level term will be. Therefore, density is considered a determining factor in the weighting strategy.
In Fig. 1a, the prediction heatmap (\(\hat{y}\) in the formulas) and ground truth segmentation masks are inputted to the Density-ASP module. The process of prioritizing loss is presented in Fig. 1b. Since there is no proven or intuitive connection between density and pixel-level term, the weight for this category remains fixed. However, the contribution of the region-level term will change in an adaptive manner, as shown in Eq. 7 and Fig. 1b. It should be noted that the weighting coefficients inside the pixel-level and region-level loss terms are not adaptive.
\(L_{Density-ASP}^i\) is the final Density-ASP loss for the \(i^{th}\) sample. \(\theta \) is the prioritizing vector consisting of the weights assigned to each density category, and \(p_i\) denotes a one-hot encoding of the density category to which the \(i^{th}\) sample belongs. \(p^i \theta ^T\) will be the weight for the region-level loss term, which determines the importance of the region-level term according to the density category of \(i^{th}\) sample.
4 Experimental Results
This section begins with a description of the datasets, and evaluation metrics. Subsequently, the experimental setting is presented, followed by a comprehensive analysis of the results on both datasets, including comparisons with the state-of-the-art approaches.
4.1 Datasets
We have conducted mass segmentation experiments using two publicly available datasets: INbreast and CBIS-DDSM. We have normalized the intensity of the images in both datasets and all images have been resized to \(256 \times 256\). No data augmentation or image enhancement were considered in our experiments. To prevent overfitting, a randomly selected validation set was utilized for hyperparameter tuning. For the baseline approach, the batch size was set to four, the learning rate was initially set to 10−e4, and a step decay policy with a decay factor of 0.5 was employed in all experiments. Irrespective of the abnormality type, all the images containing masses have been utilized in our experiments.
INbreast Dataset. The INbreast dataset contains 410 images associated with 150 cases, including various abnormality types. In the context of mass segmentation, only 107 of the images containing masses (the total number of masses across all of the images is 116) have been used in this study. A 5-fold cross-validation was employed, a commonly used setting for the measurement of the performance of methods on the INbreast dataset. The dataset was randomly divided into training (80%), validation (10%), and test (10%) sets.
CBIS-DDSM Dataset. From a total of 1944 cases in the CBIS-DDSM dataset, 1591 images containing masses were utilized in our experiments. The official split of the dataset (1231 and 360 images for train and test sets, respectively) was employed for the experimental results presented in this paper. 10% of the training data was randomly selected for the validation set. In a preprocessing stage for the CSIB-DDSM dataset, artifacts were removed, and images were cropped and resized.
4.2 Evaluation Metrics
Since mass segmentation in mammograms is characterized by a pixel class imbalance, we have selected several metrics to better illustrate the strengths and weaknesses of the proposed methods. Specifically, the Dice Similarity Coefficient (DSC), Relative Area Difference (\(\varDelta A\)), Sensitivity, and Accuracy have been selected due to the complementary information they provide. This combination of evaluation metrics highlights the performance of each method both on majority (background) and minority (masses) classes. It also reflects how accurately a method performs in terms of predicting the boundary of masses, which is crucial for mass classification.
4.3 Comparison with State-of-the-Art Methods
To assess the performance of Density-ASP loss, we have conducted a comprehensive comparison with three state-of-the-art mass segmentation approaches on whole mammograms: AU-Net (baseline), ARF-Net, and ASP. The official implementation of AU-Net, and the setting described in the AU-Net paper [6] (only the architecture was publicly available) were used in our experiments. ARF-Net is a state-of-the-art method for mass segmentation on whole mammograms. The method was implemented to the best of our understanding based on the original paper (i.e., the implementation of the approach or the trained models were not publicly available). For the ASP loss, we have used the same experimental setting and data split, so we have directly used the reported results in the original paper. The publicly available implementations of the RMI and SSIM were utilized. To ensure a fair comparison of the methods, no pre-training or data augmentation were used. The coefficients for density-based loss were \(\theta = [0.5, 0.5, 0.85, 0.95]\) and \(\theta = [0.25, 0.25, 0.85, 0.95]\) for the INbreast and CSIB-DDSM , respectively . \(\eta \), \(\gamma \), \(\beta \) were set to one; \(\alpha \) was set to 2 and 2.5 for INbreast and CBIS-DDSM. These hyperparameters were selected through experimental evaluation.
Experimental Results Using INbreast. Table 1 summarizes our experimental results for all models trained on INbreast. The best results are highlighted using bold font. The Density-ASP loss achieved better performance across all of the metrics compared to the pixel-level hybrid losses. The improvement for the Density-ASP (over using hybrid pixel-level loss in the baseline method) is as follows: (DSC: \(+9.27\%\), \(\varDelta A\): \(-12.77\%\), Sensitivity: \(+ 20.21\%\), Accuracy: +0.19 % ), which is consistent across all metrics. The Density-ASP outperformed ARF-Net in DSC, \(\varDelta A\), and sensitivity while the accuracy is \(0.06 \%\) less. It should be noted that ARF-Net is designed to incorporate different sizes and, surpasses the baseline method in DSC, sensitivity, and accuracy. Better performance of the Density-ASP (in most metrics) compared to ARF-Net, indicates that improvement in the training that Density-ASP provides for the baseline method, not only closes the gap between AU-Net and ARF-Net in most of the metrics but also makes AU-Net outperform ARF-Net. In comparison with the ASP loss variations (as the best-performing version, cluster-based ASP was selected for comparison), Density-ASP performed better in terms of DSC, \(\varDelta A\), and sensitivity. The accuracy of the cluster-based ASP variation is \(0.13 \% \) higher than the Density-ASP. The results of the Density-ASP further validate the effectiveness of sample-level losses. Moreover, the fact that Density-ASP outperforms the ASP in most of the metrics indicates that introducing the region-level losses to the loss function with the density as a weighting signal is a promising approach for mass segmentation. We attribute this improvement to the utilization of density as the prioritizing signal, which helps to distinguish the contribution of the losses for each sample, leading to better segmentation.
The first four columns in Fig. 2 show some representative results for Density-ASP, AU-Net, ARF-Net, and all ASP variations for INbreast. These examples have been selected to include instances for each density category (mentioned at the top of the columns), demonstrating the segmentation capabilities of the methods across different density categories. The green and blue lines represent the contours of the ground truth and the prediction masks, respectively. It can be observed that the segmentation results for Density-ASP are more accurate compared to state-of-the-art methods across all the density categories.
Experimental Results Using CBIS-DDSM. The performance of Density-ASP loss on the CBIS-DDSM dataset compared to state-of-the-art approaches is presented in Table 2. The improvement for the Density-ASP on the CBIS-DDSM dataset (over using hybrid pixel-level loss in the baseline method) is as follows: (DSC: \(+1.59\%\), \(\varDelta A\): \(-3.98\%\), Sensitivity: \(+ 0.66\%\), Accuracy: +0.03 % ). The Density-ASP outperformed ARF-Net (which has a different architecture but used common hybrid loss) in all metrics except for accuracy, which is \(0.02\%\) lower. When compared to quantile-based ASP loss – a version of the ASP with the best performance on CBIS-DDSM– while Density-ASP outperformed quantile-based ASP in sensitivity (\(+0.15\% \)), it under-performed in other metrics (DSC: \(- 0.4 \%\), \(\varDelta A\): \(+3.91\%\), Accuracy: \(- 0.03 \% \) ). We speculate that the reason might be related to the fact that the mass ratio in ASP loss is a data-driven factor that completely correlates with the statistics of the pixels in the image. On the other hand, density is predefined and, in some cases, might not be aligned with the visual features, which might be a more common issue in the CBIS-DDSM dataset. The fact that Density-ASP improves in all of the metrics over the baseline method shows the effectiveness of using density and region-level losses for the CBIS-DDSM dataset. The last four columns in Fig. 2 show some representative examples where Density-ASP has better performance when compared to the previous methods in different density categories.
In general, the performance of the Density-ASP is better for the INbreast dataset. One observation is that in both datasets, there are examples that the density category of the image might not be perfectly aligned with visual features (for example, the 2nd column in Fig. 2), which could cause a higher weight for the term that does not match the initial idea of the Density-ASP. We speculate that the assigned density for the INbreast is more visually aligned with the images, resulting in better weighting for loss terms. Different distributions of each category in the datasets might also be a factor in the performance of the Density-ASP on two datasets.
5 Conclusion
We have proposed a new sample-level adaptive prioritizing loss that utilizes breast tissue density as a weighting signal. Moreover, we have proposed a hybrid loss function that includes region-level losses in the training. Finally, given the observed connection between the difficulty of mass identification and the breast tissue density category, this approach focuses on using the density for weighting of the region-level loss term to highlight the importance of the region-level term according to the density category for each sample adaptively. Our experimental results demonstrate improvements in all evaluation metrics on two benchmark datasets: INbreast and CBIS-DDSM. Customizing this category of losses for other domains or tasks is an appealing direction for future work. One other promising direction could be the extraction of texture descriptors [40], from the images themselves, as the assigned category in some cases may not correlate with the complexity of the texture in the image. This becomes more vital in medical imaging datasets where density information might not be available. In those cases, using data-driven, higher-level information could provide a way to use pixel-level and region-level losses in an adaptive manner.
References
Siegel, R.L., Miller, K.D., Fuchs, H.E., Jemal, A.: Cancer statistics 2022. CA: Cancer J. Clin. 72(1), 7–33 (2022)
Batchu, S., Liu, F., Amireh, A., Waller, J., Umair, M.: A review of applications of machine learning in mammography and future challenges. Oncology 99(8), 483–490 (2021)
McKinney, S.M., et al.: International evaluation of an AI system for breast cancer screening. Nature 577(7788), 89–94 (2020)
Nyström, L., Andersson, I., Bjurstam, N., Frisell, J., Nordenskjöld, B., Rutqvist, L.E.: Long-term effects of mammography screening: updated overview of the Swedish randomised trials. Lancet 359(9310), 909–919 (2002)
Malof, J.M., Mazurowski, M.A., Tourassi, G.D.: The effect of class imbalance on case selection for case-based classifiers: an empirical study in the context of medical decision support. Neural Netw. 25, 141–145 (2012)
Sun, H., et al.: AUNet: attention-guided dense-upsampling networks for breast mass segmentation in whole mammograms. Phys. Med. Biol. 65(5), 055005 (2020)
Xu, C., Qi, Y., Wang, Y., Lou, M., Pi, J., Ma, Y.: ARF-Net: an adaptive receptive field network for breast mass segmentation in whole mammograms and ultrasound images. Biomed. Signal Process. Control 71, 103178 (2022)
Milletari, F., Nassir, N., Seyed-Ahmad, A.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
Yi-de, M., Qing, L., Zhi-Bai, Q.: Automated image segmentation using improved PCNN model based on cross-entropy. In: Proceedings of 2004 International Symposium on Intelligent Multimedia, Video and Speech Processing, pp. 743–746. IEEE (2004)
Liniya, P., Nicolescu, M., Nicolescu, M., Bebis, G.: ASP Loss: adaptive sample-level prioritizing loss for mass segmentation on whole mammography images. In: Iliadis, L., Papaleonidas, A., Angelov, P., Jayne, C. (eds.) ICANN 2023. LNCS, vol. 14255, pp. 102–114. Springer, Cham (2023). https://doi.org/10.1007/978-3-031-44210-0_9
Wang, Z., Bovik, A.C., Sheikh, H.R., Simoncelli, E.P.: Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13(4), 600–612 (2004)
Zhao, S., Wang, Y., Yang, Z., Cai, D.: Region mutual information loss for semantic segmentation. In: Advances in Neural Information Processing Systems, vol. 32 (2019)
Huang, H., et al.: Unet 3+: a full-scale connected unet for medical image segmentation. In: ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1055–1059. IEEE (2020)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Moreira, I.C., Amaral, I., Domingues, I., Cardoso, A., Cardoso, M.J., Cardoso, J.S.: Inbreast: toward a full-field digital mammographic database. Acad. Radiol. 19(2), 236–248 (2012)
Lee, R.S., Gimenez, F., Hoogi, A., Miyake, K.K., Gorovoy, M., Rubin, D.L.: A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 4(1), 1–9 (2017)
Baccouche, A., Garcia-Zapirain, B., Castillo Olea, C., Elmaghraby, A.S.: Connected-UNets: a deep learning architecture for breast mass segmentation. NPJ Breast Cancer 7(1), 151 (2021)
Long, J., Evan, S., Trevor, D.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440. (2015)
Wu, S., Wang, Z., Liu, C., Zhu, C., Wu, S., Xiao, K.: Automatical segmentation of pelvic organs after hysterectomy by using dilated convolution u-net++. In: 2019 IEEE 19th International Conference on Software Quality, Reliability and Security Companion (QRS-C), pp. 362–367. IEEE (2019)
Zhang, J., Jin, Y., Xu, J., Xu, X., Zhang, Y.: Mdu-net: multi-scale densely connected u-net for biomedical image segmentation. arXiv preprint arXiv:1812.00352 (2018)
Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: UNet++: a nested u-net architecture for medical image segmentation. In: Stoyanov, D., et al. (eds.) DLMIA/ML-CDS -2018. LNCS, vol. 11045, pp. 3–11. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00889-5_1
Li, C., et al.: Attention unet++: a nested attention-aware u-net for liver CT image segmentation. In: 2020 IEEE International Conference on Image Processing (ICIP), pp. 345–349. IEEE (2020)
Cao, H., et al.: Swin-Unet: Unet-like pure transformer for medical image segmentation. In: Karlinsky, L., Michaeli, T., Nishino, K. (eds.) ECCV 2022. LNCS, vol. 13803, pp. 205–218. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-25066-8_9
Oktay, O., et al.: Attention u-net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018)
Song, T., Meng, F., Rodriguez-Paton, A., Li, P., Zheng, P., Wang, X.: U-next: a novel convolution neural network with an aggregation u-net architecture for gallstone segmentation in CT images. IEEE Access 7, 166823–166832 (2019)
Hai, J., Qiao, K., Chen, J., Tan, H., Xu, J., Zeng, L., Shi, D., Yan, B.: Fully convolutional densenet with multiscale context for automated breast tumor segmentation. Journal of healthcare engineering, (2019)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40(4), 834–848 (2017)
Li, S., Dong, M., Du, G., Mu, X.: Attention dense-u-net for automatic breast mass segmentation in digital mammogram. IEEE Access 7, 59037–59047 (2019)
Chen, J., Chen, L., Wang, S., Chen, P.: A novel multi-scale adversarial networks for precise segmentation of x-ray breast mass. IEEE Access 8, 103772–103781 (2020)
Rajalakshmi, N.R., Vidhyapriya, R., Elango, N., Ramesh, N.: Deeply supervised u-net for mass segmentation in digital mammograms. Int. J. Imaging Syst. Technol. 31(1), 59–71 (2021)
Pihur, V., Datta, S., Datta, S.: Weighted rank aggregation of cluster validation measures: a monte carlo cross-entropy approach. Bioinformatics 23(13), 1607–1615 (2007)
Xie, S., Tu, Z.: Holistically-nested edge detection. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1395–1403, (2015)
Yeung, M., Sala, E., Schönlieb, C.B., Rundo, L.: Unified focal loss: generalising dice and cross entropy-based losses to handle class imbalanced medical image segmentation. Comput. Med. Imaging Graph. 95, 102026 (2022)
Jadon, S.: A survey of loss functions for semantic segmentation. In: 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), pp. 1–7. IEEE (2020)
Salehi, S.S.M., Erdogmus, D., Gholipour, A.: Tversky loss function for image segmentation using 3D fully convolutional deep networks. In: Wang, Q., Shi, Y., Suk, H.-I., Suzuki, K. (eds.) MLMI 2017. LNCS, vol. 10541, pp. 379–387. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67389-9_44
Zhao, S., Boxi, W., Wenqing, C., Yao, H., Deng, Cai.: Correlation maximized structural similarity loss for semantic segmentation. arXiv preprint arXiv:1910.08711 (2019)
Aliniya, P., Razzaghi, P.: Parametric and nonparametric context models: a unified approach to scene parsing. Pattern Recogn. 84, 165–181 (2018)
Alinia, P., Parvin, R.: Similarity based context for nonparametric scene parsing. In: 2017 Iranian Conference on Electrical Engineering (ICEE), pp. 1509–1514. IEEE (2017)
Taghanaki, S.A., et al.: Combo loss: handling input and output imbalance in multi-organ segmentation. In: Computerized Medical Imaging and Graphics, vol. 75, pp. 24–33 (2019)
Simon, P., Uma, V.: Review of texture descriptors for texture classification. In: Satapathy, S.C., Bhateja, V., Raju, K.S., Janakiramaiah, B. (eds.) Data Engineering and Intelligent Computing. AISC, vol. 542, pp. 159–176. Springer, Singapore (2018). https://doi.org/10.1007/978-981-10-3223-3_15
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Aliniya, P., Nicolescu, M., Nicolescu, M., Bebis, G. (2023). Hybrid Region and Pixel-Level Adaptive Loss for Mass Segmentation on Whole Mammography Images. In: Bebis, G., et al. Advances in Visual Computing. ISVC 2023. Lecture Notes in Computer Science, vol 14361. Springer, Cham. https://doi.org/10.1007/978-3-031-47969-4_1
Download citation
DOI: https://doi.org/10.1007/978-3-031-47969-4_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-47968-7
Online ISBN: 978-3-031-47969-4
eBook Packages: Computer ScienceComputer Science (R0)