Abstract
The diagnosis of prostate cancer is driven by the histopathological appearance of epithelial cells and epithelial tissue architecture. Despite the fact that the appearance of the tumor-associated stroma contributes to diagnostic impressions, its assessment has not been standardized. Given the crucial role of the tumor microenvironment in tumor progression, it is hypothesized that the morphological analysis of stroma could have diagnostic and prognostic value. However, stromal alterations are often subtle and challenging to characterize through light microscopy alone. Emerging evidence suggests that computerized algorithms can be used to identify and characterize these changes. This paper presents a deep-learning approach to identify and characterize tumor-associated stroma in multi-modal prostate histopathology slides. The model achieved an average testing AUROC of \(86.53\%\) on a large curated dataset with over 1.1 million stroma patches. Our experimental results indicate that stromal alterations are detectable in the presence of prostate cancer and highlight the potential for tumor-associated stroma to serve as a diagnostic biomarker in prostate cancer. Furthermore, our research offers a promising computational framework for in-depth exploration of the field effect and tumor progression in prostate cancer.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Prostate cancer (PCa) diagnosis and grading rely on histopathology analysis of biopsy slides [1]. However, prostate biopsies are known to have sampling error as PCa is heterogenous and commonly multifocal, meaning cancer legions can be missed during the biopsy procedure [2]. If significant PCa is detected on biopsies and the patient has organ-confined cancer with no contraindications, radical prostatectomy (RP) is the standard of care [3, 4]. Following RP, the prostate is processed and slices are mounted onto slides for analysis. Radical prostatectomy histopathology samples are essential for validating the biopsy-determined grade group [5, 6]. Analysis of whole-mount slides, meaning slides that include slices of the entire prostate, provide more precise tumor boundary detection, identification of various tumor foci, and increased tissue for identifying morphological patterns not visible on biopsy due to a larger field of view.
Field effect refers to the spread of genetic and epigenetic alterations from a primary tumor site to surrounding normal tissues, leading to the formation of secondary tumors. Understanding field effect is essential for cancer research as it provides insights into the mechanisms underlying tumor development and progression. Tumor-associated stroma, which consists of various cell types, such as fibroblasts, smooth muscle cells, and nerve cells, is an integral component of the tumor microenvironment that plays a critical role in tumor development and progression. Reactive stroma, a distinct phenotype of stromal cells, arises in response to signaling pathways from cancerous cells and is characterized by altered stromal cells and increased extracellular matrix components [7, 8]. Reactive stroma is often associated with tumor-associated stroma and is thought to be a result of field effects in prostate cancer. Altered stroma can create a pro-tumorigenic environment by producing a multitude of chemokines, growth factors, and releasing reactive oxygen species [9, 10], which can lead to tumor development and aggressiveness [11]. Therefore, investigating the histological characterization of tumor-associated stroma is crucial in gaining insights into the field effect and tumor progression of prostate cancer.
Manual review for tumor-associated stroma is time-consuming and lacks quantitative metrics [12, 13]. Several automated methods have been applied to analyze the tumor-stroma relationship; however, most of them focus on identifying a tumor-stroma ratio rather than finding reactive stroma tissue or require pathologist input. Machine learning algorithms have been used to quantify the percentage of tumor to stroma in bladder cancer patients, but required dichotomizing patients based on a threshold [14]. Software has been used to segment tumor and stroma tissue in breast cancer patient samples, but the method required constant supervision by a pathologist [15]. Similarly, Akoya Biosciences Inform software was used to quantify reactive stroma in PCa, but this method required substantial pathologist input to train the software [16]. Fully automated deep-learning methods have been developed to identify tumor-associated stroma in breast cancer biopsies, achieving an AUC of 0.962 in predicting invasive ductal cancer [13]. However, identifying tumor-associated stroma in prostate biopsies and whole-mount histopathology slides remains challenging.
Analyzing tumor-associated stroma in prostate cancer requires combining whole-mount and biopsy histopathology slides. Biopsy slides provide information on the presence of PCa, while whole-mount slides provide information on the extent and distribution of PCa, including more information on tumor-associated stroma. Combining the information from both modalities can provide a more accurate understanding of the tumor microenvironment. In this work, we explore the field effect in prostate cancer by analyzing tumor-associated stroma in multi-modal histopathological images. Our main contributions can be summarized as follows:
-
To the best of our knowledge, we present the first deep-learning approach to characterize prostate tumor-associated stroma by integrating histological image analysis from both whole-mount and biopsy slides. Our research offers a promising computational framework for in-depth exploration of the field effect and cancer progression in prostate cancer.
-
We proposed a novel approach for stroma classification with spatial graphs modeling, which enable more accurate and efficient analysis of tumor microenvironment in prostate cancer pathology. Given the spatial nature of cancer field effect and tumor microenvironment, our graph-based method offers valuable insights into stroma region analysis.
-
We developed a comprehensive pipeline for constructing tumor-associated stroma datasets across multiple data sources, and employed adversarial training and neighborhood consistency regularization techniques to learn robust multimodal-invariant image representations.
2 Method
2.1 Stroma Tissue Segmentation
Accurately analyzing tumor-associated stroma requires a critical pre-processing step of segmenting stromal tissue from the background, including epithelial tissue. This segmentation task is challenging due to the complex and heterogeneous appearance of the stroma. To address this, we propose utilizing the PointRend model [17], which can handle complex shapes and appearances and produce smooth and accurate segmentations through iterative object boundary refinement. Moreover, the model’s efficiency and ability to process large images quickly make it suitable for analyzing whole-mount slides. By leveraging the PointRend model, we can generate stromal segmentation masks for more precise downstream analysis.
Process of stroma patch graph construction: The left prostate model illustrates the locations of biopsy and whole-mount tissues in 3D space. Biopsy slides provide a targeted view while whole-mount slides offer a broader perspective of the tumor and surrounding tissue. The stroma segmentation module generates a stroma mask to isolate the stromal tissue, which is then used to construct spatial patch graphs for the proposed deep-learning model.
Overview of the proposed model for identifying tumor-associated stroma in multi-modal prostate histopathology slides: The input patches are represented as spatial graphs and passed through a feature extractor. The patch embeddings are fed into a graph attention network (GAT) module to capture inter-patch relationships and refine the features with neighborhood consistency regularization (NCR) for handling noisy labels. The source discriminator serves as adversarial multi-modal learning (AML) module to predict data source (biopsy/whole-mount). The stroma classifier and source discriminator are trained simultaneously with the goal of successfully classifying tumor-associated stroma using multimodal-invariant features.
2.2 Stroma Classification with Spatial Patch Graphs
To capture the spatial nature of field effect and analyze tumor-associated stroma, modeling spatial relationships between stroma patches is essential. The spatial relationship can reveal valuable information about the tumor microenvironment, and neighboring stroma cells can undergo similar phenotypic changes in response to cancer. Therefore, we propose using a spatial patch graph to capture the high-order relationship among stroma tissue regions. We construct the stroma patch graph using a K-nearest neighbor (KNN) graph and neighbor sampling. The KNN graph connects each stroma patch to its K nearest neighboring patches. Given a central stroma patch, we iteratively add neighboring patches to construct the patch graph until we reach a specified layer number L to control the subgraph size. This process results in a tree-like subgraph with each layer representing a different level of spatial proximity to the central patch. The use of neighbor sampling enables efficient processing of large images and allows for stochastic training of the model.
To predict tumor-associated binary labels of stroma patches, we employ a message-passing approach that propagates patch features in the spatial graph. To achieve this, we use Graph Convolutional Networks with attention, also known as Graph Attention Networks (GATs) [18]. The GAT uses an attention mechanism on node features to construct a weighting kernel that determines the importance of nodes in the message-passing process. In our case, the patch graph \(\mathcal {G}\) is constructed using the stroma patches as vertices, and we connect the nodes with edges based on their spatial proximity. Each vertex \(v_{i}\) is associated with a feature vector \(\vec {h}_{v{i}} \in \mathbb {R}^{N}\), which is first extracted by Resnet-50 model [19]. The GAT layer is defined as
where \(W \in \mathbb {R}^{M \times N}\) is a learnable matrix transforming N-dimensional features to M-dimensional features. \(\mathcal {N}^{\mathcal {E}}_{v_{i}}\) is the neighborhood of the node \(v_{i}\) connected by \(\mathcal {E}\) in \(\mathcal {G}\). GAT uses attention mechanism to construct the weighting coefficients as:
where T represents transposition, \(\Vert \) is the concatenation operation, and \(\rho \) is \(\text {LeakyReLU}\) function. The final output of GAT module is the tumor-associated probability of the input patch. And the module was optimized using the cross-entropy loss \(L_{\textrm{GAT}}\) in an end-to-end fashion.
2.3 Neighbor Consistency Regularization for Noisy Labels
The labeling of tumor-associated stroma can be affected by various factors, which can result in noisy labels. One of the reasons for noisy labels is the irregular distribution of the field effect, which makes it challenging to define a clear boundary between the tumor-associated and normal stroma regions. Additionally, the presence of tumor heterogeneity and the varied distribution of tumor foci can further complicate the labeling process.
To address this issue, we propose applying Neighbor Consistency Regularization (NCR) [20] to prevent the model from overfitting to incorrect labels. The assumption is that overfitting happens to a lesser degree before the final classifier, and this is supported by MOIT [21], which suggests that feature representations are capable of distinguishing between noisy and clean examples during model training. Based on this assumption, NCR introduces a neighbor consistency loss to encourage similar predictions of stroma patches that are similar in feature space. This loss penalizes the divergence of a patch prediction from a weighted combination of its neighbors’ predictions in feature space, where the weights are determined by their feature similarity. Specifically, the loss function is designed as follows:
where \(D_{\textrm{KL}}\) is the KL-divergence loss to quantify the discrepancy between two probability distributions, T represents the temperature and \(\textrm{NN}_k\left( \textbf{v}_i\right) \) is the set of k nearest neighbors of \({v}_i\) in the feature space.
2.4 Adversarial Multi-modal Learning
Biopsy and whole-mount slides provide complementary multi-modal information on the tumor microenvironment, and combining them can provide a more comprehensive understanding of tumor-associated stroma. However, using data from multiple modalities can introduce systematic shifts, which can impact the performance of a deep learning model. Specifically, whole-mount slides typically contain larger tissue sections and are processed using different protocols than biopsy slides, which can result in differences in image quality, brightness, and contrast. These technical differences can affect the pixel intensity distributions of the images, leading to systematic shifts in the features that the deep learning model learns to associate with tumor-associated stroma. For instance, a model trained on whole-mount slides only may not generalize well to biopsy slides due to systematic shifts, hindering model performance in the clinical application scenario.
To address the above issues, we propose an Adversarial Multi-modal Learning (AML) module to force the feature extractor to produce multimodal-invariant representations on multiple source images. Specifically, we incorporate a source discriminator adversarial neural network as auxiliary classifier. The module takes the stroma embedding as an input and predicts the source of the image (biopsy or whole-mount) using Multilayer Perceptron (MLP) with cross-entropy loss function \(L_{\textrm{AML}}\). The overall loss function of the entire model is computed as:
where hyper-parameters \(\alpha \) and \(\beta \) control the impact of each loss term. All modules were concurrently optimized in an end-to-end manner. The stroma classifier and source discriminator are trained simultaneously, aiming to effectively classify tumor-associated stroma while impeding accurate source prediction by the discriminator. The optimization process aims to achieve a balance between these two goals, resulting in an embedding space that encodes as much information as possible about tumor-associated stroma identification while not encoding any information on the data source. By adopting the adversarial learning strategy, our model can maintain the correlated information and shared characteristics between two modalities, which will enhance the model’s generalization and robustness.
3 Experiment
3.1 Dataset
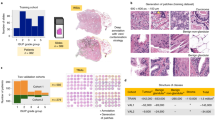
In our study, we utilized three datasets for tumor-associated stroma analysis. (1) Dataset A comprises 513 tiles extracted from the whole mount slides of 40 patients, sourced from the archives of the Pathology Department at Cedars-Sinai Medical Center (IRB# Pro00029960). It combines two sets of tiles: 224 images from 20 patients featuring stroma, normal glands, low-grade and high-grade cancer [22], along with 289 images from 20 patients with dense high-grade cancer (Gleason grades 4 and 5) and cribriform/non-cribriform glands [23]. Each tile measures 1200\(\times \)1200 pixels and is extracted from whole slide images captured at 20x magnification (0.5 microns per pixel). The tiles were annotated at the pixel-level by expert pathologists to generate stroma tissue segmentation masks and were cross-evaluated and normalized to account for stain variability. (2) Dataset B included 97 whole mount slides with an average size of over 174,000\(\times \)142,000 pixels at 40x magnification. The prostate tissue within these slides had an average tumor area proportion of 9%, with an average tumor area of 77 square mm. An expert pathologist annotated the tumor region boundaries at the region-level, providing exhaustive annotations for all tumor foci. (3) Dataset C comprised 6134 negative biopsy slides obtained from 262 patients’ biopsy procedures, where all samples were diagnosed as negative. These slides are presumed to contain predominantly normal stroma tissues without phenotypic alterations in response to cancer.
Dataset A was utilized for training the stroma segmentation model. Extensive data augmentation techniques, such as image scaling and staining perturbation, were employed during the training process. The model achieved an average test Dice score of \(95.57 \pm 0.29\) through 5-fold cross-validation. This model was then applied to generate stroma masks for all slides in Datasets B and C. To precisely isolate stroma tissues and avoid data bleeding from epithelial tissues, we only extracted patches where over 99.5% of the regions were identified as stroma at 40X magnification to construct the stroma classification dataset.
For positive tumor-associated stroma patches, we sampled patches near tumor glands within annotated tumor region boundaries, as we presumed that tumor regions represent zones in which the greatest amount of damage has progressed. For negative stroma patches, we calculated the tumor distance for each patch by measuring the Euclidean distance from the patch center to the nearest edge of the labeled tumor regions. Negative stroma patches were then sampled from whole mount slides with a Gleason Group smaller than 3 and a tumor distance larger than 5 mm. This approach aims to minimize the risk of mislabeling tumor-associated stroma as normal tissue. Setting a 5mm threshold accounts for the typically minimal inflammatory responses induced by prostate cancers, particularly in lower-grade cases. To incorporate multi-modal information, we randomly sampled negative stroma patches from all biopsy slides in Dataset C. Overall, we selected over 1.1 million stroma patches of size 256\(\times \)256 pixels at 40x magnification for experiments. During model training and testing, we performed stain normalization and standard image augmentation methods.
3.2 Model Training and Evaluation
For constructing KNN-based patch graphs, we limited the graph size by setting \(K=4\) and layer number \(L=3\). We controlled the strength of the NCR and AML terms by setting \(\alpha =0.25\) and \(\beta =0.5\), respectively. The Adam optimizer with a learning rate of 0.0005 was used for model training. All models were implemented using PyTorch on a single Tesla V100 GPU. To evaluate the model performance, we perform 5-fold cross-validation, where all slides are stratified by source origin and divided into 5 subsets. In each cross-validation trial, one subset was taken as the test set while the remaining subsets constituted the training set. We measure the prediction performance using the area under the receiver operating characteristic (AUROC), F1 score, precision, and recall.
4 Results and Discussions
To evaluate the effectiveness of our proposed method, we conducted an ablation study by comparing the performance of different model variants presented in Table 1. Specifically, the base model is the ResNet-50 feature extractor for tumor-associated stroma classification. Each model variant included a different combination of modules presented in method sections. We systematically add one or more modules to the base model to evaluate their performance contribution. The results show that the full model outperforms the base model by a large margin with \(10.04\%\) in AUROC and \(10.97\%\) in F1 score, and each module contributes to the overall performance. Compared to the base model, the addition of the GAT module resulted in a significant improvement in all metrics, suggesting spatial information captured by the patch graph was valuable for stroma classification. The most notable performance improvement was achieved by the AML module, with a \(5.72\%\) increase in AUROC and \(5.55\%\) increase in Recall. This improvement indicates that AML helps the model better capture the multimodal-invariant features that are associated with tumor-associated stroma while reducing the false negative prediction by eliminating the influence of systematic shift cross modalities. Finally, the addition of the NCR module further increased the average model performance and improved the model robustness across 5 folds. This suggests that NCR was effective in handling noisy labels and improving model’s generalization ability.
In conclusion, our study introduced a deep learning approach to accurately characterize the tumor-associated stroma in multi-modal prostate histopathology slides. Our experimental results demonstrate the feasibility of using deep learning algorithms to identify and quantify subtle stromal alterations, offering a promising tool for discovering new diagnostic and prognostic biomarkers of prostate cancer. Through exploring field effect in prostate cancer, our work provides a computational system for further analysis of tumor development and progression. Future research can focus on validating our approach on larger and more diverse datasets and expanding the method to a patient-level prediction system, ultimately improving prostate cancer diagnosis and treatment.
References
Merriel, S.W.D., Funston, G., Hamilton, W.: Prostate cancer in primary care. Adv. Therapy 35, 1285–1294 (2018)
Montironi, R., Beltran, A.L., Mazzucchelli, R., Cheng, L., Scarpelli, M.: Handling of radical prostatectomy specimens: total embedding with large-format histology. Inter. J. Breast Cancer, vol. 2012 (2012)
Han, W., et al.: Histologic tissue components provide major cues for machine learning-based prostate cancer detection and grading on prostatectomy specimens. Sci. Rep. 10(1), 9911 (2020)
Kirby, R.S., Patel, M.I., Poon, D. M.C.: Fast facts: prostate cancer: If, when and how to intervene. Karger Medical and Scientific Publishers (2020)
Nayyar, R., et al.: Upgrading of gleason score on radical prostatectomy specimen compared to the pre-operative needle core biopsy: an indian experience. Indian J. Urology: IJU: J. Urological Soc. India 26(1), 56 (2010)
Jang, W.S., et al.: The prognostic impact of downgrading and upgrading from biopsy to radical prostatectomy among men with gleason score 7 prostate cancer. Prostate 79(16), 1805–1810 (2019)
Bonollo, F., Thalmann, G.N., Julio, M.K.-d, Karkampouna, S.: The role of cancer-associated fibroblasts in prostate cancer tumorigenesis. Cancers 12(7):1887, 2020
Barron, D.A., Rowley, D.R.: The reactive stroma microenvironment and prostate cancer progression. Endocr. Relat. Cancer 19(6), R187–R204 (2012)
Levesque, C., Nelson, P.S.: Cellular constituents of the prostate stroma: key contributors to prostate cancer progression and therapy resistance. Cold Spring Harbor Perspect. Med. 8(8), a030510 (2018)
Liao, Z., Chua, D., Tan, N.S.: Reactive oxygen species: a volatile driver of field cancerization and metastasis. Mol. Cancer 18, 1–10 (2019)
Hayward, S.W., et al.: Malignant transformation in a nontumorigenic human prostatic epithelial cell line. Cancer Res. 61(22), 8135–8142 (2001)
Vivar, A.D.D., et al.: Histologic features of stromogenic carcinoma of the prostate (carcinomas with reactive stroma grade 3). Hum. Pathol. 63, 202–211 (2017)
Bejnordi, B.E., et al.: Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 31(10), 1502–1512 (2018)
Zheng, Q., et al.: Machine learning quantified tumor-stroma ratio is an independent prognosticator in muscle-invasive bladder cancer. Int. J. Mol. Sci. 24(3), 2746 (2023)
Millar, E.K.A., et al.: Tumour stroma ratio assessment using digital image analysis predicts survival in triple negative and luminal breast cancer. Cancers 12(12), 3749 (2020)
Ruder, S., et al.: Development and validation of a quantitative reactive stroma biomarker (qrs) for prostate cancer prognosis. Hum. Pathol. 122, 84–91 (2022)
Kirillov, A., Wu, Y., He, K., Girshick, R.: Pointrend: image segmentation as rendering. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9799–9808, 2020
Veličković, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., Bengio, Y.: Graph attention networks. arXiv preprint arXiv:1710.10903 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Iscen, A., Valmadre, J., Arnab, A., Schmid, C.: Learning with neighbor consistency for noisy labels. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4672–4681 (2022)
Ortego, D., Arazo, E., Albert, P., O’Connor, N.E., McGuinness, K.: Multi-objective interpolation training for robustness to label noise. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 6606–6615 (2021)
Gertych, A., et al.: Machine learning approaches to analyze histological images of tissues from radical prostatectomies. Comput. Med. Imaging Graph. 46, 197–208 (2015)
Ing, N., et al.: Semantic segmentation for prostate cancer grading by convolutional neural networks. In: Medical Imaging 2018: Digital Pathology, vol. 10581, pp. 343–355. SPIE (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, Z. et al. (2023). Deep Learning for Tumor-Associated Stroma Identification in Prostate Histopathology Slides. In: Greenspan, H., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. Lecture Notes in Computer Science, vol 14225. Springer, Cham. https://doi.org/10.1007/978-3-031-43987-2_62
Download citation
DOI: https://doi.org/10.1007/978-3-031-43987-2_62
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43986-5
Online ISBN: 978-3-031-43987-2
eBook Packages: Computer ScienceComputer Science (R0)