Abstract
Self-supervised learning (SSL) has led to important breakthroughs in computer vision by allowing learning from large amounts of unlabeled data. As such, it might have a pivotal role to play in biomedicine where annotating data requires a highly specialized expertise. Yet, there are many healthcare domains for which SSL has not been extensively explored. One such domain is endoscopy, minimally invasive procedures which are commonly used to detect and treat infections, chronic inflammatory diseases or cancer. In this work, we study the use of a leading SSL framework, namely Masked Siamese Networks (MSNs), for endoscopic video analysis such as colonoscopy and laparoscopy. To fully exploit the power of SSL, we create sizable unlabeled endoscopic video datasets for training MSNs. These strong image representations serve as a foundation for secondary training with limited annotated datasets, resulting in state-of-the-art performance in endoscopic benchmarks like surgical phase recognition during laparoscopy and colonoscopic polyp characterization. Additionally, we achieve a 50% reduction in annotated data size without sacrificing performance. Thus, our work provides evidence that SSL can dramatically reduce the need of annotated data in endoscopy.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Endoscopic operations are minimally invasive medical procedures which allow physicians to examine inner body organs and cavities. During an endoscopy, a thin, flexible tube with a tiny camera is inserted into the body through a small orifice or incision. It is used to diagnose and treat a variety of conditions, including ulcers, polyps, tumors, and inflammation. Over 250 million endoscopic procedures are performed each year globally and 80 million in the United States, signifying the crucial role of endoscopy in clinical research and care.
A cardinal challenge in performing endoscopy is the limited field of view which hinders navigation and proper visual assessment, potentially leading to high detection miss-rate, incorrect diagnosis or insufficient treatment. These limitations have fostered the development of computer-aided systems based on artificial intelligence (AI), resulting in unprecedented performance over a broad range of clinical applications [10, 11, 17, 23,24,25]. Yet the success of such AI systems heavily relies on acquiring annotated data which requires experts of specific knowledge, leading to an expensive, prolonged process. In the last few years, Self-Supervised Learning (SSL [5,6,7,8]) has been shown to be a revolutionary strategy for unsupervised representation learning, eliminating the need to manually annotate vast quantities of data. Training large models on sizable unlabeled data via SSL leads to powerful representations which are effective for downstream tasks with few labels. However, research in endoscopic video analysis has only scratched the surface of SSL which remains largely unexplored.
This study introduces Masked Siamese Networks (MSNs [2]), a prominent SSL framework, into endoscopic video analysis where we focus on laparoscopy and colonoscopy. We first experiment solely on public datasets, Cholec80 [32] and PolypsSet [33], demonstrating performance on-par with the top results reported in the literature. Yet, the power of SSL lies in large data regimes. Therefore, to exploit MSNs to their full extent, we collect and build two sizable unlabeled datasets for laparoscopy and colonoscopy with 7, 700 videos (>23M frames) and 14, 000 videos (>2M frames) respectively. Through extensive experiments, we find that scaling the data size necessitates scaling the model architecture, leading to state-of-the-art performance in surgical phase recognition of laparoscopic procedures, as well as in polyp characterization of colonoscopic videos. Furthermore, the proposed approach exhibits robust generalization, yielding better performance with only 50% of the annotated data, compared with standard supervised learning using the complete labeled dataset. This shows the potential to reduce significantly the need for expensive annotated medical data.
2 Background and Related Work
There exist a wide variety of endoscopic applications. Here, we focus on colonoscopy and laparoscopy, which combined covers over 70% of all endoscopic procedures. Specifically, our study addresses two important common tasks, described below.
Cholecystectomy Phase Recognition. Cholecystectomy is the surgical removal of the gallbladder using small incisions and specialized instruments. It is a common procedure performed to treat gallstones, inflammation, or other conditions affecting the gallbladder. Phase recognition in surgical videos is an important task that aims to improve surgical workflow and efficiency. Apart from measuring quality and monitoring adverse event, this task also serves in facilitating education, statistical analysis, and evaluating surgical performance. Furthermore, the ability to recognize phases allows real-time monitoring and decision-making assistance during surgery, thus improving patient safety and outcomes. AI solutions have shown remarkable performance in recognizing surgical phases of cholecystectomy procedures [17, 18, 32]; however, they typically require large labelled training datasets. As an alternative, SSL methods have been developed [12, 28, 30], however, these are early-days methods that based on heuristic, often require external information and leads to sub-optimal performance. A recent work [27] presented an extensive analysis of modern SSL techniques for surgical computer vision, yet on relatively small laparoscopic datasets.
Optical Polyp Characterization. Colorectal cancer (CRC) remains a critical health concern and significant financial burden worldwide. Optical colonoscopy is the standard of care screening procedure for preventing CRC through the identification and removal of polyps [3]. According to colonoscopy guidelines, all identified polyps must be removed and histologically evaluated regardless of their malignant nature. Optical biopsy enables practitioners to remove pre-cancerous adenoma polyps or leave distal hyperplastic polyps in situ without the need for pathology examination, by visually predicting histology. However, this technique is highly dependent on operator expertise [14]. This limitation has motivated the development of AI systems for automatic optical biopsy, allowing non-experts to also effectively perform optical biopsy during polyp management. In recent years, various AI systems have been developed to this end [1, 19]. However, training such automatic optical biopsy systems relies on a large body of annotated data, while SSL has not been investigated in this context, to the best of our knowledge.
3 Self-supervised Learning for Endoscopy
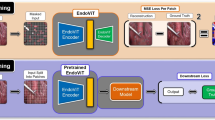
SSL approaches have produced impressive results recently [5,6,7,8], relying on two key factors: (i) effective algorithms for unsupervised learning and (ii) training on large-scale datasets. Here, we first describe Masked Siamese Networks [2], our chosen SSL framework. Additionally, we present our large-scale data collection (see Fig. 2). Through extensive experiments in Sect. 4, we show that training MSNs on these substantial datasets unlocks their potential, yielding effective representations that transfer well to public laparoscopy and colonoscopy datasets.
3.1 Masked Siamese Networks
SSL has become an active research area, giving rise to efficient learning methods such as SimCLR [7], SwAV [5] and DINO [6]. Recently, Masked Siamese Networks [2] have set a new state-of-the-art among SSL methods on the ImageNet benchmark [29], with a particular focus on the low data regime. This is of great interest for us since our downstream datasets are typically of small size [32, 33]. We briefly describe MSNs below and refer the reader to [2] for further details.
During pretraining, on each image \({\textbf{x}}_i\in {\mathbb {R}}^n\) of a mini-batch of \(B\ge 1\) samples (e.g. laparoscopic images) we apply two sets of random augmentations to generate anchor and target views, denoted by \({\textbf{x}}_i^a\) and \({\textbf{x}}_i^t\) respectively. We convert each view into a sequence of non-overlapping patches and perform an additional masking (“random” or “focal” styles) step on the anchor view by randomly discarding some of its patches. The resultant anchor and target sequences are used as inputs to their respective image encoders \(f_{\theta ^a}\) and \(f_{\theta ^t}\). Both encoders share the same Vision Transformer (ViT [16]) architecture where the parameters \(\theta ^t\) of the target encoder are updated via an exponential moving average of the anchor encoder parameters \(\theta ^a\). The outputs of the networks are the representation vectors \({\textbf{z}}_i^a\in {\mathbb {R}}^d\) and \({\textbf{z}}_i^t\in {\mathbb {R}}^d\), corresponding to the [CLS] tokens of the networks. The similarity between each view and a series of \(K>1\) learnable prototypes is then computed, and the results undergo a softmax operation to yield the following probabilities \({\textbf{p}}_i^a= softmax\left( \frac{{\textbf{Q}}{\textbf{z}}_i^a}{\tau ^a}\right) \) and \({\textbf{p}}_i^t= softmax\left( \frac{{\textbf{Q}}{\textbf{z}}_i^t}{\tau ^t}\right) \) where \(0<\tau ^t<\tau ^a<1\) are temperatures and \({\textbf{Q}}\in {\mathbb {R}}^{K\times d}\) is a matrix whose rows are the prototypes. The probabilities are promoted to be the same by minimizing the cross-entropy loss \(H(p_i^t,p_i^a)\), as illustrated in Fig. 1.
In practice, a sequence of \(M\ge 1\) anchor views are generated, leading to multiple probabilities \(\{{\textbf{p}}_{i,m}^a\}_{m=1}^M\). Furthermore, to prevent representation collapse and encourage the model to fully exploit the prototypes, a mean entropy maximization (me-max) regularizer [2, 22] is added, aiming to maximize the entropy \(H(\bar{\textbf{p}}^a)\) of the average prediction across all the anchor views \(\bar{\textbf{p}}^a\triangleq \frac{1}{MB}\sum _{i=1}^B\sum _{m=1}^M{\textbf{p}}^a_{i,m}\). Thus, the overall training objective to be minimized for both \(\theta ^a\) and \({\textbf{Q}}\) is where \(\lambda >0\) is an hyperparameter and the gradients are computed only with respect to the anchor predictions \({\textbf{p}}^a_{i,m}\) (not the target predictions \({\textbf{p}}_i^t\)). Applying MSNs on the large datasets described below, generates representations that serve as a strong basis for various downstream tasks, as shown in the next section.
3.2 Private Datasets
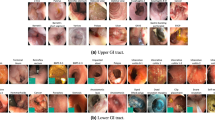
Laparoscopy. We compiled a dataset of laparoscopic procedures videos exclusively performed on patients aged 18 years or older. The dataset consists of 7,877 videos recorded at eight different medical centers in Israel. The dataset predominantly consists of the following procedures: cholecystectomy (35%), appendectomy (20%), herniorrhaphy (12%), colectomy (6%), and bariatric surgery (5%). The remaining 21% of the dataset encompasses various standard laparoscopic operations. The recorded procedures have an average duration of 47 min, with a median duration of 40 min. Each video recording was sampled at a rate of 1 frame per second (FPS), resulting in an extensive dataset containing 23.3 million images. Further details are given in the supplementary materials.
Colonoscopy. We have curated a dataset comprising 13,979 colonoscopy videos of patients aged 18 years or older. These videos were recorded during standard colonoscopy procedures performed at six different medical centers between the years 2019 and 2022. The average duration of the recorded procedures is 15 min, with a median duration of 13 min. To identify and extract polyps from the videos, we employed a pretrained polyp detection model [21, 25, 26]. Using this model, we obtained bounding boxes around the detected polyps. To ensure high-quality data, we filtered out detections with confidence scores below 0.5. For each frame, we cropped the bounding boxes to generate individual images of the polyps. This process resulted in a comprehensive collection of 2.2 million polyp images.
4 Experiments
In this section, we empirically demonstrate the power of SSL in the context of endoscopy. Our experimental protocol is the following: (i) first, we perform SSL pretraining with MSNs over our unlabeled private dataset to learn informative and generic representations, (ii) second we probe these representations by utilizing them for different public downstream tasks. Specifically, we use the following two benchmarks. (a) Cholec80 [32]: 80 videos of cholecystectomy procedures resulting in nearly 200k frames at 1 FPS. Senior surgeons annotated each frame to one out of seven phases. (b) PolypsSet [33]: A unified dataset of 155 colonoscopy videos (37,899 frames) with labeled polyp classes (hyperplastic or adenoma) and bounding boxes. We use the provided detections to perform binary classification.
Downstream Task Evaluation Protocols. (a) Linear evaluation: A standard protocol consisting in learning a linear classifier on top of frozen SSL features [6, 20]. (b) Temporal evaluation: A natural extension of the linear protocol where we learn a temporal model on top of the frame-level frozen features. We specifically use Multi-Stage Temporal Convolution Networks (MS-TCN) as used in [13, 27]. This incorporates the temporal context which is crucial for video tasks such as phases recognition. (c) Fine-tuning: An end-to-end training of a classification head on top of the (unfrozen) pretrained backbone. We perform an extensive hyperparameter grid search for all downstream experiments and report the test results for the models that exceed the best validation results. We report the Macro F1 (F-F1) as our primary metric. For phase recognition we also report the per-video F1 (V-F1), computed by averaging the F1 scores across all videos [27].
Implementation Details. For SSL we re-implemented MSNs in JAX using Scenic library [15]. As our image encoders we train Vision Transformer (ViT [16]) of different sizes, abbreviated as ViT-S/B/L, using 16 TPUs. Downstream experiments are implemented in TensorFlow where training is performed on 4 Nvidia Tesla V100 GPUs. See the supplementary for further implementation details.Footnote 1
4.1 Results and Discussion
Scaling Laws of SSL. We explore large scale SSL pretraining for endoscopy videos. Table 1 compares the results of pretraining with different datasets (public and private) and model sizes. We pretrain the models with MSN and then report their downstream performances. We present results for the cholecystectomy phase recognition task based on fine-tuned models and for the optical polyp characterization task based on linear evaluation, due to the small size of the public dataset. As baselines, we report fully-supervised ResNet50 results, trained on public datasets. We find that replacing ResNet50 with ViT-S, despite comparable number of parameters, yields sub-optimal performance.
SSL pretraining on public datasets (without labels) provides comparable or better results than fully supervised baselines. The performance in per-frame phase recognition is comparable with the baseline. Phase recognition per-video results improve by 1.3 points when using the MSN pretraining, while polyp characterization improve by 2.2 points. Importantly, we see that the performance gap becomes prominent when using the large scale private datasets for SSL pretraining. Here, per-frame and per-video phase recognition performances improve by \(6.7\%\) and \(8.2\%\), respectively. When using the private colonoscopy dataset the Macro F1 improves by \(11.5\%\) compared to the fully supervised baseline. Notice that the performance improves with scaling both model and private data sizes, demonstrating that both factors are crucial to achieve optimal performance.
Low-Shot Regime. Next, we examine the benefits of using MSNs to improve downstream performance in a low-shot regime with few annotated samples. Note that MSNs have originally been found to produce excellent features for low data regime [2]. We train a linear classifier on top of the extracted features and report the test classification results. Figure 3 shows the low-shot performance for the two endoscopic tasks. We report results using a fraction \(k=\{12\%, 25\%, 50\%, 75\%, 100\%\}\) of the annotated public videos. We also report results for fully-supervised baselines trained on the same fraction of annotated samples. Each experiment is repeated three times with a random sample of train videos, and we report the mean and standard deviation (shaded area).
As seen, SSL-based models provide enhanced robustness to limited annotations. When examining the cholecystectomy phase recognition task, it is evident that we can achieve comparable frame-level performance by using only \(12\%\) of the annotated videos. Using \(25\%\) of the annotated videos yields comparable results to the fully supervised temporal models. Optical polyp characterization results show a similar trend, but with a greater degree of variability. Using small portions of PolypSet (12% and 25%) hindered the training process and increased sensitivity to the selected portions. However, when using more than 50% of PolypSet, the training process stabilized, yielding results comparable to the fully supervised baseline. This feature is crucial for medical applications, given the time and cost involved in expert-led annotation processes.
4.2 Ablation Study
Table 2 details different design choices regarding our SSL pretraining. Ablations are done on ViT-S trained over the public Cholec80. We report results on the validation set after linear evaluation. In Table 2a), we see that the method is robust to the number of prototypes, though over-clustering [4] with 1k prototypes is optimal. In Table 2b) and Table 2c), we explore the effect of random and focal masking. We see that 50% random masking (i.e. we keep 98 tokens out of 196 for the global view) and using 4 local views gives the best of performance. In Table 2d) we study the effect of data augmentation. SSL augmentation pipelines have been developed on ImageNet-1k [7], hence, it is important to re-evaluate these choices for medical images. Surprisingly, we see that augmentations primarily found to work well on ImageNet-1k are also effective on laparoscopic videos (e.g. color jiterring and horizontal flips). In Table2e), we look at the effect of the training length when starting from scratch or from a good SSL pretrained checkpoint on ImageNet-1k. We observe that excellent performance is achieved with only 10 epochs of finetuning on medical data when starting from a strong DINO checkpoint [6]. Table 2g) shows that ImageNet-1k DINO is a solid starting point compared to other alternatives [9, 20, 31, 34]. Finally, Table2f) confirms the necessity of regularizing with Sinkhorn-Knopp and me-max to avoid representation collapse by encouraging the use of all prototypes.
5 Conclusion
This study showcases the use of Masked Siamese Networks to learn informative representations from large, unlabeled endoscopic datasets. The learnt representations lead to state-of-the-art results in identifying surgical phases of laparoscopic procedures and in optical characterization of colorectal polyps. Moreover, this methodology displays strong generalization, achieving comparable performance with just 50% of labeled data compared to standard supervised training on the complete labeled datasets. This dramatically reduces the need for annotated medical data, thereby facilitating the development of AI methods for healthcare.
Notes
- 1.
For reproducibility purposes, code and model checkpoints are available at https://github.com/RoyHirsch/endossl.
References
Antonelli, G., Rizkala, T., Iacopini, F., Hassan, C.: Current and future implications of artificial intelligence in colonoscopy. Ann. Gastroenterol. 36(2), 114–122 (2023)
Assran, M., et al.: Masked siamese networks for label-efficient learning. In: ECCV (2022). https://doi.org/10.1007/978-3-031-19821-2_26
Byrne, M.F., Shahidi, N., Rex, D.K.: Will computer-aided detection and diagnosis revolutionize colonoscopy? Gastroenterology 153(6), 1460–1464 (2017)
Caron, M., Bojanowski, P., Joulin, A., Douze, M.: Deep clustering for unsupervised learning of visual features. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) Computer Vision – ECCV 2018. LNCS, vol. 11218, pp. 139–156. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01264-9_9
Caron, M., Misra, I., Mairal, J., Goyal, P., Bojanowski, P., Joulin, A.: Unsupervised learning of visual features by contrasting cluster assignments. In: NeurIPS (2020)
Caron, M., et al.: Emerging properties in self-supervised vision transformers. In: ICCV (2021)
Chen, T., Kornblith, S., Norouzi, M., Hinton, G.: A simple framework for contrastive learning of visual representations. In: ICML (2020)
Chen, X., He, K.: Exploring simple siamese representation learning. In: CVPR (2021)
Chen, X., Xie, S., He, K.: An empirical study of training self-supervised vision transformers. In: ICCV (2021)
Cohen, R., Blau, Y., Freedman, D., Rivlin, E.: It has potential: gradient-driven denoisers for convergent solutions to inverse problems. Adv. Neural. Inf. Process. Syst. 34, 18152–18164 (2021)
Cohen, R., Elad, M., Milanfar, P.: Regularization by denoising via fixed-point projection (RED-PRO). SIAM J. Imag. Sci. 14(3), 1374–1406 (2021)
da Costa Rocha, C., Padoy, N., Rosa, B.: Self-supervised surgical tool segmentation using kinematic information. In: 2019 International Conference on Robotics and Automation (ICRA), pp. 8720–8726. IEEE (2019)
Czempiel, T., et al.: TeCNO: surgical phase recognition with multi-stage temporal convolutional networks. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12263, pp. 343–352. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59716-0_33
Dayyeh, B.K.A., et al.: Asge technology committee systematic review and meta-analysis assessing the asge pivi thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest. Endosc. 81(3), 502.e1–502.e16 (2015)
Dehghani, M., Gritsenko, A., Arnab, A., Minderer, M., Tay, Y.: Scenic: a jax library for computer vision research and beyond. In: CVPR, pp. 21393–21398 (2022)
Dosovitskiy, A., et al.: An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
Golany, T., et al.: AI for phase recognition in complex laparoscopic cholecystectomy. Surgical Endoscopy, 1–9 (2022)
Goldbraikh, A., Avisdris, N., Pugh, C.M., Laufer, S.: Bounded future MS-TCN++ for surgical gesture recognition. In: ECCV 2022 Workshops, October 23–27, 2022, Proceedings, Part III, pp. 406–421. Springer (2023). https://doi.org/10.1007/978-3-031-25066-8_22
Hassan, C., et al.: Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest. Endosc. 93(1), 77–85 (2021)
He, K., Chen, X., Xie, S., Li, Y., Dollár, P., Girshick, R.: Masked autoencoders are scalable vision learners. In: CVPR (2022)
Intrator, Y., Aizenberg, N., Livne, A., Rivlin, E., Goldenberg, R.: Self-supervised polyp re-identification in colonoscopy. arXiv preprint arXiv:2306.08591 (2023)
Joulin, A., Bach, F.: A convex relaxation for weakly supervised classifiers. arXiv preprint arXiv:1206.6413 (2012)
Katzir, L., et al.: Estimating withdrawal time in colonoscopies. In: ECCV, pp. 495–512. Springer (2022). https://doi.org/10.1007/978-3-031-25066-8_28
Kutiel, G., Cohen, R., Elad, M., Freedman, D., Rivlin, E.: Conformal prediction masks: visualizing uncertainty in medical imaging. In: ICLR 2023 Workshop on Trustworthy Machine Learning for Healthcare (2023)
Livovsky, D.M., et al.: Detection of elusive polyps using a large-scale artificial intelligence system (with videos). Gastrointest. Endosc. 94(6), 1099–1109 (2021)
Ou, S., Gao, Y., Zhang, Z., Shi, C.: Polyp-YOLOv5-Tiny: a lightweight model for real-time polyp detection. In: International Conference on Information Technology, Big Data and Artificial Intelligence (ICIBA), vol. 2, pp. 1106–1111 (2021
Ramesh, S., et al.: Dissecting self-supervised learning methods for surgical computer vision. Med. Image Anal. 88, 102844 (2023)
Ross, T., et al.: Exploiting the potential of unlabeled endoscopic video data with self-supervised learning. Int. J. Comput. Assist. Radiol. Surg. 13, 925–933 (2018)
Russakovsky, O., et al.: Imagenet large scale visual recognition challenge. In: IJCV (2015)
Sestini, L., Rosa, B., De Momi, E., Ferrigno, G., Padoy, N.: A kinematic bottleneck approach for pose regression of flexible surgical instruments directly from images. IEEE Robotics Autom. Lett. 6(2), 2938–2945 (2021)
Touvron, H., Cord, M., Jégou, H.: DeIT III: Revenge of the ViT. arXiv preprint arXiv:2204.07118 (2022)
Twinanda, A.P., Shehata, S., Mutter, D., Marescaux, J., De Mathelin, M., Padoy, N.: Endonet: a deep architecture for recognition tasks on laparoscopic videos. IEEE Trans. Med. Imaging 36(1), 86–97 (2016)
Wang, G.: Replication data for: colonoscopy polyp detection and classification: dataset creation and comparative evaluations. Harvard Dataverse (2021). https://doi.org/10.7910/DVN/FCBUOR
Zhou, J., et al.: ibot: image bert pre-training with online tokenizer. arXiv preprint arXiv:2111.07832 (2021)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Hirsch, R. et al. (2023). Self-supervised Learning for Endoscopic Video Analysis. In: Greenspan, H., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. Lecture Notes in Computer Science, vol 14224. Springer, Cham. https://doi.org/10.1007/978-3-031-43904-9_55
Download citation
DOI: https://doi.org/10.1007/978-3-031-43904-9_55
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43903-2
Online ISBN: 978-3-031-43904-9
eBook Packages: Computer ScienceComputer Science (R0)