Abstract
Recent developments in the context of semantic technologies have given rise to ontologies for modelling scientific information in various fields of science. Over the past years, we have been engaged in the development of the Science Knowledge Graph Ontologies (SKGO), a set of ontologies for modelling research findings in various fields of science. This paper introduces the Modern Science Ontology (ModSci), an upper ontology for modelling relationships between modern science branches and related entities, including scientific discoveries, phenomena, prominent scientists, instruments, etc. ModSci provides a unifying framework for the various domain ontologies that make up the Science Knowledge Graph Ontology suite. Well-known ontology development guidelines and principles have been followed in the development and publication of the resource. We present several use cases and motivational scenarios to express the motivation behind developing the ontology and, therefore, its potential uses. We deem that within the next few years, a science knowledge graph is likely to become a crucial component for organizing and exploring scientific work.
S. Fathalla—The majority of the research presented in this work was carried out at the University of Bonn.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
Resource type: Ontology
License: CC BY 4.0 International
1 Introduction
Ontologies have become widely used due to their ability to define relationships between different types of data, thus, improving data exploration strategies and enabling efficient data management and analysis. Ontologies provide an essential foundation for making data FAIR [32], primarily Interoperable and Reusable. For instance, the representation of scientific events metadata, including historical data about the publications, and submissions, in RDF format in EVENTS [8] and EVENTSKG datasets [9]. Knowledge-based representations of scientific data, which motivates the development of data models, ontologies, and knowledge graphs, will support a richer representation of this data, which makes it easier to query and process [2]. This greatly supports the analysis and exploration of scientific data, for example in digital libraries [8].
In this work, we present the Modern Science Ontology (ModSci), an upper ontology for providing a taxonomy of research fields, or fields of science. ModSci is a poly-hierarchical ontology that provides a hierarchical classification of various entities such as publications, events and scientists’ research fields. Besides, classification allows research and experimental development activities to be categorized by field of study. Furthermore, it models the relationships between modern science branches and related entities, such as scientific discoveries, phenomena, prominent scientists, instruments, and common interlinking relationships. ModSci is a part of the Science Knowledge Graph Ontology Suite (SKGO) [7], which comprises ontologies describing scientific data in Physics [25], pharmaceutical science [26] and computer science [10]. Thus, the project is embedded within a wider setting of knowledge representation efforts covering diverse scientific disciplines aimed at making scientific knowledge FAIR. Indeed, ModSci provides a unifying framework for the various domain ontologies that make up the SKGO suite.
Motivation. The ModSci ontology is motivated by real-life requirements that we encounter during day-to-day research and supervision work: 1) finding fields of science that best match the interests of researchers in the early stages and what the applications of this field are, 2) gaining an insight into the instruments used in, and applications of, a particular field of science, 3) deriving a comprehensive overview of other fields of science that study a given phenomenon, and 4) indeed, the classification of research topics supports a diversity of research areas, such as information exploration (e.g., in digital libraries), scholarly data analytics and integration, and modelling research dynamics [19]. Therefore, this resource can be used in practice, for example, it helps editorial teams of multidisciplinary journals in positioning submissions according to the taxonomy of research topics, thus avoiding direct out-of-scope rejections. To the best of our knowledge, there is yet no semantic model that organizes major fields and related sub-fields of science and emerging areas of study. More details and four motivating scenarios are presented in Subsect. 3.1.
Potential Impact. The potential impacts of this work include but are not limited to the following: 1) ModSci can be used for internal classification by scholarly publishers, e.g., Springer Nature, for suggesting books, journals, and conference proceedings to readers, i.e., researchers interested in scholarly articles in a specific domain, 2) Cross-disciplinary indexing, and 3) ontology-based recommendation system for scholarly events as well as research papers, and classification of authors and organizations in digital libraries according to their research topics. ModSci is designed to afford high modelling capability and elasticity to deal with a wide variety of modern science branches and associated entities, which makes it applicable also to other areas besides research where the classification of science is an important aspect. ModSci powers two projects for semantically representing scholarly information: the Open Research Knowledge Graph [15] and the OpenResearch.org collaboration platform [28] (more details in Sect. 3).
2 Related Data Models
In the following, we present research efforts on developing ontologies for modelling research findings in different fields of science. Conversely, research efforts to develop taxonomies for modelling Computer Science subfields/subtopics are limited.
In computer science, one of the earliest efforts, dating back to 1998, is the traditional version of the ACM Computing Classification System (CCS) of the Association for Computing Machinery and its latest version in 2012, which is based on SKOS. The ACM context ontology [21] has been developed by ACM to provide a cognitive map of the computing space from the most common computer science fields, such as Applied Computing, to the most specific ones, such as Electronic commerce. In 2019, the large-scale Computer Science Ontology (CSO) [24] had been developed in order to represent scientific publications, mainly in Computer Science. In CSO, the skos:broaderGeneric property is used to express that a topic is a super-area of another one (e.g., the Information systems area is a super-area of Data management systems).
In the field of Environmental Science, the Semantic Web for Earth and Environmental Terminology (SWEET) ontology [22] models knowledge about Earth system science and related concepts, such as “Phenomena” and “RadiationalCooling”. In Mathematics, the Mathematics Subject Classification (MSC) is an alphanumerical classification scheme consisting of 63 macro-areas in mathematics, which is used by many mathematics journals for classifying articles; in an earlier work, we have proposed an implementation in SKOS. The latest version has been released in 2010; a revision is in progressFootnote 1.
In Economics, the Journal of Economic Literature (JEL)Footnote 2 classification system is a standard. JEL is available as a classification tree in a custom XML format (i.e., not implemented as an ontology); the latest update at the time of writing was performed at the end of 2018. Fields of Research (FoR) classification [23], last updated 2008, is one of the three classifications in the Australian and New Zealand Standard Research Classification (ANZSRC) for classifying major sub-fields of research. The main disadvantage is that FoR is not available in a machine-readable format. The Dewey Decimal Classification (DDC) system is a general knowledge hierarchy in various disciplines, involving Computer science, Philosophy, and Social sciences [17]. Arabic numerals are used to represent each class in the DDC, e.g., 300 represents the Social Sciences class, and 320 represents the Political science subclass. The Library of Congress Classification (LCC), a classification system which organizes the book collections of the Library [16], is available in various machine-oriented formats including SKOS and the related MADS representation.
Despite these continuous efforts, none of the existing data models provides a complete view of the taxonomy of the various fields of science and their subfields, but rather focuses on the classification, in plain taxonomies, i.e., models with weak semantics, of knowledge belonging to a particular research area regardless of the overlap between them. What additionally distinguishes our work from the related work mentioned above is 1) the inclusion of related entities, 2) the representation of relationships between fields of science, and 3) the publication of the ontology considering FAIR principles and W3C standards and best practices.
3 Motivation and Usage Scenarios
Each of the modern science branches comprises various specialized yet overlapping scientific disciplines that often possess their own nomenclature and expertise [5]. For example, astrometrical studies use statistical methods to compute data estimates and error ranges; hence, an overlap between astrometry and statistics occurs here. In addition, there are collaborations between scientists from different fields of science. For example, biologists require mathematics to process, analyze and report experimental research data and to represent relationships between some biological phenomena. Statistics are also used in economics in the measurement of correlation, analyzing demand and supply, and forecasting through regression, interpolation, and time series analysis.
3.1 Motivating Scenarios
The objective of presenting the following scenarios is to express the motivation behind developing the ModSci ontology and, therefore, its potential uses.
Cross-Disciplinary Indexing: Cross-disciplinary research refers to research that embraces efforts conducted by researchers from two or more academic disciplines. Publications from this kind of research place obstacles to cross-disciplinary indexing and searching in digital libraries. Therefore, the classification of scholarly articles based on a rigid classification scheme is crucial.
Scholarly Information Classification: Classification of information is an important issue in wiki-based content management systems, such as CatawikiFootnote 3, WikispeciesFootnote 4, and WikiAnswersFootnote 5. In particular, developing a universal classification scheme of the various fields of science will greatly support information management in wikis devoted to research fields, such as nLabFootnote 6, Gene WikiFootnote 7 and SNPediaFootnote 8. The aforementioned motivation scenarios showed that such a classification makes a difference.
3.2 Real-World and Potential Use Cases
Several concrete real-world uses are presented to illustrate the added value of ModSci in various application areas, including interdisciplinary indexing, enriched bibliographic data, and network analysis within interdisciplinary scientific fields.
Open Research Knowledge GraphFootnote 9: ModSci is being integrated into the Open Research Knowledge Graph (ORKG) [15] to support the classification of research papers. ORKG is a step towards the next generation of digital libraries for semantic scientific knowledge communicated in scholarly literature [15]. ModSci is being integrated into the step of selecting the research field of the research papers added to the knowledge graph, which provides more than 200 research fields in various fields of modern science. Besides, it can be used in browsing the research papers by fields through the “Browse by research field” feature.
Publication Classification: OpenResearch.org contains scholarly information in several fields of science, i.e., not restricted to particular fields. This semantic wiki aims at making scholarly information more accessible and shareable. ModSci is used to categorize information about scientific events, research projects, scientific papers, publishers, and journals.
Support Domain Ontologies Development: To name just a few, several classes and properties are in use by several emerging ontologies developed for consortia of the German National Research Data Infrastructure NFDI, including NFDI4CultureFootnote 10 and NFDI-MatWerkFootnote 11.
Publications and Scholarly Events Classification: ModSci can be used to classify research projects, research results, papers submitted to multidisciplinary journals and course contents. Poly-hierarchical ontologies can be used in digital libraries for categorizing published research articles as well as scholarly events. Furthermore, it supports exploring new features and unknown relationships between articles belonging to different fields of science to provide recommendations to end users [9].
4 Ontology Development
In the following, we present the decisions made during the development of the ontology.
-
The Systematic Approach for Building Ontologies (SABiO) [1] has been followed in the development process of ModSci. It comprises five phases ontology requirements elicitation, ontology capture and formalization, ontology design, ontology implementation, and finally ontology evaluation.
-
We have chosen a top-down approach because it makes more sense to start with the main branches of modern science and then classify them into specific hierarchies.
-
The ontology is being developed in an iterative process which involves cross-disciplinary interaction between ontology engineers and researchers belonging to the respective fields of science. This process was continuing through the entire lifecycle of the ontology
-
In the very beginning, we decided to define an initial version of the ontology and then to assess what we have at hand by discussing raised issues with the scientists involved, and finally performing changes accordingly. The assessment was done by drafting a set of competency questions that a knowledge base based on the ontology should answer to determine the usefulness of the ontology (i.e., whether it satisfies functional requirements). This helps the ontology engineer to identify relevant concepts and their properties, as well as constraints.
-
The creation of classes’ definitions and their properties are closely interlaced to better ingest the new class to the ontology. In addition, it also helps to define the scope of knowledge that the ontology encapsulates effectively.
-
To make ModSci compatible with well-known classifications, we decided to reuse them.
4.1 Reusing External Vocabularies
Building the ontology hierarchy has been bootstrapped from the following resources: 1) reusing terms from existing models developed for describing the scientific work in various fields of science, such as BioAssay Ontology (BAO) [29], and the SWEET ontologies [22], FOAF, hence achieving FAIR’s Interoperability (I2 and I3), 2) several taxonomies of research fields, such as the Field of Research (FoR) by ANZSRC [23], Dewey Decimal [17], DFGFootnote 12 structure of research areas, and Library of Congress Classification [16] have been integrated with ModSci for expanding various science branches, including mathematical, physical, and chemical sciences, 3) interviews with domain experts have been conducted in order to validate, remove or update identified concepts as well as add missing ones, and 4) research area classifications by universities (i.e. divisions of their research disciplines) have been considered.
4.2 Core Concepts
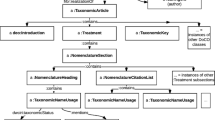
The pivotal concepts of ModSci are the branches of modern science and its sub-branches. Several concepts (we follow the definitions found in [31]) related to such concepts, including scientific discovery, phenomenon, scientists, and scientific instruments, have been defined. Where possible, these concepts are mapped to well-known ontologies such as SWEET, SKOS and FOAF, and Role from Basic Formal Ontology (BFO) as well. Concretely, these entities are represented in ModSci as owl:Class as shown in Fig. 1.
We observed a great extent of collaboration between various fields of science, which in turn gave rise to new fields of science. For example, ecology, a branch of science that studies the distribution and interactions between living things and the physical environment, is a new field of science that combines methods and techniques from both biology and earth sciences. Thus, the Ecology class is defined as a subclass of both the Biology and the EarthSciences classes. Another example is Biochemistry, a subclass of both Biology and Chemistry. Class specialization: one example is the creation of Ethology, Psychology, SocialPsychology, and Sociobiology as sub-classes of BehavioralSciences. Class Disjointness: adding disjointness axioms to ontologies enables a wide range of noteworthy applications [30]. We explicitly asserted the pairwise disjointness of various classes in ModSci, for instance, the AstronomicalPhenomena class is disjoint with BiologicalPhenomena. Class equivalence: an example is the LaboratoryInstrument class which is equivalent to ScientificInstrument.
4.3 Semantic Relations
A full view of the properties defined in ModSci, including their domains and ranges, is shown in Fig. 1. Some properties have complex ranges and domains (i.e. logical disjunction), e.g., the domain of discoveredByScience is (Phenomenon \(\sqcup \) ScientificDiscovery), which means that a Phenomenon or a Scientific Discovery can be discovered by a particular Science.
Property Restrictions. A property restriction provides a type of logic-based constructor for complex classes by defining a particular type of class description, which is a class of all individuals that satisfy the restriction. OWL defines two kinds of property restrictions: value constraints (restricting the range of the property) and cardinality constraints (restricting the number of values a property can take). One example of a property restriction in ModSci is the use of owl:minCardinality for restricting discoveredByScientist to assure that a phenomenon is discovered by at least one scientist (owl:someValuesFrom). Another kind of property restriction is the owl:allValuesFrom constraint, which restricts the individuals used as objects with a given property to be either a member of a certain class or data values within a specified set of values. For instance, the property discoveredByScientist has been restricted by owl:allValuesFrom to the class Scientist.
4.4 Design Patterns
Patterns provide a well-proven solution to a specific engineering problem, so they are recurrent solutions to design problems that can be reused when developing ontologies [4]. Several ontology design patterns (ODPs) [11], involving content, alignment and logical ODPs, have been applied to represent, for example, such as inverse relations and composition of relations. A full list of the ODPs can be found in the official catalogueFootnote 13 of ontology design patterns. Here, we list some examples of the used patterns. The TimePeriod content ontology design pattern (CP) [20] is used to represent the time periods in which the renowned scientists lived, as illustrated in Fig. 2a. An example of the Alignment ODPs is the Class Union pattern, which is used to define a class in one ontology as the union of two or more classes in another one(s). For instance, the ScientificOrganization class is defined as the union of both ScientificAgent and foaf:Organization. One common problem in ontology engineering is representing the N-ary relations (\(N\ge 3\)). An ordinary solution is to use the N-ary relation pattern [13]. In this pattern, the N-ary relation is reified by creating a class rather than a property and uses N properties to point to the related entities [18]. Individuals of such classes are individuals of the N-ary relation and additional properties can provide binary links to each argument of the relation, i.e., an individual of the relation linking the N individuals. For example, consider the case of representing that Biology facilitated Physics in the discovery of Energy conservation phenomenon. This case can only be represented as an N-ary relation. As shown in Fig. 2b, the individual _:helpInDiscoveryOfEnergyConservation is an individual of helpInDiscovery, which represents a single object encapsulating both sciences that helped in the discovery of the phenomenon Energy Conservation via the functional property helpInDiscoveryOfPhenomenon.
4.5 Reasoning
To maximize ModSci’s inference capability, several property characteristics, including reflexivity, symmetry, inverse, and transitivity, have been asserted [14]. To support the inference process, several symmetric relations have been defined. For instance, hasCloseRelationshipTo is a symmetric relation where Statistics is connected to Mathematics via this property, meaning the opposite also holds. Moreover, all corresponding inverse properties are created, where here possible to support bidirectional traversal between two concepts in the ontology network. For instance, isApplicationOfScience property being an inverse of hasApplication is an example of an inverse relation. Thus, if an application of science A, e.g., a Biochip, isApplicationOfScience S, then it can be inferred that S hasApplication A. Furthermore, some properties have the same domain and range, e.g., hasCollaborationWith has ModernScience as its domain and range, thus providing the information that there exist collaborations between two modern sciences. This property is additionally defined as a reflexive relation, i.e., scientists in a particular field of science have collaborations with themselves. An example of functional properties is the inspiredBy property, whereas a particular scientific method is inspired by either a phenomenon or a scientific discovery. For instance, Deep Learning is inspired by Biomedical Signals, the observations of the physiological activities of organisms. Finally, a set of SWRL rules have been defined for discovering new relationships and inferring new knowledge that is not explicitly given in the ontology. These rules have been semantically validated using the HermiT reasoner.
5 Technical Specifications
Ontology Publishing: ModSci is published (following ontology publication best practices [3]) via a persistent identifier and dereferenced in HTML and OWL (both in RDF/XML and Turtle serialisations), hence achieving the FAIR’s Findability (F1 and F4). Content negotiation is enabled via its PID in a way that requests from browsers get the HTML while others from semantic web applications or ontology editors (e.g. Protegé) get the requested representation (i.e. RDF serialization) of the ontology.
Interoperability: we implemented our ontology using OWL, hence achieving FAIR’s Interoperability (I1).
Indexing and Availability: The ontology is licensed under the open CC-BY 4.0 license and its source is available from a GitHub repositoryFootnote 14, hence achieving FAIR’s Reusability (R1). It can be browsed through a web-based repository front-end for browsing and visualizing published ontologies, such as BioPortalFootnote 15, and Linked Open VocabulariesFootnote 16. Furthermore, these services also store the metadata of the ontology, hence achieving FAIR’s Accessibility (A2).
Announcement: several mailing lists, such as the W3C LOD list (public-lod@w3.org), the discussion list of the open science community (open-science@lists.okfn.org), and discussion forums, such as those of the Open Knowledge Foundation (OKFN)Footnote 17 have been used for announcing the latest release of the ontology. We received valuable feedback, involving suggesting existing ontologies for reuse, presenting the ontology by explaining different parts of it and the composing concepts and improving the documentation from several parties (e.g., researchers in our community).
Logical Correctness: We validated the ontologies against inconsistencies using the HermiT reasoner, and OOPS! Ontology Pitfall ScannerFootnote 18.
Documentation: Widoco wizard for documenting ontologies [12] is used to create HTML documentation, thus enabling human understanding of the ontology and increasing its reusability. The documentation is available online through the persistent identifier of the ontology. The rdfs:comment property is used to provide a human-readable description of each resource.
Metadata Completion: A checklistFootnote 19 for completing the vocabulary metadata proposed has been used to complete the ontology’s metadata (FAIR’s Findability (F2 and F3)), e.g., authorship information in terms of Dublin Core and license. This makes it easier for academia and industry to identify and reuse the ontology effectively and efficiently.
Ontology Maintenance: Ontology maintenance includes fixing bugs (i.e. inconsistencies and inefficient implementation) and enhancing (i.e. improving coverage and integration with other models). The maintenance process is performed through the GitHub issue tracker with the possibility of submitting issues for either suggesting improvements, e.g., reusing related ontologies that may appear in the future, or reports of problems via Improvement request and Problem report issue templates (see Community collaboration part in the documentation page. Thus, enabling external collaboration in the development of the ontology to maintain its future sustainability.
6 Data-Driven Evaluation
The evaluation of the ontology has been carried out in two directions, 1) evaluating the success of the ontology in modelling a real-world domain (Formative evaluation) in which we use the verification and validation approach and 2) evaluating the quality of the ontology (Summative evaluation) in which we used a metric-based ontology quality analysis approach.
6.1 Test Data
To aid the development and testing of ModSci, we have created +150 individuals (including, ScientificInstrumentManufacturer (17), ScientificInstrument (35), AtmosphericPhenomena (5), Scientist (10), and ScientificOrganization (8)). These individuals have been created into a separate file to make it more modular. Figure 3 depicts the relationship between a sample of individuals in ModSci. These individuals help to assist in characterizing core concepts within the ontology and to provide links (where available) between ModSci and the reused ontologies. Even though some of these individuals are not required for evaluating the ontology, they are essential for understanding the domain; hence they help in the development process. Individuals are defined with individual axioms, also called “facts”; green circles in Fig. 1 and Fig. 3 present some of these individuals. Two types of facts have been created: 1) facts about class membership and property values of individuals: for example, deep learning algorithms (an individual of algorithms), or Non-Negative Matrix Factorization (NMF), are based on biological data called “biomedical signals” (also called Biosignals), and 2) facts about identical individuals. The OWL owl:sameAs construct is used to establish the identity of individuals, i.e., states that two URIs refer to the same individual.
6.2 Formative Evaluation
We performed ontology verification and validation (V &V) following the guidelines proposed in [1]. Ontology verification aims at ensuring that the ontology is being built correctly, while ontology validation (using test cases) aims at ensuring that the correct ontology is being built, i.e. it fulfills its intended purpose. After identifying motivational scenarios in a use-case fashion, the next step is to derive a set of competency questions (CQs)Footnote 20 from these scenarios. Competency questions can serve as a kind of functional requirement specification for an ontology. Therefore, a set of functional requirements have been identified from the CQs identified by domain experts and from the data sources (cf. subsection 4.1).
The verification is performed mainly to justify that the ontology being developed has adhered to these requirements, i.e it should be able to answer all CQs correctly. Some of these questions are defined at a high level of abstraction to help determine the scope of the ontology and its potential uses and others are more specific to cover potential use cases.
This evaluation has been conducted by means of expert judgment (ontology engineering experts), in which the concepts, relations and axioms defined in the ontology have been checked regarding whether they are able to answer the defined CQs [6] (cf. Table 1). Ontology engineers and scientists from different research fields, including Dentistry, Engineering and chemistry, have been recruited while developing both the ontology and CQs to validate, remove, add missing ones or update identified concepts. This approach enabled us not only to check whether the ontology is built correctly but also efficiently. For this reason, we performed this evaluation in parallel with the ontology development in an iterative manner, which significantly helped in improving the ontology. In addition, it saves a lot of time by detecting defects at an early stage of the development process. After each iteration, a set of SPARQL queries have been run against the ontology to ensure that it meets the functional requirements. After five complete iterations (i.e. development-to-evaluation and vice versa), the final version of ModSci is obtained.
In Table 1, we present a sample of the CQs. These CQs have been derived from a set of facts either collected from interviewing researchers from various fields of science, including chemistry, biology and pharmaceutical science or have been collected from scientific articles. Some of these facts are (F1) The production of psychiatric drugs is a result of studying the relationship between chemistry and psychology, (F2) Organic chemistry has a close relationship to biology since it supplies its substances and (F5) Biology applies natural physical laws since all living matter is composed of atoms. Then, the CQs are translated into SPARQL queries, considering producing results which should be somehow informative for both non-experts and expert participants.
Overall, 25 queries were run against the ontology. The results have got 100% accuracy which means that ModSci fulfils all the specified functional requirements. This verified that ModSci is able to answer all competency questions defined. Table 2 illustrates a part of the verification process of ModSci, showing matched entities corresponding to the CQs.
Ontology Validation. Generally, validation is a one-time process that starts after verification is completed to make sure that the ontology is suitable for its intended uses (i.e. the correctness). In this phase, the participation of domain experts and ontology engineers is essential. The validation is accomplished by preparing several test cases (derived from the predefined competency questions) in a competency question-driven approach for ontology testing. In order to design test cases, we derived more specific questions from the predefined CQs. For example, we have rewritten CQ01 more specifically as: “CQ01.01: What are the main branches of Social Sciences and their sub-branches?” and “CQ01.02: What are the sub-fields of Astronomy?”. In addition, we have rewritten CQ5 more specifically as: “CQ05.01: List all phenomena discovered by Physics along with the scientists who discovered them?”. Inspired by the white box testing method in software testing, we have prepared test cases so that each test case comprises three variables (i.e. input, actual output, and expected results). The objective is used to verify if the actual output of the CQs meets the anticipated output. Because of the space limit, we present sample test cases shown in Table 3 and we omitted the output column because it is identical to the expected results. The listing below shows the SPARQL query corresponding to CQ04.01, which is used in T04.

After executing each test case, the returned results have been compared with the expected results, and the recall is computed. If the recall was less than 1.0, which means that not all required results (identified by experts) were returned, we analyzed the reason, iteratively adapted the ontology and re-executed the test case until all expected results were returned, i.e., we obtained a recall of 1.0. In this case, we marked the test case as passed. Algorithm 1 summarises the whole procedure. In the end, all the test cases are executed and results are reported.
6.3 Summative Evaluation
In this evaluation, we assess the richness/quality of the ontology by using OntoQA [27] evaluation model, a metric-based ontology quality analysis model. OntoQA evaluates the ontology using schema metrics and population/instance metrics. In this model, various metrics are calculated to asses different richness within the ontology. For ModSci, we found the most interesting metric is the Inheritance Richness (IR) describes the distribution of information across different levels of the ontology inheritance tree. IR indicates how knowledge is grouped into different classes and sub-classes in the ontology. Formally, IR is defined by
where H is the number of inheritance relationships and C is the number of classes. Strikingly, ModSci got a relatively high inheritance richness of 0.99, which indicates that knowledge/data can be well classified into different categories and subcategories in the ontology. In addition, it indicates that the ontology represents a wide range of general knowledge with a low level of detail.

7 Conclusions and Future Work
This paper presents the Modern Science Ontology, which models relationships between modern science branches and related entities, such as scientific discoveries, prominent scientists, instruments, phenomena, etc. Several design principles have been taken into consideration in the development of ModSci, such as configuration to support semantic web applications, registration in online services for ontology visualization and exploration, syntactic and semantic validation, human-readable documentation, and sustainability. The SABiO methodology has been followed when developing the ontologies, as well as FAIR principles for data publication. To maximize reasoning capability, 1) several property characteristics, such as reflexivity, symmetry, and transitivity, have been asserted, 2) disjointness of roles, and 3) several logic rules have been added to the ontologies. Motivating examples affirmed the usefulness and potential uses of ModSci ontologies. Two evaluation strategies have been carried out to assure the success of the ontology in modelling a real-world domain (Formative evaluation) and the quality of the ontology (Summative evaluation).
Our future work has three main directions: refining the formal representation of science in the ModSci ontology itself, covering further fields of science by dedicated ontologies, and realizing services on top of these ontologies. Regarding the formal representation of the scientific process and its entities, we aim at aligning ModSci’s own model with existing formal models of science whose processes and structures have already been investigated in depth, i.e., Mathematics. Furthermore, we are studying the applicability of ModSci in cross-disciplinary indexing, enriched bibliographic data, and network analysis within cross-disciplinary scientific communities, among others. Finally, we intend to release a new version of ModSci that supports multilingualism and we plan to incorporate all the relevant catalogue information for more instruments, applications and scientific discoveries.
Notes
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
- 7.
- 8.
- 9.
- 10.
- 11.
- 12.
- 13.
- 14.
- 15.
- 16.
- 17.
- 18.
- 19.
- 20.
The final set of competency questions is available at the GitHub repository.
References
de Almeida Falbo, R.: SABiO: systematic approach for building ontologies. In: 1st Joint Workshop Onto. Com/ODISE on Ontologies in Conceptual Modeling and Information Systems Engineering (2014)
Auer, S., Kovtun, V., Prinz, M., Kasprzik, A., Stocker, M., Vidal, M.E.: Towards a knowledge graph for science. In: Proceedings of the 8th International Conference on Web Intelligence, Mining and Semantics. ACM, p. 1 (2018)
Berrueta, D., Phipps, J., Miles, A., Baker, T., Swick, R.: Best practice recipes for publishing RDF vocabularies. In: Working Draft, W3C (2008). http://www.w3.org/TR/swbp-vocab-pub/
Blomqvist, E.: Ontology patterns: typology and experiences from design pattern development. In: The Swedish AI Society Workshop 20–21 May 2010. Uppsala University. 048, pp. 55–64. Linköping University Electronic Press (2010)
Boyack, K., Klavans, D., Paley, W., Börner, K.: Scientific method: relationships among scientific paradigms. Seed Mag. 9, 36–37 (2007)
Brank, J., Grobelnik, M., Mladenic, D.: A survey of ontology evaluation techniques. In: Proceedings of the Conference on Data Mining and Data Warehouses, pp. 166–170. Citeseer Ljubljana, Slovenia (2005)
Fathalla, S., Auer, S., Lange, C.: Towards the semantic formalization of science. In: Proceedings of the 35th Annual ACM Symposium on Applied Computing, pp. 2057–2059 (2020)
Fathalla, S., Lange, C.: EVENTS: a dataset on the history of top-prestigious events in five computer science communities. In: González-Beltrán, A., Osborne, F., Peroni, S., Vahdati, S. (eds.) SAVE-SD 2017-2018. LNCS, vol. 10959, pp. 110–120. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01379-0_8
Fathalla, S., Lange, C., Auer, S.: EVENTSKG: a 5-star dataset of top-ranked events in eight computer science communities. In: Hitzler, P., et al. (eds.) ESWC 2019. LNCS, vol. 11503, pp. 427–442. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-21348-0_28
Fathalla, S., Vahdati, S., Auer, S., Lange, C.: SemSur: a core ontology for the semantic representation of research findings. Procedia Comput. Sci. 137, 151–162 (2018)
Gangemi, A., Presutti, V.: Ontology design patterns. In: Staab, S., Studer, R. (eds.) Handbook on Ontologies. IHIS, pp. 221–243. Springer, Heidelberg (2009). https://doi.org/10.1007/978-3-540-92673-3_10
Garijo, D.: WIDOCO: a wizard for documenting ontologies. In: d’Amato, C., et al. (eds.) ISWC 2017. LNCS, vol. 10588, pp. 94–102. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-68204-4_9
Giunti, M., Sergioli, G., Vivanet, G., Pinna, S.: Representing n-ary relations in the semantic web. Logic J. IGPL 29(4), 697–717 (2021)
Grau, B.C., Horrocks, I., Motik, B., Parsia, B., Patel-Schneider, P., Sattler, U.: OWL 2: the next step for OWL. Web Seman. 6(4), 309–322 (2008)
Jaradeh, M.Y., et al.:Open research knowledge graph: next generation infrastructure for semantic scholarly knowledge. In: Proceedings of the 10th International Conference on Knowledge Capture, pp. 243–246. ACM (2019)
Library of Congress contributors. Library of Congress Classification (2014). https://www.loc.gov/catdir/cpso/lcc.html. Accessed December 2022
Mitchell, J.S.: Relationships in the dewey decimal classification system. In: Bean, C.A., Green, R. (eds.) Relationships in the Organization of Knowledge. Information Science and Knowledge Management, vol 2, pp. 211–226. Springer, Dordrecht (2001). https://doi.org/10.1007/978-94-015-9696-1_14
Noy, N., Rector, A., Hayes, P., Welty, C.: Defining n-ary relations on the semantic web. In: W3C Working Group Note, vol. 12, no. 4 (2006)
Osborne, F., Salatino, A., Birukou, A., Motta, E.: Automatic classification of springer nature proceedings with smart topic miner. In: Groth, P., et al. (eds.) ISWC 2016. LNCS, vol. 9982, pp. 383–399. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46547-0_33
Presutti, V., Gangemi, A.: Content ontology design patterns as practical building blocks for web ontologies. In: Li, Q., Spaccapietra, S., Yu, E., Olivé, A. (eds.) ER 2008. LNCS, vol. 5231, pp. 128–141. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-87877-3_11
Priya, M., Aswani Kumar, C.: Construction and merging of ACM and sciencedirect ontologies. In: Abraham, A., Cherukuri, A.K., Melin, P., Gandhi, N. (eds.) ISDA 2018 2018. AISC, vol. 941, pp. 238–252. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-16660-1_24
Raskin, R.G., Pan, M.J.: Knowledge representation in the semantic web for earth and environmental terminology (SWEET). Comput. Geosci. 31(9), 1119–1125 (2005)
Rousseau, R., Ecoom, F.O.: The Australian and New Zealands fields of research (FoR) codes. ISSI Newslet. 14(3), 59–61 (2018)
Salatino, A.A., Thanapalasingam, T., Mannocci, A., Osborne, F., Motta, E.: The computer science ontology: a large-scale taxonomy of research areas. In: Vrandečić, D., et al. (eds.) ISWC 2018. LNCS, vol. 11137, pp. 187–205. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00668-6_12
Say, A., Fathalla, S., Vahdati, S., Lehmann, J., Auer, S.: Semantic representation of physics research data. In: 12th International Conference on Knowledge Engineering and Ontology Development (KEOD 2020), pp. 64–75. Science and Technology Publications. LDA Setúbal, Portugal (2020)
Say, Z., Fathalla, S., Vahdati, S., Lehmann, J., Auer, S.: Ontology design for pharmaceutical research outcomes. In: Hall, M., Merčun, T., Risse, T., Duchateau, F. (eds.) TPDL 2020. LNCS, vol. 12246, pp. 119–132. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-54956-5_9
Tartir, S., Arpinar, I.B., Moore, M., Sheth, A.P., Aleman-Meza, B.: OntoQA: metric- based ontology quality analysis. In: IEEE ICDM Workshop on Knowledge Acquisition from Distributed, Autonomous, Semantically Heterogeneous Data and Knowledge Sources (2005)
Vahdati, S., Arndt, N., Auer, S., Lange, C.: OpenResearch: collaborative management of scholarly communication metadata. In: EKAW (2016)
Visser, U., Abeyruwan, S., Vempati, U., Smith, R.P., Lemmon, V., Schürer, S.C.: BioAssay Ontology (BAO): a semantic description of bioassays and high-throughput screening results. BMC Bioinf. 12(1), 257 (2011)
Völker, J., Fleischhacker, D., Stuckenschmidt, H.: Automatic acquisition of class disjointness. Web Seman. Sci. Serv. Agents World Wide Web 35, 124–139 (2015)
Wikipedia contributors. Science - Wikipedia, The Free Encyclopedia (2019). http://en.wikipedia.org/w/index.php?title=Science &oldid=918085492. Accessed December 2022
Wilkinson, M.D., et al.: The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 1–9 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Fathalla, S., Lange, C., Auer, S. (2023). An Upper Ontology for Modern Science Branches and Related Entities. In: Pesquita, C., et al. The Semantic Web. ESWC 2023. Lecture Notes in Computer Science, vol 13870. Springer, Cham. https://doi.org/10.1007/978-3-031-33455-9_26
Download citation
DOI: https://doi.org/10.1007/978-3-031-33455-9_26
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-33454-2
Online ISBN: 978-3-031-33455-9
eBook Packages: Computer ScienceComputer Science (R0)